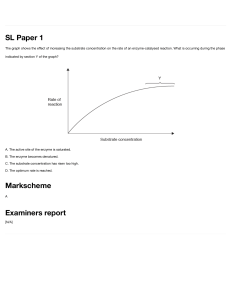
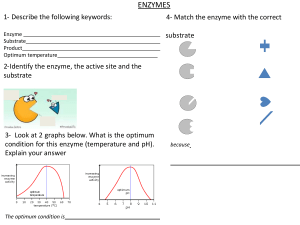
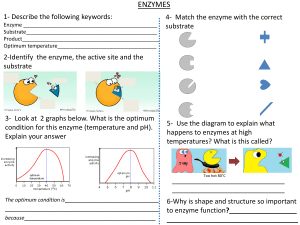
Enzymes Introduction All enzymes are protein. Catalytic activity of the enzyme depends on the integrity of their native protein conformation. Catalytic activity will be lost/destroyed by enzymes that will be broken or denatured. Primary, secondary, tertiary, and quaternary structures of proteins are essential for catalytic activity. Some enzymes require cofactors or coenzymes or both for their activity. Cofactor Cu2+ Fe2+/Fe3+ K+ Mg2+ Mn2+ Mo Ni2+ Se Zn2+ Coenzyme Biocytin Coenzyme A Enzyme Cytochrome oxidase Cytochrome oxidase Catalase Peroxidase Pyruvate kinase Pyruvate kinase Hexokinase Glucose-6-phosohatase Arginase Ribonucleotide reductase Dinitrogenase Urease Glutathione peroxidase Carboxy peptidase A & B Carbonic anhydrase Alcohol dehydrogenase Table 1.0: Some inorganic elements serve as cofactors for enzymes. Chemical group transferred CO2 Acyl groups Dietary precursor in mammals Biotin Pantothenic acid and other compounds Vitamin B12 5’-Deoxyadenosylcobalamin H atoms and alkyl groups (Coenzyme B12) Flavin adenine dinucleotide Electrons Riboflavin (Vitamin B2) Lipoate Electrons and acyl groups Not required in diet Nicotinamide adenine Hydride ion (:H-) Nicotinic acid (Niacin) dinucleotide Pyridoxal phosphate Amino group Pyridoxine (Vitamin B6) Tetrahydrofolate One-carbon group Folate Thiamine pyrophosphate Aldehydes Thiamine (Vitamin B1) Table 2.0: Some coenzymes that serve as transient carriers of specific atoms or functionals groups Figure 1.0: Prosthetic group (ie. Heam group) A coenzyme/metal ion is very tightly/covalently bound to the enzyme protein is called “prosthetic group”. Figure 2.0: Simple diagram for holoenzyme. A complete, catalytically active enzyme together with its bound coenzyme/metal ion is called “holoenzyme”. The protein part of such an enzyme is called “Apoprotein”. Apoprotein Coenzyme No. 1 Class Oxidoreductase 2 3 Transferases Hydrolases 4 Lyases 5 Isomerases 6 Ligases Table 3.0: International Classification of Enzymes Type of reaction catalyzed Transfer of electrons (Hydride ions/H atoms) Group transfer reaction Hydrolysis reactions (Transfer of functionals groups to water) Addition of groups of double bonds/formation of double bonds by removal of groups Transfer of groups within molecules to yield isomeric forms Formation of C-C, C-S, C-O, and C-N bonds by condensation reactions coupled to ATP cleavage Chemical transformation Chemical transformation is the surface of the active group is lined with amino acid residues with substituent groups that bind the substrate and catalyze. Active site The distinguish feature of enzyme-catalyzed reaction takes place within the confines of a pocket on the enzyme is called active site. Substrate The molecules is bound with active site and acted upon by the enzyme. Commonly the reaction rate of enzymes (highly effective catalysts) 105 - 1017. Enzyme-Catalyzed Reaction = Enzyme + Substrate (ES Complex) Function of enzymes and other catalysts depend on the lower activation energy for a reaction and enhance the reaction rate. Enzyme does not affect the equilibrium of a reaction. Enzymatic rate enhancements are derived from weak interactions (H bonds and hydrophobic and ionic interactions) between substrate and enzyme. Enzyme active site and substrate specificity Enzymes have active sites. Active sites are specific and have unique amino acid residue to bind the specific substrate. There may be one/more substrates for each type of enzyme depending on the chemical reaction. Sometimes, a single reactant substrate is broken down into multiple products and 2 substrates may come together to create one large molecule. However, 2 reactants might enter the reaction, both become modified, and leave the reaction as 2 products. The active site binds with substrate. Therefore, this site is composed by unique combination of amino acid residues (side chains/R groups). Each amino acid residues can be large/small; weakly acidic/base; hydrophilic/hydrophobic; positively charged/negatively charged/neutral. Specific chemical environment is created by the position, sequences, structures, and properties of these residues in the active site. A specific chemical substrate makes the enzyme specific to its substrate. Active site and environmental condition Enzyme’s active site is affected by environmental conditions. Increasing the environmental temperature increases reaction rates. Because when the temperature is rising, the molecules are moving more quickly and come to contact with each other. Increasing/decreasing the temperature outside of an optimal range can affect chemical bonds and change the shape of enzyme. If the enzyme changes shape, the active site may no longer bind with the appropriate substrate and the rate of reaction will decrease. Temperature and pH cause enzymes to denature. Induces fit and enzyme function. For long years, scientists believe that enzyme-substrate binding took place in “Lock and Key” fashion. According to the modern asserted that the enzyme and substrate fit together perfectly in one instantaneous step. Current research supports a more refined view which is called induced fit. When enzyme and substrate come together, their interaction causes a mild shift in the enzyme’s structure that confirms an ideal binding arrangement between the enzyme and the substrate. This dynamic binding maximizes the enzyme’s ability to catalyze its reaction. Enzyme-Substrate Complex Enzyme-substrate complex is formed when an enzyme binds its substrate. This complex is lower the activation energy and promotes rapid progression by providing certain ions/chemical groups that form covalent bonds with molecules. Enzymes promote chemical reactions by bringing substrates together in an optimal orientation, lining up the atoms and bonds of one molecule with the atoms and bonds of the molecule. It can contort the substrate molecules and facilitate bond breaking. The active site of an enzyme creates an ideal environment (slightly acidic/non-polar environment). After the completion of the reaction, the enzyme will return to its original state. One of the important properties of enzymes is that they remain ultimately unchanged by the reactions they catalyze. After an enzyme is done catalyzing a reaction, it releases its products (substrates). Competitive and non-competitive inhibition The cell uses specific molecules to regulate enzymes to promote/inhibit certain chemical reactions. It is necessary to inhibit an enzyme to reduce a reaction rate, and there is more than one way for this inhibition to occur. In competitive inhibition, an inhibitor molecule is similar enough to a substrate that it can bind to the enzyme’s active site to stop it from binding to the substrate. It competes with the substrate to bind the enzyme. In non-competitive inhibition, an inhibitor molecule binds to the enzyme at a location other than the active site (an allosteric site). The substrate can still bind to the enzyme, but the inhibitor changes the shape of the enzyme, so it is no longer in optimal position to catalyze the reaction. Allosteric inhibition and activation In non-competitive allosteric inhibition, inhibitor molecules bind to an enzyme at the allosteric site. Their binding induces a conformational change that reduces the affinity of the enzyme’s active site for its substrate. The binding of this allosteric inhibitor changes the conformation of the enzyme and its active site, so the substrate is not able to bind. It prevents the enzyme from lowering the activation energy of the reaction and the reaction rate is reduced. Allosteric inhibitors are not the only molecules that bind to allosteric sites. Allosteric activators can increase reaction rates. They bind to an allosteric site which induces a conformational change that increases the affinity of the enyme’s active site for its substrate. It increases the reaction rate.