combindedAronsMyxoNoSim
advertisement
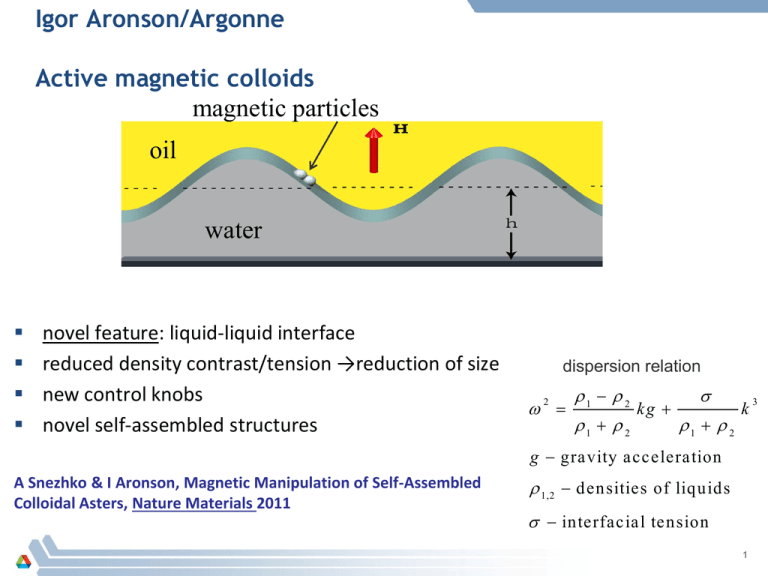
Igor Aronson/Argonne Active magnetic colloids magnetic particles oil water novel feature: liquid-liquid interface reduced density contrast/tension →reduction of size new control knobs novel self-assembled structures dispersion relation 2 1 2 1 2 kg 1 2 k g gravity acceleration A Snezhko & I Aronson, Magnetic Manipulation of Self-Assembled Colloidal Asters, Nature Materials 2011 1,2 densities of liquids interfacial tension 1 3 Localized self-assembled magnetic asters two flavors: asters & anti-asters remotely controlled locomotion speed/direction/shape controlled by dc in-plane field (not by the field gradient) asters anti-asters self-generated flow aster/anti-aster pair speed (cm/s) aster’s speed vs in-plane dc magnetic field Hdc magnetic field (Oe) 2 Self-assembled colloidal robots at liquid-liquid interface perform simple robotic functions (gripper) remotely controlled by dc in-plane magn field shape change and self-repair “soft” colloidal robot: grip, move, release Materials opportunities for soft robotics, George Whitesides, Ang Chem 2011: materials with actively tunable compliance … enable fundamentally new strategies for manipulation 4 aster-array: new functionality 1 mm YouTube Movie YouTube Movie Energy.gov: Tiny Terminators: New Micro-Robots Assemble, Repair Themselves and Are Surprisingly Strong youtube views>100,000 3 Unique GPU Modeling Capabilities at Argonne hive.msd.anl.gov GPU cluster Nvidia GTX 480 ngene, 4 GPUs cluster, 3D hydrostatic Navier-Stokes equation liquid-liquid system -novel features: toroidal flows in the bulk -modeling of asters and array of asters MSD GPU cluster, BES DOE capital equipment (42 Fermi GPU cards, 50 TFlops, single precision) 4 Igor Aronson, Argonne: Simple Bacterial Machines second law of thermodynamics prohibits rectification of Brownian motion bacteria live in out-of-equilibrium world and thus can extract useful energy important step for the design of hybrid bio-mechanical systems powered by microorganisms or synthetic swimmers 1 mm YouTube Movie •1-2 rotations per minute •mass of the gear ~ 106 mass of bacterium •power of about 1 femtowatt=10-15 Watt •about 300 bacteria power the gear YouTube Movie featured in NY Times,Forbes, Wired, SciAmerican highlighted as Argonne’s major success in 2010 YouTube > 100,000 views Sokolov, Apodaca, Grzybowski, I. Aronson, PNAS, 2010 5 Next step: swimmers-assisted assembly preliminary results swimming bacteria spin microscopic gears self-assembled primitive machines – gears assembly can bacteria assemble gears from properly functionalized units? massive parallel approach to assemble thousands of microscopic structure magnetically-controlled micro-shuttle powered by bacteria gears assembly 500 mm 100 mm 6 Alber Research group bacteria Experiments and Simulations Image analysis of bacteria movies – Cell Segmentation and tracking – Illumination of Cell “slime tracks” Observations include: – Cell division – Slime Track navigation by cells • Cells Turning to follow tracks • Cells bending as the move along tracks – Cell Clutsers Dynamics • Bending and Cell Oreintation in groups of cells Simulations of rod-shaped bacteria moving on surfaces Cell Division Preliminary data suggests that cells stall during division. (Yellow arrow highlight additional time between frames during division. Red arrow point to septum forming) Questions: How does polarity of mother cell relate to daughter cells? Does phase of reversal period get passed to daughter cells? Cell Turn on Slime Tracks M. xanthus is known to produce slime tracks when gliding on agar. By highlighting cell trajectories, cell-slime track locations can be visualized and cell-track interactions can be analyzed Questions: How much turning can cells undergo to get onto track? Detailed Segmentation can analyze cell bending on Slime Tracks Segmentation of raw image data is processed to form meshes on each cell. The mesh can be used to extract a central line that can be analyzed for bending of cells during collisions or during cell-trail interaction. Cell Bending and Orientation in Clusters We are also interested in cell bending and cell orientation and spatial ordering as cells dynamical form and move as groups. Sub-Cellular Elements Simulations Simulation Movie Link (http://biomath.math.nd.edu/compbio/CellSim4.gif) Simulations are being run to study flexibility, adhesion and cell reversals on cell clustering dynamics. Also, simulations that look at cell–slime track interaction are being run by coupling cells to a 2D substrate that simulates slime trails left by cells. (Slime not shown, but the 2d grid represents the scaler field discretization) Simulations prospects for Aneromyxobacter Model for Rod Shaped bacteria moving on surfaces is established. The substrate can be created to simulated Iron-hydroxide substrate or other environments to simulate chemical reactions at positions of cells. 13