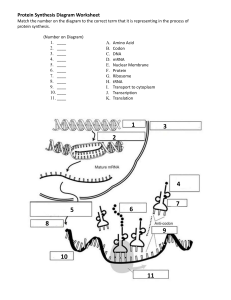
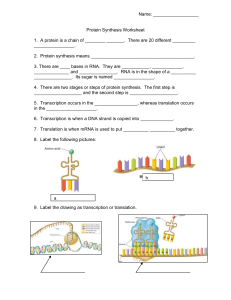
Protein Synthesis Dr. Nalin S Gama-Arachchige Reference: Campbell NA, Reece JB. 2002. Biology (6th ed). Benjamin Cummings, San Francisco, CA. 1 Topic 04: Protein Synthesis 1 hour Course Objectives: Identify different plant and animal cells/tissues and explain their functions Learning Objectives: Upon completion of this lesson, students will be able to: • • • Describe the process of protein synthesis Explain the function of different enzymes in protein synthesis Given a code, students will be able to ascertain the amino acid sequence 2 www.witnesswell.net Central Dogma of Molecular Biology The central dogma of molecular biology explains the flow of information within a biological system. DNA Transcription RNA Protein Translation 3 4 The Flow of Genetic Information RR/Rr Rr Round Wrinkled RR/Rr Round • The information content of DNA is in the form of specific sequences of nucleotides • The DNA inherited by an organism leads to specific traits by dictating the synthesis of proteins • Proteins are the links between genotype and phenotype • Gene expression: the process by which DNA directs protein synthesis (or, in some cases, just RNAs) rr Wrinkled 5 DNA molecule Gene 2 Gene 1 Gene 3 DNA template strand TRANSCRIPTION mRNA Codon TRANSLATION Protein Amino acid 6 Gene • A region of specific nucleotide sequence in a chromosome that codes for a specific polypeptide chain • The coding sequences (exons) are interrupted by noncoding DNA (introns) 7 Protein Synthesis: Four stages 1. Transcription 2. RNA processing 3. Translation 4. Post-translation processing 8 TRANSCRIPTION The THREE stages of transcription 1. Initiation of Transcription 2. Elongation of the RNA Strand 3. Termination of Transcription 9 TRANSCRIPTION 1. Initiation of Transcription • Promoters signal the initiation of RNA synthesis • Transcription factors mediate the binding of RNA polymerase and the initiation of transcription • The completed assembly of transcription factors and RNA polymerase II bound to a promoter is called a transcription initiation complex • A promoter called a TATA box is crucial in forming the initiation complex in eukaryotes 10 TRANSCRIPTION Transcription Factors • Transcription factors are proteins that bind to DNA near the start of transcription of a Gene. • Transcription factors either inhibit or assist RNA polymerase in initiation and maintenance of transcription. 11 TRANSCRIPTION 2. Elongation of Transcription • As RNA polymerase moves along the DNA, it untwists the double helix, 10 to 20 bases at a time • Transcription progresses at a rate of 40 nucleotides per second in eukaryotes • A gene can be transcribed simultaneously by several RNA polymerases 12 Promoter Transcription unit 5’ 3’ Start point RNA polymerase 3’ 5’ DNA 1 Initiation 5’ 3’ 3’ 5’ RNA transcript Unwound DNA Template strand of DNA 2 Elongation Rewound DNA 5’ 3’ 3’ 5’ 3’ 5’ RNA transcript 13 Nontemplate strand of DNA Elongation RNA polymerase 3’ RNA nucleotides 3’ end 5’ 5’ Direction of transcription (“downstream”) Newly made RNA Template strand of DNA 14 TRANSCRIPTION 3. Termination of Transcription • In eukaryotes, the polymerase continues transcription after the pre-mRNA is cleaved from the growing RNA chain; the polymerase eventually falls off the DNA 15 Promoter Transcription unit 5’ 3’ Start point RNA polymerase 3’ 5’ DNA 1 Initiation 5’ 3’ 3’ 5’ RNA transcript Unwound DNA Template strand of DNA 2 Elongation Rewound DNA 5’ 3’ 3’ 5’ 3’ 5’ RNA transcript 3 Termination 5’ 3’ 3’ 5’ 3’ 5’ Completed RNA transcript (Pre-mRNA) 16 RNA PROCESSING • Enzymes in the eukaryotic nucleus modify pre-mRNA before the genetic messages are dispatched to the cytoplasm • During RNA processing, both ends of the primary transcript are usually altered • Also, usually some interior parts of the molecule are cut out, and the other parts spliced together 17 RNA PROCESSING • Each end of a pre-mRNA molecule is modified in a particular way: – The 5 end receives a modified nucleotide 5 cap – The 3 end gets a poly-A tail • These modifications share several functions – They seem to facilitate the export of mRNA – They protect mRNA from hydrolytic enzymes – They help ribosomes attach to the 5 end 18 RNA PROCESSING Protein-coding segment Polyadenylation signal 5’ G P P 5’ Cap 3’ P AAUAAA 5’ UTR Start codon A modified guanine nucleotide added to the 5′ end Stop codon 3’ UTR AAA … AAA Poly-A tail 50–250 adenine nucleotides added to the 3′ end RNA processing: addition of the 5 cap and poly-A tail 19 RNA PROCESSING • Most eukaryotic genes and their RNA transcripts have long noncoding stretches of nucleotides that lie between coding regions • These noncoding regions are called intervening sequences, or introns • The other regions are called exons because they are eventually expressed, usually translated into amino acid sequences • RNA splicing removes introns and joins exons, creating an mRNA molecule with a continuous coding sequence 20 RNA PROCESSING 5’ Exon Intron Pre-mRNA Exon Exon Intron 3’ Poly-A tail 5’ Cap 1 30 31 Coding segment mRNA 5’ Cap 1 5’ UTR 104 146 105 Introns cut out and exons spliced together Poly-A tail 146 3’ UTR RNA processing: RNA splicing 21 RNA PROCESSING • RNA splicing is carried out by spliceosomes • Spliceosomes consist of a variety of proteins and several small nuclear ribonucleoproteins (snRNPs) that recognize the splice sites 22 5’ RNA transcript (pre-mRNA) Exon 1 Intron Exon 2 Protein snRNA Other proteins snRNPs Spliceosome 5’ Spliceosome components 5’ mRNA Exon 1 Exon 2 Cut-out intron 23 TRANSLATION Translation is the process by which ribosomes read the genetic message in the mRNA and produce a protein product according to the instruction that message 24 Ribosomes • Ribosomes facilitate specific coupling of tRNA anticodons with mRNA codons in protein synthesis • The two ribosomal subunits (large and small) are made of proteins and ribosomal RNA (rRNA) Growing polypeptide Exit tunnel tRNA molecules Large subunit EP A Small subunit 5’ mRNA 3’ 25 Computer model of functioning ribosome Ribosomes P site (Peptidyl-tRNA binding site) A site (Aminoacyl-tRNA binding site) tRNA The ribosome has multiple binding sites: – P site – binds the tRNA attached to the growing peptide chain E P – A site – binds the tRNA carrying the next amino acid – E site – binds the tRNA that carried the last amino acid mRNA binding site Large subunit A Large subunit E site (Exit site) 26 Schematic model showing binding sites Ribosomes The ribosome has two primary functions – decode the mRNA – form peptide bonds Peptidyl transferase is the enzymatic component of the ribosome which forms peptide bonds between amino acids 27 tRNA tRNA molecules carry amino acids to the ribosome for incorporation into a polypeptide – aminoacyl-tRNA synthetases add amino acids to the amino acid attachment site of tRNA – the anticodon loop contains 3 nucleotides complementary to mRNA codons 28 3’ Amino acid attachment site PO4 5’ Amino acid attachment site 5’ 3’ T loop Hydrogen bonds D loop Hydrogen bonds Variable loop 5’ 3’ Anticodon Anticodon Anticodon Two-dimensional structure of tRNA Three-dimensional structure of tRNA 29 Twenty Amino Acids 30 The codon table for31mRNA Growing polypeptide chain Next amino acid to be added to polypeptide chain Amino end Large subunit tRNA E mRNA 5’ Codons 3’ Small subunit Schematic model with mRNA and tRNA 32 TRANSLATION The THREE stages of translation 1. Initiation of Translation 2. Elongation of the Polypeptide Chain 3. Termination of Translation 33 TRANSLATION 1. Initiation of Translation • The initiation stage of translation brings together mRNA, a tRNA with the first amino acid, and the two ribosomal subunits • First, a small ribosomal subunit binds with mRNA and a special initiator tRNA • Then the small subunit moves along the mRNA until it reaches the start codon (AUG) • Proteins called initiation factors bring in the large subunit that completes the translation initiation complex 34 Large ribosomal subunit 3’ U A C 5’ 5’ A U G 3’ Initiator tRNA P site GTP GDP E mRNA 5’ Start codon mRNA binding site 3’ Small ribosomal subunit 5’ A 3’ Translation initiation complex 35 TRANSLATION 2. Elongation of Translation • During the elongation stage, amino acids are added one by one to the preceding amino acid • Each addition involves proteins called elongation factors and occurs in three steps: codon recognition, peptide bond formation, and translocation 36 Amino end of polypeptide E 3’ mRNA Ribosome ready for next aminoacyl tRNA P A site site 5’ GTP GDP E E P A P A GDP GTP E P A 37 Amino acids Polypeptide Ribosome tRNA with amino acid attached tRNA Anticodon Codons 5’ mRNA 3’ 38 TRANSLATION 3. Termination of Translation • Termination occurs when a stop codon in the mRNA reaches the A site of the ribosome • The A site accepts a protein called a release factor • The release factor causes the addition of a water molecule instead of an amino acid • This reaction releases the polypeptide, and the translation assembly then comes apart 39 Release factor Free polypeptide 5’ 3’ 5’ 5’ Stop codon (UAG, UAA, or UGA) 3’ 2 GTP 3’ 2 GDP 40 41 TRANSLATION Polyribosomes • A number of ribosomes can translate a single mRNA simultaneously, forming a polyribosome (or polysome) • Polyribosomes enable a cell to make many copies of a polypeptide very quickly 42 TRANSLATION Polyribosomes Growing polypeptides Completed polypeptide Incoming ribosomal subunits Start of mRNA (5’ end) End of mRNA (3’ end) 43 POST TRANSLATION PROCESSING • Often translation is not sufficient to make a functional protein • Polypeptide chains are modified after translation • Completed proteins are targeted to specific sites in the cell 44 POST TRANSLATION PROCESSING • During and after synthesis, a polypeptide chain spontaneously coils and folds into its three-dimensional shape • Proteins may also require post-translational modifications before doing their job • Some polypeptides are activated by enzymes that cleave them • Other polypeptides come together to form the subunits of a protein 45 Targeting Polypeptides to Specific Locations • Free ribosomes (in the cytosol) mostly synthesize proteins that function in the cytosol • Bound ribosomes (attached to the ER) make proteins of the endomembrane system and proteins that are secreted from the cell • Ribosomes are identical and can switch from free to bound 46 Targeting Polypeptides to Specific Locations • Polypeptide synthesis always begins in the cytosol • Synthesis finishes in the cytosol unless the polypeptide signals the ribosome to attach to the ER • Polypeptides destined for the ER or for secretion are marked by a signal peptide 47 Ribosome mRNA Signal peptide Signal peptide removed Signalrecognition particle (SRP) CYTOSOL ER LUMEN SRP receptor protein ER membrane Protein Translocation complex 48 RR/Rr Round rr Wrinkled The Flow of Genetic Information The pea plant can produce smooth or wrinkled seeds. Wrinkled peas taste sweet. • When functioning normally, the SBE1(starch-branching enzyme 1) enzyme builds starch. • A mutation took placed in the coding sequence of the SBE1 gene. disables the protein. • The job of SBE1 is to add branches to the chains of sugar. • The peas can't make branched starch, and sugar accumulates. • Branches help starch grow very large, and break down quickly when the plant needs energy. • The sugar attracts water, causing the peas to swell and then wrinkle as they mature and dry. 49 KEY TERMS: Genes to Proteins • Trascription • mRNA • Translation • RNA Processing • TATA box • Pre-mRNA • Template Strand • Codon • RNA polymerase • Transcription factors • Terminator • 5’ cap • Poly (A) tail • RNA splicing • Intron • Exon • Splicesome • tRNA • Anti codon • P A E sites 50