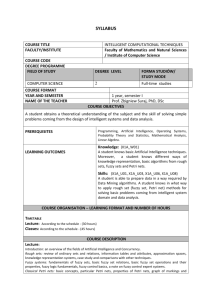
Formal Methods in Systems Biology: Presentation
advertisement

Special Topics in Computational
Biology:
Formal Methods in Systems
Biology
Spring, 2008
Chris Langmead
Department of Computer Science
Carnegie Mellon University
James Faeder
Department of Computational Biology
University of Pittsburgh School of Medicine
General Info
• Course Numbers:
– CMU 15-872(A)
– CMU 02-730
– Pitt CMPBIO 2045(Arts & Sciences)
– Pitt MSCBIO 2045 (School of Medicine)
• Location: Newell-Simon Hall (NSH) 3002 - OK?
• Time: Tu, Th 1:30-2:50 PM
• Instructors
– Chris Langmead (cjl@cs.cmu.edu)
– Jim Faeder (faeder@pitt.edu)
• Office Hours: By appointment (please email)
• Course Wiki:
http://bionetgen.org/index.php/Formal_Methods_in_S
ystems_Biology (email Jim for account)
Course Format: An Informal
Course about Formal Methods
• Introductory lectures (two weeks)
• Students will read and present research
papers
– Sign up for open dates on the wiki (25 - projects)
• Students will design and complete a course
project on a subject of special interest
• Grading is based on completion of work
• Flexibility depending on course enrollment
– Journal club
– Focused project
– Review article
Encouragement
• Opportunity to learn about new areas and
methods that will be of direct interest in your
research.
• (True for the “instructors” as well)
• We will operate as a multi-disciplinary team
– Computer Scientists, Physicists, Chemists,
Engineers, Mathematicians, …, Biologists
– Good communication essential
Products of the Course
• Comprehensive bibliography in wiki format
• Research projects leading to publishable
results in the field
• Review article (?)
• Improved organization and presentation skills
• Participation on a multi-disciplinary team
Introductions
• Your name
• Your university, department, research
area(s) and research advisor
• Your educational background
– Computer Science, Math, Physics, etc.
• Goals taking the course
Outline of Today’s Lecture
• Definition of terms
• Goals
• Examples of Successful Abstractions
– Flux Balance Analysis
– Mass Action Kinetics
• Brief survey of topics
Importance of Symbols
• Invention of symbol for zero and decimal
system for writing numbers “among the
greatest human inventions.”
• 3 known independent inventions
• In each case, development took
centuries
• Major impact on trade, culture, and
philosophy.
• Celebration of zero dot in Sanskrit
poetry
“The dot on her forehead / Increases her
beauty tenfold,/ Just as a zero dot [sunyabindu] /Increases a number tenfold. Biharilal
QuickTime™ and a
PNG decompressor
are needed to see this picture.
Key Definitions - Formal Methods
• In computer science and software engineering, formal methods
are mathematically-based techniques for the specification,
development and verification of software and hardware systems.
• The use of formal methods for software and hardware design is
motivated by the expectation that, as in other engineering
disciplines, performing appropriate mathematical analyses can
contribute to the reliability and robustness of a design.
• However, the high cost of using formal methods means that they
are usually only used in the development of high-integrity
systems, where safety or security is important.
- WIKIPEDIA
Expanded View of Formal Methods
• Formal abstractions that may be used to
model system of interest
• In addition to sytems that can be
formally analyzed, we will consider
representations that can only be fully
explored by simulations.
Key Definitions - Systems Biology
• Systems biology is a relatively new biological study field that
focuses on the systematic study of complex interactions in
biological systems, thus using a new perspective (integration
instead of reduction) to study them.
• Particularly from 2000 onwards, the term is used widely in the
biosciences, and in a variety of contexts.
• Because the scientific method has been used primarily toward
reductionism, one of the goals of systems biology is to discover
new emergent properties that may arise from the systemic view
used by this discipline in order to understand better the entirety
of processes that happen in a biological system.
- WIKIPEDIA
Origin of Systems Biology
• Completion of genome projects is major
inspiration
• Provided “parts list” for the cell
• Next obvious step is to ask how parts
work together to carry out function?
Vision for Role of Computer
Science in Systems Biology
• “Computer science could provide the
abstraction[s] needed for consolidating
knowledge of biomolecular systems”
• “...the abstractions, tools and methods
used to specify and study computer
systems should illuminate our
accumulated knowledge about
biomolecular systems.”
Regev and Shapiro, “Cells as Computation,” Nature (2002).
Abstract Representations in
Biology
• DNA sequence represented by strings
with 4 letter alphabet (ATGC)
• Protein sequence and structure
– Strings with 20 letter alphabet
– Set of 3D atomic coordinates (PDB file)
The KaiC hexamer, a
Circadian clock protein. From
pdb.org.
(Some) Desirable Properties of an
Abstract Representation
1.
2.
3.
4.
5.
Relevant / accurate
Computable
Understandable
Extensible
Scalable
Modular
Hierarchical
1-4 from Regev and Shapiro, “Cells as Computation,” Nature (2002).
An Irony
• CS community aims to provide powerful
abstract representations to improve
understanding of systems.
• Manner of reporting results - technical reports
in conference proceedings - presents major
barrier to wider adoption by science and
engineering communities.
• There is a need for better communication
among disciplines!
Sometimes formalism creates a
barrier
Example: Red blood cell model
Agenda
• We are looking for useful abstractions
that can improve our understanding of
how biological systems behave
Goals
• Language(s) for constructing whole-cell
models (comprehensive, system-wide)
• Formal analysis (reasoning) of such models
• Simulation of models on distributed systems
• Combination of analysis and simulation to
predict behavior of models
– genotype phenotype
Challenges
• Accuracy
– Missing interactions
• Computability
– Requirement to perform simulations for many
properties of interest
– Poor scaling of simulations
• Understanding
– Problem of network visualization
• Extensibility
– Missing biophysics
• Scalability
– Need to compute behavior on multiple scales, e.g.
tissuecellcytoplasmnucleus
Mathematical vs. Computational
Models
Consider an elementary chemical reaction
r1: A + B -> C
Mathematical
d[A] / dt k[A][B]
Computational
module A
: [0..N] init N;
[r1] (A > 0) -> k*A*B: (A’ = A - 1);
…
endmodule
How important is this distinction?
Fisher & Henzinger, Nat. Biotechnol. (2007).
Tension between Accuracy and
Computability
• Application of formal methods requires that elements
of representation be relatively simple.
• For example, a representation that includes all
analytical functions in mathematics might not be
useful - impossible to make predictions.
• In general, increasing the complexity of the
representation limits ability for analysis.
• Representations are sometimes chosen for
amenability to analysis rather than realism - e.g.
boolean networks.
• Computational (“executable”) models tend to make
restrictions explicit.
Some successful abstractions in
systems biology
• Flux Balance Analysis
– Genome-wide models of metabolism
• Mass Action Kinetics
– Cell-cycle model
– Growth factor signaling model
Network Reconstruction (2D
Annotation)
B. O. Palsson, Nature Biotechnology 22, 1218 - 1219 (2004)
Network Reconstruction (cont.)
• Wiring diagram for the components in a cell
• Elements are
– Molecular Components (Species)
– Interactions (Reactions)
• Additional detail can be added.
• Genome-wide reconstructions for metabolism
are available for many model organisms
(including Homo Sapiens!)
• “All such interactions are ultimately represented by a
genome-scale stoichiometric matrix—a twodimensional genome annotation.”
B. O. Palsson, Nature Biotechnology 22, 1218 - 1219 (2004)
Overview of Flux Balance Analysis
• Genome-wide reconstruction of metabolic
network
v1
ri : s1 s2
s3
S1i S2i 1; S3i 1; S ji 0, j {1, 2, 3}
• Assume steady state
S v b, where bi are known transport fluxes.
• Assume optimal growth (biomass production)
maximize f (v) v v out
Genome-Wide Reconstruction of
Haemophilus influenzae
Edwards, J. S. et al. J. Biol. Chem. 1999;274:17410-17416
Single and double deletion in the
central metabolic pathways of H.
Influenzae
Edwards, J. S. et al. J. Biol. Chem.
1999;274:17410-17416
What Accounts for Success?
• Knowledge Base
– Metabolic chemistry known from >50 years
biochemistry and genome sequence
• Simple Abstraction
– Biochemistry reduced to list of reaction
stoiochimetries
• Powerful Computation Method
– Highly optimized solvers for Linear Programming
problem
• Extensibility
– Non-optimal growth in mutants
– Constraints arising from molecular crowding
Cellular Signal Transduction
ligand
ligand-receptor
transphosphorylation
binding
aggregation
signaling complex
receptor
plasma
membrane
SH2
domain
kinase
adaptor
SH3
domain
Mass Action Kinetics
Q u ic k T im e ™ a n d a
T I F F ( Un c o m p r e s s e d ) d e c o m p r e s s o r
a r e n e e d e d t o s e e t h is p ic t u r e .
Q u ic k T im e ™ a n d a
T I F F ( Un c o m p r e s s e d ) d e c o m p r e s s o r
a r e n e e d e d t o s e e t h is p ic t u r e .
à àà àÜ
R Lá
RL
à
à
k
ka
d
Differential Equations
d[R]
ka [R][L] kd [RL]
dt
d[L]
ka [R][L] kd [RL]
dt
d[RL]
ka [R][L] kd [RL]
dt
Reaction Network Model of
Signaling
Kholodenko et al., J. Biol. Chem. 274, 30169 (1999)
Comparing Model and Experiment
Experimental Data
Simulation Results
Benefits of Mass Action Kinetic
Modeling
• Large knowledge base of signaling
biochemistry
• Models dynamical behavior
• Computational Methods Well Established
– ODE solvers for continuous systems
• Nonlinear Dynamics Theory
• Extensibility
– Stochastic Simulation Algorithm for discrete
systems
– Spatially-resolved models can be built on same
mass action equations
Limitations of Mass Action Kinetic
Modeling
• Rapidly expanding knowledge base
– Many components and interactions unknown
• Lack of precision
– ad hoc assumptions to limit combinatorial
explosion (next lecture)
• Large sets of nonlinear ODE’s are difficult to
simulate or analyze
• No comprehensive models yet
Map of Signaling Initiated by a
Single Family of Receptors
Qu ickTime™ and a
TIFF (Uncompressed) decompressor
are need ed to see this picture .
Oda and Kitano (2006) Mol. Syst. Biol.
Map of Signaling Initiated by a
Single Family of Receptors
Analysis is limited to
simple graph theoretic
measures and
qualitative discussions
of architecture.
Qu ickTime™ and a
TIFF (Uncompressed) decompressor
are need ed to see this picture .
Oda and Kitano (2006) Mol. Syst. Biol.
(Partial) List of Topics
•
•
•
•
•
•
•
•
Boolean Networks
Petri Nets
Statecharts
Process Algebras
Agent-Based Modeling
Hybrid Systems
Model Checking
Simulation Algorithms
Brief Overview of Two Useful
Abstractions
•
•
•
•
•
•
•
•
Boolean Networks
Petri Nets
Statecharts
Process Algebras
Agent-Based Modeling
Hybrid Systems
Model Checking
Simulation Algorithms
Boolean Networks
BN model of cell cycle in budding yeast
G1
Li, F., et al. PNAS 101, 4781–4786 (2004).
Boolean Networks
BN model of cell cycle in budding yeast
Update:
b(t 1) a1 (t) a2 (t) a3 (t) a4 (t)
Li, F., et al. PNAS 101, 4781–4786 (2004).
G1
Boolean Networks
BN model of cell cycle in budding yeast
Update:
b(t 1) a1 (t) a2 (t) a3 (t) a4 (t)
Li, F., et al. PNAS 101, 4781–4786 (2004).
G1
Blue arrows form
stable basin of
attraction
Balance Sheet for BNs
Pro
• Models may be
constructed on basis of
scant data*
• Fast computation
• Strong analysis tools (?)
• Good for reasoning
about stability and
robustness
Con
• Two levels may not be
enough
• Lack of compositionality
• Not hierarchical, but
may be embedded in
more complex models.
*Li S, Assmann SM, Albert R (2006) Predicting Essential
Components of Signal Transduction Networks: A Dynamic Model of
Guard Cell Abscisic Acid Signaling. PLoS Biol 4(10): e312
Petri Nets
Tokens
Places
Transition
Transition
Chaouiya, C. Petri net modelling of biological networks. Brief. Bioinform.
8, 210–219 (2007).
Petri Nets
Time Evolution
Tokens
Places
Transition
Transition
Chaouiya, C. Petri net modelling of biological networks. Brief. Bioinform.
8, 210–219 (2007).
Petri Nets Generalize Network
Reconstruction
p3
t2
p4
C corresponds to S
Chaouiya, C. Brief. Bioinform. 8, 210–219 (2007).
Some useful formal properties of
PNs
• P-invariants (C T x 0 ) ~ Mass Conservation
• T-invariants ( C y 0 ) ~ Loops / Ele. Modes
• Reachability - whether a state can be
reached
• Liveness - whether a transition can be fired
Overview of PNs
• PNs are graphs, and provide tight connection
between visualization and modeling
• PN formalism is isomorphic to network
reconstruction formalism (reaction networks)
• Many extensions are possible to overcome
limitations
– Colored Petri Nets, Hierarchical CPNs, Multi-level
PN, Stochastic PNs, etc.
• Extensions provide further modeling
capabilities at the expense of analysis.
Concluding Remarks
• Goal of course is to explore various
representations from CS literature that
can be used to model biomolecular
systems.
• What opportunities do these
representations offer in terms of
analysis, simulation, understanding, and
scalability?