GCAT-SEEK - Lycoming College
advertisement
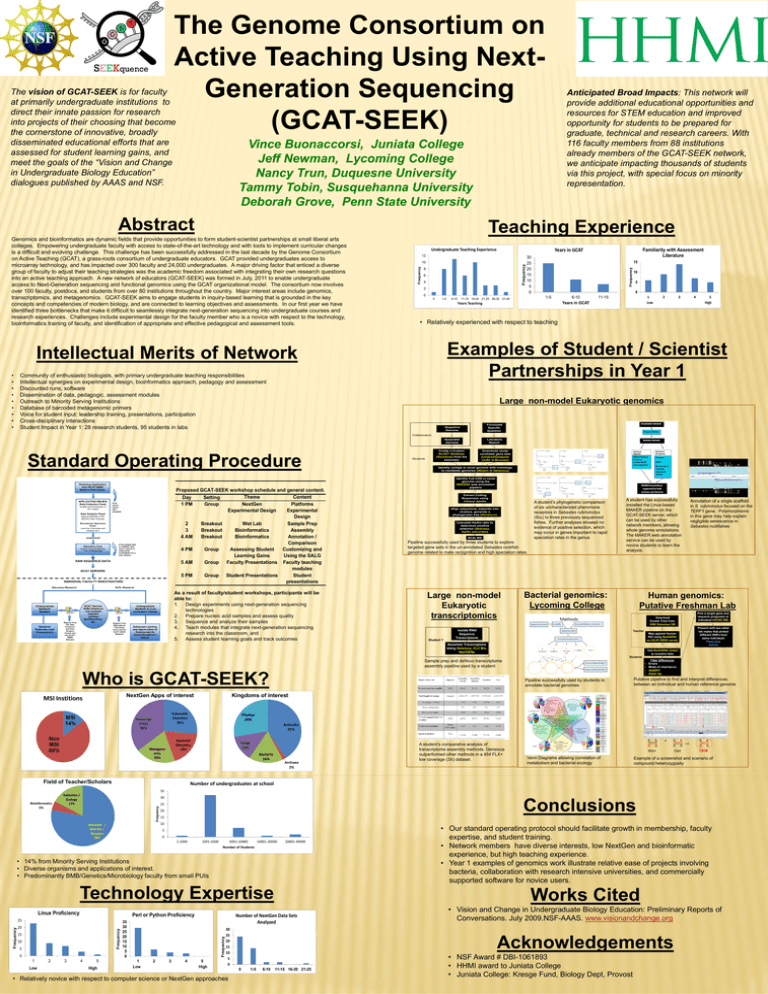
The Genome Consortium on Active Teaching Using NextGeneration Sequencing (GCAT-SEEK) The vision of GCAT-SEEK is for faculty at primarily undergraduate institutions to direct their innate passion for research into projects of their choosing that become the cornerstone of innovative, broadly disseminated educational efforts that are assessed for student learning gains, and meet the goals of the “Vision and Change in Undergraduate Biology Education” dialogues published by AAAS and NSF. Vince Buonaccorsi, Juniata College Jeff Newman, Lycoming College Nancy Trun, Duquesne University Tammy Tobin, Susquehanna University Deborah Grove, Penn State University Abstract Teaching Experience Undergraduate Teaching Experience Years in GCAT Frequency 10 8 6 4 2 0 0 1-5 6-10 Familiarity with Assessment Literature 30 25 20 15 10 5 0 15 Frequency 12 Frequency Genomics and bioinformatics are dynamic fields that provide opportunities to form student-scientist partnerships at small liberal arts colleges. Empowering undergraduate faculty with access to state-of-the-art technology and with tools to implement curricular changes is a difficult and evolving challenge. This challenge has been successfully addressed in the last decade by the Genome Consortium on Active Teaching (GCAT), a grass-roots consortium of undergraduate educators. GCAT provided undergraduates access to microarray technology, and has impacted over 300 faculty and 24,000 undergraduates. A major driving factor that enticed a diverse group of faculty to adjust their teaching strategies was the academic freedom associated with integrating their own research questions into an active teaching approach. A new network of educators (GCAT-SEEK) was formed in July, 2011 to enable undergraduate access to Next-Generation sequencing and functional genomics using the GCAT organizational model. The consortium now involves over 100 faculty, postdocs, and students from over 80 institutions throughout the country. Major interest areas include genomics, transcriptomics, and metagenomics. GCAT-SEEK aims to engage students in inquiry-based learning that is grounded in the key concepts and competencies of modern biology, and are connected to learning objectives and assessments. In our first year we have identified three bottlenecks that make it difficult to seamlessly integrate next-generation sequencing into undergraduate courses and research experiences. Challenges include experimental design for the faculty member who is a novice with respect to the technology, bioinformatics training of faculty, and identification of appropriate and effective pedagogical and assessment tools. 1-5 11-15 16-20 21-25 26-30 31-40 10 5 0 Years Teaching 6-10 Years in GCAT 1 Low 11-15 2 3 4 5 High • Relatively experienced with respect to teaching Examples of Student / Scientist Partnerships in Year 1 Intellectual Merits of Network • • • • • • • • • Anticipated Broad Impacts: This network will provide additional educational opportunities and resources for STEM education and improved opportunity for students to be prepared for graduate, technical and research careers. With 116 faculty members from 88 institutions already members of the GCAT-SEEK network, we anticipate impacting thousands of students via this project, with special focus on minority representation. Community of enthusiastic biologists, with primary undergraduate teaching responsibilities Intellectual synergies on experimental design, bioinformatics approach, pedagogy and assessment Discounted runs, software Dissemination of data, pedagogic, assessment modules Outreach to Minority Serving Institutions Database of barcoded metagenomic primers Voice for student input: leadership training, presentations, participation Cross-disciplinary interactions Student Impact in Year 1: 28 research students, 95 students in labs Large non-model Eukaryotic genomics Sequence Genome Formulate Specific Question Assemble Genome Literature Search Create a Custom BLAST database (Geneious) from the assembly Download, study candidate gene sets (Uniprot/Genbank/ UCSC G.Browser) Collaborators Standard Operating Procedure Proposed GCAT-SEEK workshop schedule and general content. Theme Content Day Setting 1 PM Group NextGen Platforms Experimental Design Experimental Design 2 Breakout Wet Lab Sample Prep 3 Breakout Bioinformatics Assembly 4 AM Breakout Bioinformatics Annotation / Comparison 4 PM Group Assessing Student Customizing and Learning Gains Using the SALG 5 AM Group Faculty Presentations Faculty teaching modules 5 PM Group Student Presentations Student presentations As a result of faculty/student workshops, participants will be able to: 1. Design experiments using next-generation sequencing technologies 2. Prepare nucleic acid samples and assess quality 3. Sequence and analyze their samples 4. Teach modules that integrate next-generation sequencing research into the classroom, and 5. Assess student learning goals and track outcomes Students Identify contigs in novel genome with homology to candidate genomes (tBlastn in Geneious) Identify Full CDS in novel genome using the MAKER2 web annotation pipeline Extract Coding Sequences using Galaxy/ Apollo A student’s phylogenetic comparison of six uncharacterized pheromone receptors in Sebastes rubrivinctus (Sru) to three previously sequenced fishes. Further analyses showed no evidence of positive selection, which may occur in genes important to rapid speciation rates in the genus. Align sequences, separate into clusters, generate a phylogenetic tree (Geneious) Calculate Ka/Ks ratio to determine positive selection (Selecton, Ka/Ks calculator) Write MS Pipeline successfully used by three students to explore targeted gene sets in the un-annotated Sebastes rockfish genome related to mate recognition and high speciation rates. Large non-model Eukaryotic transcriptomics Student 1 A student has successfully installed the Linux-based MAKER pipeline on the GCAT-SEEK server, which can be used by other network members, allowing whole genome annotations. The MAKER web annotation service can be used by novice students to learn the analysis. Bacterial genomics: Lycoming College Annotation of a single scaffold in S. rubrivinctus focused on the TERF1 gene. Polymorphisms in this gene may help explain negligible senescence in Sebastes rockfishes Human genomics: Putative Freshman Lab Download Exome Trios from 1000 Genomes DB Isolate RNA/ Sequence Transcriptome Teacher Map against Human Ref using NextGENe on GCAT-SEEK server Assemble Transcriptome Using Geneious, CLC Bio, NextGENe Pick a single gene and research prognosis of individual (HUGO DB) Present with two other lab mates that picked different SNPs from same individual: Prognosis Advice Use NextGENe viewer to examine data Students Sample prep and deNovo transcriptome assembly pipeline used by a student Who is GCAT-SEEK? NextGen Apps of interest MSI Institions MSI 14% Non MSI 86% Animalia 41% Fungi 13% Bacteria 16% Field of Teacher/Scholars Putative pipeline to find and interpret differences between an individual and human reference genome. Plantae 28% Bacterial Genomics 18% Metageno mics 20% Pipeline successfully used by students to annotate bacterial genomes Filter differences Errors Mode of inheritance dbNSFP Allele fqs Kingdoms of interest Eukaryotic Genomics 26% Transcript omics 36% • • • • Archaea 2% A student’s comparative analysis of transcriptome assembly methods. Geneious outperformed other methods in a 454 FLX+ low coverage (3X) dataset. A G C G Mom Venn Diagrams allowing correlation of metabolism and bacterial ecology x A A A G Dad A A C G Child Example of a screenshot and scenario of compound heterozygosity Number of undergraduates at school 35 30 Frequency Bioinformatics 5% Evolution / Ecology 17% Conclusions 25 20 15 10 Biochem / Mol Bio / Genetics 78% 5 0 1-1000 1001-5000 5001-10000 10001-20000 Number of Students 20001-30000 • 14% from Minority Serving Institutions • Diverse organisms and applications of interest. • Predominantly BMB/Genetics/Microbiology faculty from small PUIs Technology Expertise Linux Proficiency Perl or Python Proficiency 15 10 5 0 1 Low 2 3 4 5 High 35 30 25 20 15 10 5 0 1 Low 2 3 4 Works Cited Number of NextGen Data Sets Analyzed Frequency 20 Frequency Frequency 25 5 High 30 25 20 15 10 5 0 • Relatively novice with respect to computer science or NextGen approaches • Our standard operating protocol should facilitate growth in membership, faculty expertise, and student training. • Network members have diverse interests, low NextGen and bioinformatic experience, but high teaching experience. • Year 1 examples of genomics work illustrate relative ease of projects involving bacteria, collaboration with research intensive universities, and commercially supported software for novice users. • Vision and Change in Undergraduate Biology Education: Preliminary Reports of Conversations. July 2009.NSF-AAAS. www.visionandchange.org Acknowledgements 0 1-5 6-10 11-15 16-20 21-25 • NSF Award # DBI-1061893 • HHMI award to Juniata College • Juniata College: Kresge Fund, Biology Dept, Provost



![9_Komlenac - start [kondor.etf.rs]](http://s2.studylib.net/store/data/005352037_1-bdc91b0717c49a75493200bca431c59c-300x300.png)