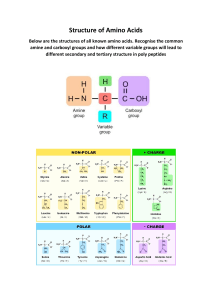
Huynh 1 Bioinformation Lab Report Name: Tony Huynh Course: BIO3810 Date: April 12th 2024 Instructor: Toba Omotosho Huynh 2 Result In this experiment, 9 different organisms were conducted including: Streptococcus suis, Streptococcus pneumoniae, Streptococcus mutans, Staphylococcus haemolyticus, Staphylococcus aureus, Thermoplasma acidophilum, Clostridium botulinum, Clostridium tetani, and Clostridium butyricum. After using NCBI to pick for the DNA topoisomerases of each organisms, Clustal Omega were used to determine the Phylogenetic Tree. After, Phylogenetic Tree were used to determine the two closest related organisms which were Clostridium tetani and Clostridium butyricum. Both sequences of the two organisms were then used to create alignment to determine the factors of its. There were several identical amino acids between C. Tetani and C. Butyricum ( Table 1). We used the colon “:” to determine the strongly similar molecular properties, which we were able to find at least three amino acid in a row. (Table 2) Weekly similar molecular properties were able to be found having 3 amino acids in a row as well, which indicate that their molecular functions were less likely to be linked. ( Table 4). And lastly, there were very few dissimilar amino acid chain due to the fact that these two organisms are closely related to each other. Although that, we were able to find 2 amino acids in a row ( Table 4). Questions 1. FIND IDENTICAL AMINO ACIDS Can you find anywhere in the alignment where 2 amino acids in a row are identical? What are the amino acids and their corresponding numbers? Yes. Table 1. Huynh 3 Species Identical Amino Acid Sequence Numbers Clostridium tetani 664-666 SFG or SerinePhenylalanine- Glycine Clostridium butyricum 664-666 SFG or SerinePhenylalanine- Glycine 2. FIND STRONGLY SIMILAR AMINO ACIDS Can you find anywhere in the alignment where 2 amino acids in a row are strongly similar? What are the amino acids and their corresponding numbers? Construct a table of your findings. Yes Table 2. Species Strongly Similar Amino Sequence Acid Numbers (:) Clostridium tetani 618-670 TKD or Threonine- LysineAspartic Acid Clostridium butyricum 618-670 SRE or Serine- ArginineGlutamic Acid Huynh 4 3. FIND WEAKLY SIMILAR AMINO ACIDS Can you find anywhere in the alignment where 2 amino acids in a row are weakly similar? What are the amino acids and their corresponding numbers? Construct a table of your findings. Yes Table 3. Species Weakly Similar Amino Acid Sequence Numbers (.) Clostridium tetani 361-363 FNK or PhenylalaineAsparagine- Lysine Clostridium butyricum 361-363 FTT or Phenylalanine- 4. FIND DISSIMILAR AMINO ACIDS Can you find anywhere in the alignment where 2 amino acids in a row are different and not similar (not alike at all)? What are the amino acids and their corresponding numbers? Construct a table of your findings. Yes Table 4. Species Dissimiliar Amino Acid Numbers Sequence Huynh 5 Clostridium tetani 253-254 KI or Lysine-Isoleucine Clostridium butyricum 253-254 SK or Serine- Lysine 5. WHICH END (N- TERMINUS OR C-ERMINUS) OF THE DNA TOPOISOMERASES IS THE MOST SIMILAR BETWEEN THE 2 DNA TOPOISOMERASES? HOW DO YOU KNOW? N-Terminus of these two DNA Topoisomerases is the most similar because N-Terminus side has longer sequences of identical amino acids than the C-Terminus. 6. Of the 9 total DNA topoisomerase sequences, which 2 are the most homologous (most alike). HOW DO YOU KNOW? Clostridium tetani and Clostridium butyricum are the most homologous (most alike) to each other because of their alignment scores, which indicate the similarity of those two DNA topoisomerases. C. Tetani has score of 0.13373 and C. Butyricum has score of 0.13646 and their difference are only 0.0027 which the smallest difference compared to the rest of DNA topoisomerases.