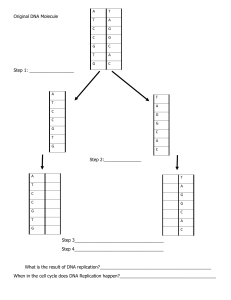
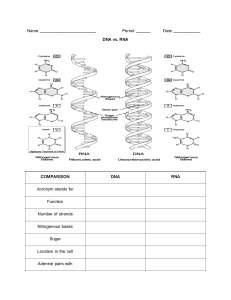
test-bank-for-essential-cell-biology-4th-edition-alberts
advertisement

lOMoARcPSD|30659517 Test Bank for Essential Cell Biology 4th Edition Alberts Nursing Care of the Child (Alabama State University) Scan to open on Studocu Studocu is not sponsored or endorsed by any college or university Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Chapter 1: CELLS: THE FUNDAMENTAL UNITS OF LIFE Unity and Diversity of Cells 1-1 Living systems are incredibly diverse in size, shape, environment, and behavior. It is estimated that there are between 10 million and 100 million different species. Despite this wide variety of organisms, it remains difficult to define what it means to say something is alive. Which of the following can be described as the smallest living unit? (a) DNA (b) cell (c) organelle (d) protein 1-2 Indicate whether the following statements are true or false. If the statement is false, explain why it is false. A. The Paramecium is a multicellular microorganism covered with hair-like cilia. B. Cells of different types can have different chemical requirements. C. The branchlike extensions that sprout from a single nerve cell in a mammalian brain can extend over several hundred micrometers. 1-3 For each of the following sentences, fill in the blanks with the best word or phrase selected from the list below. Not all words or phrases will be used; each word or phrase should be used only once. Cells can be very diverse: superficially, they come in various sizes, ranging from bacterial cells such as Lactobacillus, which is a few in length, to larger cells such as a frog’s egg, which has a diameter of about one .Despite the diversity, cells resemble each other to an astonishing degree in their chemistry. For example, the same 20 are used to make proteins. Similarly, the genetic information of all cells is stored in their . Although Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 contain the same types of molecules as cells, their inability to reproduce themselves by their own efforts means that they are not considered living matter. amino acids micrometer(s) viruses DNA millimeter(s) yeast fatty acids plants meter plasma membranes 1-4 How does cellular specialization serve multicellular organisms and how might a high degree of specialization be detrimental? 1-5 The flow of genetic information is controlled by a series of biochemical reactions that Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 result in the production of proteins, each with its own specific order of amino acids. Choose the correct series of biochemical reactions from the options presented here. (a) replication, transcription, translation (b) replication, translation, transcription (c) translation, transcription, replication (d) translation, replication, transcription 1-6 Proteins are important architectural and catalytic components within the cell, helping to determine its chemistry, its shape, and its ability to respond to changes in the environment. Remarkably, all of the different proteins in a cell are made from the same 20 . By linking them in different sequences, the cell can make protein molecules with different conformations and surface chemistries, and therefore different functions. (a) nucleotides. (b) sugars. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (c) amino acids. (d) fatty acids. 1-7 Which statement is NOT true about mutations? (a) A mutation is a change in the DNA that can generate offspring less fit for survival than their parents. (b) A mutation can be a result of imperfect DNA duplication. (c) A mutation is a result of sexual reproduction. (d) A mutation is a change in the DNA that can generate offspring that are as fit for survival as their parents are. 1-8 Changes in DNA sequence from one generation to the next may result in offspring that are altered in fitness compared with their parents. The process of change and selection over the course of many generations is the basis of . (a) mutation. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (b) evolution. (c) heredity. (d) reproduction. 1-9 Select the option that best finishes the following statement: “Evolution is a process .” (a) that can be understood based on the principles of mutation and selection. (b) that results from repeated cycles of adaptation over billions of years. (c) by which all present-day cells arose from 4–5 different ancestral cells. (d) that requires hundreds of thousands of years. 1-10 Select the option that correctly finishes the following statement: “A cell’s genome .” (a) is defined as all the genes being used to make protein. (b) contains all of a cell’s DNA. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (c) constantly changes, depending upon the cell’s environment. (d) is altered during embryonic development. Cells Under the Microscope 1-11 Which statement is NOT true about the events/conclusions from studies during the mid- 1800s surrounding the discovery of cells? (a) Cells came to be known as the smallest universal building block of living organisms. (b) Scientists came to the conclusion that new cells can form spontaneously from the remnants of ruptured cells. (c) Light microscopy was essential in demonstrating the commonalities between plant and animal tissues. (d) New cells arise from the growth and division of previously existing cells. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-12 What unit of length plant or animal cell? would you generally use to measure atypical (a) centimeters (b) nanometers (c) millimeters (d) micrometers 1-13 Match the type of microscopy on the left with the corresponding description provided below. There is one best match for each. A. confocal B. transmission electron C. fluorescence D. phase-contrast E. scanning electron F. bright-field Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 uses a light microscope with an optical component to take advantage of the different refractive indices of light passing through different regions of the cell. employs a light microscope and requires that samples be fixed and stained in order to reveal cellular details. requires the use of two sets of filters. The first filter narrows the wavelength range that reaches the specimen and the second blocks out all wavelengths that pass back up to the eyepiece except for those emitted by the dye in the sample. scans the specimen with a focused laser beam to obtain a series of two-dimensional optical sections, which can be used to reconstruct an image of the specimen in three dimensions. The laser excites a fluorescent dye molecule, and the emitted light from each illuminated point is captured through a pinhole and recorded by a detector. has the ability to resolve cellular components as small as 2 nm. requires coating the sample with a thin layer of a heavy metal to produce three- Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 dimensional images of the surface of a sample. 1-14 Indicate whether the following statements are true or false. If the statement is false, explain why it is false. A. The nucleus of an animal cell is round, small, and difficult to distinguish using light microscopy. B. The presence of the plasma membrane can be inferred by the well-defined boundary of the cell. C. The cytosol is fairly empty, containing a limited number of organelles, which allows room for rapid movement via diffusion. 1-15 Cell biologists employ targeted fluorescent dyes or modified fluorescent proteins in both standard fluorescence microscopy and confocal microscopy to observe specific details in the cell. Even though fluorescence permits better visualization, the resolving power is Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 essentially the same as that of a standard light microscope because the resolving power of a microscope is limited by the of light. (a) absorption (b) intensity (c) filtering (d) wavelength 1-16 What is the smallest distance two points can be separated and still resolved using light microscopy? (a) 20 nm (b) 0.2 µm (c) 2 µm (d) 200 µm The Prokaryotic Cell Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-17 By definition, prokaryotic cells do not possess . (a) a nucleus. (b) replication machinery. (c) ribosomes. (d) membrane bilayers. 1-18 Although there are many distinct prokaryotic species, most have a small range of shapes, sizes, and growth rates. Which of the following characteristics are not observed in prokaryotes? (a) a highly structured cytoplasm (b) endoplasmic reticulum (c) the ability to divide rapidly (d) a cell wall Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-19 Indicate whether the following statements are true or false. If the statement is false, explain why it is false. A. The terms “prokaryote” and “bacterium” are synonyms. B. Prokaryotes can adopt several different basic shapes, including spherical, rod- shaped, and spiral. C. Some prokaryotes have cell walls surrounding the plasma membrane. 1-20 Prokaryotic cells are able to evolve very fast, which helps them to rapidly adapt to new food sources and develop resistance to antibiotics. Which of the options below lists the three main characteristics that support the rapid evolution of prokaryotic populations? (a) microscopic, motile, anaerobic (b) aerobic, motile, rapid growth (c) no organelles, cell wall, can exchange DNA (d) large population, rapid growth, can exchange DNA Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-21 Indicate whether the following statements are true or false. If the statement is false, explain why it is false. A. Oxygen is toxic to certain prokaryotic organisms. B. Mitochondria are thought to have evolved from anaerobic bacteria. C. Photosynthetic bacteria contain chloroplasts. 1-22 Some prokaryotes can live by utilizing entirely inorganic materials. Which of the following inorganic molecules would you predict to be the predominant building block for fats, sugars, and proteins? (a) O2 (b) N2 (c) CO2 (d) H2 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 The Eukaryotic Cell 1-23 Use the list of structures below to label the schematic drawing of an animal cell in Figure Q1-23. Figure Q1-23 A. plasma membrane B. nuclear envelope C. cytosol D. Golgi apparatus E. endoplasmic reticulum F. mitochondrion G. transport vesicles Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-24 For each of the following sentences, fill in the blanks with the best word or phrase selected from the list below. Not all words or phrases will be used; each word or phrase should be used only once. Eukaryotic cells are bigger and more elaborate than prokaryotic cells. By definition, all eukaryotic cells have a , usually the most prominent organelle. Another organelle found in essentially all eukaryotic cells is the for the cell. In , which generates the chemical energy contrast, the is a type of organelle found only in the cells of plants and algae, and performs photosynthesis. If we were to strip away the plasma membrane from a eukaryotic cell and remove all of its membrane- enclosed organelles, we would be left with the , which Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 contains many long, fine filaments of protein that are responsible for cell shape and structure and thereby form the cell’s chloroplast cytosol . nucleus chromosome endoplasmic reticulum ribosomes cytoskeleton mitochondrion 1-25 The is made up of two concentric membranes and is continuous with the membrane of the endoplasmic reticulum. (a) plasma membrane (b) Golgi network (c) mitochondrial membrane (d) nuclear envelope Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-26 The nucleus, an organelle found in eukaryotic cells, confines the , keeping them separated from other components of the cell. (a) lysosomes (b) chromosomes (c) peroxisomes (d) ribosomes 1-27 Which of the following organelles has both an outer and an inner membrane? (a) endoplasmic reticulum (b) mitochondrion (c) lysosome (d) peroxisome 1-28 Mitochondria perform cellular respiration, a process that uses oxygen, generates carbon Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 dioxide, and produces chemical energy for the cell. Which answer below indicates a correct pairing of material “burned” and the form of energy produced during cellular respiration? (a) fat, ADP (b) sugar, fat (c) sugar, ATP (d) fat, protein 1-29 You fertilize egg cells from a healthy plant with pollen (which contains the male germ cells) that has been treated with DNA-damaging agents. You find that some of the offspring have defective chloroplasts, and that this characteristic can be passed on to future generations. This surprises you at first because you happen to know that the male germ cell in the the egg cell and thus pollen grain contributes no chloroplasts to fertilized to the offspring. What can you deduce from these results? Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-30 Mitochondria contain their own genome, are able to duplicate, and actually divide on a different time line from the rest of the cell. Nevertheless, mitochondria cannot function for long when isolated from the cell because they are . (a) viruses. (b) parasites. (c) endosymbionts. (d) anaerobes. 1-31 The mitochondrial proteins found in the inner membrane are involved in the conversion of ADP to ATP, a source of energy for the cell. This process consumes which of the following substances? (a) oxygen (b) nitrogen Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (c) sulfur (d) carbon dioxide 1-32 Indicate whether the following statements are true or false. If the statement is false, explain why it is false. A. With respect to cellular respiration, the only organelles used by animal cells are mitochondria, while plant cells use both mitochondria and chloroplasts. B. The number of mitochondria inside a cell remains constant over the life of the cell. 1-33 Chloroplasts are found only in eukaryotic cells that carry out photosynthesis: plants and algae. Plants and algae appear green as a result of the presence of chlorophyll. Where is chlorophyll located in the chloroplast? (a) in the first, outer membrane Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (b) in the space between the first and second membranes (c) in the second, inner membrane (d) in the third, innermost membrane 1-34 Photosynthesis enables plants to capture the energy from sunlight. In this essential process, plants incorporate the carbon from CO2 into high-energy molecules, which the plant cell mitochondria use to produce ATP. (a) fat (b) sugar (c) protein (d) fiber 1-35 Indicate whether the following statements are true or false. If the statement is false, explain why it is false. A. Membrane components in the cell are made in the endoplasmic reticulum. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 B. The Golgi apparatus is made up of a series of membrane-enclosed compartments through which materials destined for secretion must pass. C. Lysosomes are small organelles where fatty acid synthesis occurs. 1-36 Circle the appropriate cell type in which the listed structure or molecule can be found. Note that the structure or molecule can be found in more than one type of cell. 1-37 Which of the following choices best describes the role of the lysosome? (a) transport of material to the Golgi (b) clean-up, recycling, and disposal of macromolecules (c) sorting of transport vesicles (d) the storage of excess macromolecules Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-38 The protozoan Didinium feeds on other organisms by engulfing them. Why are bacteria, in general, unable to feed on other cells in this way? 1-39 The cell constantly exchanges materials by bringing nutrients in from the external environment and shuttling unwanted by-products back out. Which term describes the process by which external materials are captured inside vesicles and brought into the cell? (a) degradation (b) exocytosis (c) phagocytosis (d) endocytosis 1-40 Eukaryotic cells are able to trigger the release of material from secretory vesicles to the Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 extracellular space using a process called exocytosis. An example of materials commonly released this way is . (a) hormones. (b) nucleic acids. (c) sugars. (d) cytosolic proteins. 1-41 are fairly small organelles that provide a safe place within the cell to carry out certain biochemical reactions that generate harmful, highly reactive oxygen species. These chemicals are both generated and broken down in the same location. (a) N ucleosomes (b) Lysosomes (c) Peroxisomes (d) Endosomes Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-42 The cytoskeleton provides support, structure, motility, and organization, and it forms tracks to direct organelle and vesicle transport. Which of the cytoskeletal elements listed below is the thickest? (a) actin filaments (b) microtubules (c) intermediate filaments (d) none of the above (all the same thickness) 1-43 Despite the differences between eukaryotic and prokaryotic cells, prokaryotes have proteins that are distantly related to eukaryotic actin filaments and microtubules. What is likely to be the most ancient function of the cytoskeleton? (a) cell motility (b) vesicle transport Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (c) membrane support (d) cell division 1-44 Which of the following characteristics would not support the idea that the ancestral eukaryote was a predator cell that captured and consumed other cells? (a) dynamic cytoskeleton (b) large cell size (c) ability to move (d) rigid membrane 1-45 Choose the phrase that best completes this sentence: Microtubules required to pull duplicated chromosomes to opposite poles of dividing cells. (a) generate contractile forces (b) are intermediate in thickness Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) and are lOMoARcPSD|30659517 (c) can rapidly reorganize (d) are found in especially large numbers in muscle cells 1-46 Indicate whether the following statements are true or false. If the statement is false, explain why it is false. A. Plants do not require a cytoskeleton because they have a cell wall that lends structure and support to the cell. B. The cytoskeleton is used as a transportation grid for the efficient, directional movement of cytosolic components. C. Thermal energy promotes random movement of proteins, vesicles, and small molecules in the cytosol. 1-47 Which pair of values best fills in the blanks in this statement: On average, eukaryotic cells are times longer and have times more volume than prokaryotic cells. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (a) 5, 100 (b) 10, 200 (c) 10, 100 (d) 10, 1000 1-48 Indicate whether the following statements are true or false. If the statement is false, explain why it is false. A. Primitive plant, animal, and fungal cells probably acquired mitochondria after they diverged from a common ancestor. B. Protozoans are single-celled eukaryotes with cell morphologies and behaviors that can be as complex as those of some multicellular organisms. C. The first eukaryotic cells on Earth must have been aerobic; otherwise, they would not have been able to survive when the planet’s atmosphere became oxygen-rich. Model Organisms Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-49 Given what you know about the differences between prokaryotic cells and eukaryotic cells, rate the following things as “good” or “bad” processes to study in the model organism E. coli. A. formation of the endoplasmic reticulum B. DNA replication C. how the actin cytoskeleton contributes to cell shape D. how cells decode their genetic instructions to make proteins E. how mitochondria get distributed to cells during cell division 1-50 Scientists learned that cell death is a normal and even important part of life by studying the development of the nematode worm C. elegans. What was the most important feature of C. elegans for the study of programmed cell death? (a) The nematode is smaller and simpler than the fruit fly. Page 10 of 21 (b) 70% of C. elegans genes have homologs in humans. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (c) The developmental pathway of each cell in the adult worm was known. (d) Its genome was partially sequenced. 1-51 Biologists cannot possibly study all living species. Instead, they try to understand cell behavior by studying a select subset of them. Which of the following characteristics are useful in an organism chosen for use as a model in laboratory studies? (a) amenability to genetic manipulation (b) ability to grow under controlled conditions (c) rapid rate of reproduction (d) all of the above 1-52 Many of the mechanisms that cells use for maintenance and reproduction were first studied at the molecular level in bacteria. Which bacterial species had a central role in advancing the field of molecular biology? (a) E. coli Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (b) D. melanogaster (c) S. pombe (d) C. elegans 1-53 Brewer’s yeast, apart from being an irreplaceable asset in the brewery and in the bakery, is an experimental organism used to study eukaryotic cells. However, it does have some limitations. Which of the processes below cannot be studied in yeast? (a) DNA replication (b) cell motility (c) exocytosis (d) cell division 1-54 For each process (A–D), circle the simplest model organism from the list of three that would be best used for investigation: Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-55 A. thaliana, or Arabidopsis, is a common weed. Biologists have selected it over hundreds of thousands of other flowering plant species to serve as an experimental model organism because . (a) it can withstand extremely cold climates. (b) it can reproduce in 8–10 weeks. (c) it produces thousands of offspring per plant. (d) Both (b) and (c) are true. 1-56 Drosophila melanogaster is a/an . This type of animal is the most abundant of all animal species, making it an appropriate choice as an experimental model. (a) insect Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (b) bird (c) amphibian (d) mammal 1-57 Caenorhabditis elegans is a nematode. During its development, it produces more than 1000 cells. However, the adult worm has only 959 somatic cells. The process by which 131 cells are specifically targeted for destruction is called . (a) directed cell pruning. (b) programmed cell death. (c) autophagy. (d) necrosis. 1-58 Zebrafish (Danio rerio) are especially useful in the study of early development because their embryos . (a) are exceptionally large. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (b) develop slowly. (c) are transparent. (d) are pigmented. 1-59 You wish to explore how mutations in specific genes affecting sugar metabolism might alter tooth development. Which organism is likely to provide the best model system for your studies, and why? (a) horses (b) mice (c) E. coli (d) Arabidopsis 1-60 Indicate whether the following statements are true or false. If the statement is false, explain why it is false. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 A. The human genome is roughly 30 times larger than the Arabidopsis genome, but contains approximately the same number of protein-coding genes. B. The variation in genome size among protozoans is larger than that observed across all species of mammals, birds, and reptiles. C. The vast majority of our genome encodes functional RNA molecules or proteins and most of the intervening DNA is nonfunctional. 1-61 Genes that have homologs in a variety of species have been discovered through the analysis of genome sequences. In fact, it is not uncommon to find a family of homologous genes encoding proteins that are unmistakably similar in amino acid sequence in organisms as diverse as budding yeast, archaea, plants, and humans. Even more remarkably, many of these proteins can substitute functionally for their homologs in other organisms. Explain what it is about the origins of cells that makes it possible for proteins expressed by homologous genes to be functionally interchangeable in different Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 organisms. 1-62 Match each biological process with the model organism that is best suited or most specifically useful for its study, based on information provided in your textbook. You may list individual processes more than once. A. cell division B. development (multicellular) C. programmed cell death D. photosynthesis E. immunology A. thaliana (Arabidopsis) M. musculus (mouse) S. pombe C. elegans Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 S. cerevisiae D. rerio (zebrafish) D. melanogaster Testing the Concepts 1-63 Employ the principles of evolution discussed in this chapter to explain how the specific features and predatory behaviors of some primitive eukaryotes may have given them a selective advantage over others 1.5 billion years ago. 1-64 Evolutionary biologists have always used a broad range of modern organisms to infer the characteristics that ancestral organisms may have possessed. Genomic sequences are now available for an increasing number of species, and scientists studying evolutionary processes can take advantage of this enormous amount of data to bring evolution into the arena of molecular studies. By aligning the sequences of homologous genes and looking Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 for regions of similarity and where changes have occurred, it is possible to infer the sequence of the ancestral gene. A. What term is used to describe the changes in gene sequences that have occurred? How can we use what we know about this process to construct a time line showing when various sequence changes occurred and when they led to the modern sequences that we know today? B. It is possible to express an ancestral gene sequence in modern organisms and subsequently compare the function of its product with that of the modern protein. Why might this approach give misleading conclusions? 1-65 The antibiotic streptomycin inhibits protein synthesis in bacteria. If this antibiotic is added to a culture of animal cells, protein synthesis in the cytosol continues normally. However, over time, the population of mitochondria in the cell becomes depleted. Specifically, it is observed that the protein-synthesis machinery inside the mitochondria is Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 inhibited. A. Explain this observation based on what you know about the origins of the modern eukaryote. B. What do you expect to observe if, in a new experiment, animal cells are treated with diphtheria toxin, a compound that is known to block cytosolic protein synthesis but does not have any impact on bacterial growth? 1-66 You have been following the recent presidential elections and have heard some candidates disparaging excessive and “unnecessary” federal government expenditures. One particular candidate asks: “Why are we spending millions of dollars studying fruit flies? How can that possibly help us find a cure for cancer?” Use your knowledge of model organisms to explain why studies in D. melanogaster (the fruit fly) are actually an excellent use of research funding. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-67 Cellular processes are often regulated by unknown mechanisms. In many cases, biologists work in which they are backward in an attempt to understand a process interested. This was the case when Nurse and Hartwell were trying to understand how cell division is controlled in yeast. Describe the process by which they “broke” the system and then supplied the “missing parts” to get the cell cycle running again. What further evidence did they collect to show that human cells and yeast cells regulate the cell cycle using a similar mechanism? 1-68 Your friend has just returned from a deep-sea mission and claims to have found a new single-celled life-form. He believes this new life-form may not have descended from the common ancestor that all types of life on Earth share. You are convinced that he must be wrong, and you manage to extract DNA from the cells he has discovered. He says that the mere presence of DNA is not enough to prove the point: his cells might have adopted DNA as a useful molecule quite independently of all other known life-forms. What could you do to provide additional evidence to support your argument? Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 ANSWERS 1-1 (b) 1-2 A. False. The Paramecium is a single-celled organism. B. True. C. True. 1-3 Cells can be very diverse: superficially, they come in various sizes, ranging from bacterial cells such as Lactobacillus, which is a few micrometers in length, to larger cells such as a frog’s egg, which has a diameter of about one millimeter. Despite the diversity, cells resemble each other to an astonishing degree in their chemistry. For example, the same 20 amino acids are used to make proteins. Similarly, the genetic information of all cells is stored in their DNA. Although viruses contain the same types of molecules as cells, their inability to reproduce themselves by their own efforts means that they are not considered living matter. 1-4 In a multicellular organism, the specialization of cells creates a division of labor and each type of cell relies on the activities of other cell types for survival. This cooperation between specialized cells is essential for the organism as a whole. If one of these overly specialized cells were removed from the context of the organism, it would not have the capabilities needed to generate offspring and would probably not even live very long. 1-5 (a) 1-6 (c) 1-7 (c) 1-8 (b) 1-9 (a) 1-10 (b) 1-11 (b) 1-12 (d) 1-13 F D Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 C A B E 1-14 A. False. The nucleus is one of the largest organelles and is the easiest organelle to discern within a typical cell. B. True. C. False. The cytosol is actually brimming with individual proteins, protein fibers, extended membrane systems, transport vesicles, and small molecules. And although cellular components do move by diffusion, the rate of movement is limited by the space available and the size of the component in question. 1-15 (d) 1-16 (b) 1-17 (a) Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-18 (a), (b) 1-19 A. False. Archaea that are significantly make up a class of prokaryotic organisms different from bacteria. B. True. C. True. 1-20 (d) 1-21 (a) True. (b) False. Mitochondria use oxygen to generate energy and are thought to have evolved from aerobic bacteria. (c) False. Photosynthetic bacteria have enzyme systems similar to those found in chloroplasts, which allow them to harvest light energy to fix carbon dioxide. 1-22 (c) 1-23 A. Plasma membrane—3 B. Nuclear envelope—5 C. Cytosol—1 D. Golgi apparatus—2 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 E. Endoplasmic reticulum—4 F. Mitochondrion—7 G. Transport vesicles—6 1-24 Eukaryotic cells are bigger and more elaborate than prokaryotic cells. By definition, all eukaryotic cells have a nucleus, usually the most prominent organelle. Another organelle found in essentially all eukaryotic cells is the mitochondrion, which generates the chemical energy for the cell. In contrast, the chloroplast is a type of organelle found only in the cells of plants and algae, and performs photosynthesis. If we were to strip away the plasma membrane from a eukaryotic cell and remove all of its membrane-enclosed organelles, we would be left with the cytosol, which contains many long, fine filaments of protein that are responsible for cell shape and structure and thereby form the cell’s cytoskeleton. 1-25 (d) 1-26 (b) 1-27 (b) Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-28 (c) 1-29 Your results show that not all of the information required for making a chloroplast is encoded in the chloroplast’s own DNA; some, at least, must be encoded in the DNA carried in the nucleus. The reasoning is as follows. Genetic information is carried only in DNA, so the defect in the chloroplasts must be due to a mutation in DNA. But all of the chloroplasts in the offspring (and thus all of the chloroplast DNA) must derive from those in the female egg other Hence, all of cell, since chloroplasts only arise from chloroplasts. the chloroplasts contain undamaged DNA from the female parent’s chloroplasts. In all the cells of the offspring, however, half of the nuclear DNA will have come from the male germ-cell nucleus, which combined with the female egg nucleus at fertilization. Since this DNA has been treated with DNA-damaging agents, it must be the source of the heritable chloroplast defect. Thus, some of the information required for making a chloroplast is encoded by the nuclear DNA. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-30 (c) 1-31 (a) 1-32 A. False. In plants, only mitochondria perform cellular respiration (using oxygen to break down organic molecules to produce carbon dioxide) just as in animal cells. Chloroplasts perform photosynthesis in which water molecules are split to generate oxygen and fix carbon dioxide molecules. B. False. Mitochondria have their own division cycle and their numbers change based on the rate of division. 1-33 (d) 1-34 (b) Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-35 A. True. B. True. C. False. Lysosomes house enzymes that break down nutrients for use by the cell and help recycle materials that cannot be used, which will later be excreted from the cell. 1-36 1-37 (b) 1-38 Didinium engulfs prey by changing its shape, and for this it uses its cytoskeleton. Bacteria have no cytoskeleton and cannot easily change their shape because they are generally surrounded by a tough cell wall. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-39 (d) 1-40 (a) 1-41 (c) 1-42 (b) 1-43 (d) 1-44 (d) 1-45 (c) 1-46 A. False. Although plant cells do have a cell wall that lends structure and support, they still need a cytoskeleton, which also helps with connections between cells Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 and the transport of vesicles inside the cell. B. True. C. True. 1-47 (d) 1-48 A. False. The mitochondria in modern plant, animal, and fungal cells are very similar, implying that these lines diverged after the mitochondrion was acquired by the ancestral eukaryote. B. True. C. False. The first eukaryotic cells likely contained a nucleus but no mitochondria. These ancestral eukaryotes subsequently adapted to survive in a world filled with oxygen by engulfing primitive aerobic prokaryotic cells. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-49 A. bad B. good C. bad D. good E. bad 1-50 (c) This is the best answer because it was the prior developmental studies tracing cell lineages from the egg to the adult that allowed scientists to identify the precise time and location of cells that were being targeted for cell death. It was observed that this cell death was a normal and necessary part of the developmental pathway in the worm. Programmed cell death has since become known to be an important process in all multicellular eukaryotic organisms. 1-51 (d) Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-52 (a) 1-53 (b) AC. elegans 1-54 BArabidopsis Cmouse DDrosophila 1-55 (b) 1-56 (a) 1-57 (b) Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1-58 (c) 1-59 (b) Mice are likely to provide the best model system. Mice have teeth and have long been used as a model organism. Mice reproduce relatively rapidly and the extensive scientific community that works with mice has developed techniques to facilitate genetic manipulations. E. coli (a bacterium) and Arabidopsis (a plant) do not have teeth. Horses like sugar and have big teeth, but they would not be a good model organism. There is not an extensive scientific community working on the molecular and biochemical mechanisms of cell behaviors in horses; they are expensive and have a long reproduction time, which makes genetic studies costly and slow; and tools for genetic manipulation (other than traditional breeding) have not been developed. 1-60 A. True. B. True. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 C. False. It is a relatively small proportion of our DNA that encodes RNA and protein molecules. The majority of nonencoding sequences is probably involved in critical regulatory processes. 1-61 All living beings on Earth (and thus, all cells) are thought to be derived from a common ancestor. Solutions to many of the essential challenges that face a cell (such as the synthesis of proteins, lipids, and DNA) seem to have been achieved in this ancient common ancestor. The ancestral cell therefore possessed sets of proteins to carry out these essential functions. Many of the essential challenges facing modern-day cells are the same as those facing the ancestral cell, and the ancient solutions are often still effective. Thus, it is not uncommon for organisms to use proteins and biochemical pathways inherited from their ancestors. Although these proteins usually show some species-specific diversification, they still retain the basic biochemical characteristics of the ancestral protein. For example, homologous proteins often retain their ability to interact with a specific protein target, even in cells of diverse species. Because the basic biochemical characteristics are retained, homologous proteins are often capable of functionally substituting for one another. 1-62 B, D A. thaliana (Arabidopsis) B, E M. musculus (mouse) A S. pombe C C. elegans A S. cerevisiae B D. rerio (zebrafish) Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 B D. melanogaster 1-63 The Earth’s atmosphere became oxygen-rich roughly 1.5 billion years ago. If some primitive predatory eukaryotic cells were similar to modern-day protozoans, they may have been mobile and able to engulf other cells. These characteristics would have been advantageous in the face of a changing atmosphere, and the establishment of a symbiotic relationship with an engulfed aerobe would have been selected for in the eukaryotic cell populations. 1-64 A. Changes in gene sequence occur through mutation. Mutations accumulate over time, occurring independently and at different sites in each gene lineage. Homologous genes that diverged recently will differ only slightly; genes that diverged long ago will differ more. Knowing the average mutation rate, you can estimate the time that has elapsed since the different versions of the gene diverged. By seeing how closely the various members of the family of homologous genes resemble one another, you can draw up a family tree, showing the sequence of lineage splits that lead from the ancestral gene to its many modern descendants. Suppose this family tree shows that family members A and B diverged from one another long ago, but that C diverged from B more recently; and suppose that at a certain site in the gene, A and B have the same sequence but Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 C is different. Then, it is likely that the sequence of A and B is ancestral, while that of C reflects a recent mutation that has occurred in the lineage of C alone. B. Although an inferred ancestral sequence can be reconstructed and the protein expressed, you would be placing an inferred, ancient protein in the context of a modern cell. If there are important interacting partners for the modern protein, there is a chance they may not recognize the ancestral protein, and therefore any information about its function may be inaccurate. 1-65 A. If the mitochondria originated from an ancient aerobic bacterium that was engulfed by an ancient eukaryote, as postulated, it is possible that an antibiotic that inhibits protein synthesis in bacteria could also block that process in mitochondria. B. We would expect that although cytosolic protein synthesis would stop, mitochondrial protein synthesis should still occur normally (at least for a little while). This result would lend further support to the idea that mitochondria are Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 derived from a noneukaryotic organism. If this were not the case, these compounds would be expected to affect protein synthesis at both locations. 1-66 Funding research o a for several reasons: (1) n D. melanogasteris worthwhile investment working with insect animal models is relatively inexpensive; (2) fruit flies have historically proven useful in helping understand eukaryotic chromosome behavior; and (3) many of the genes in Drosophila are highly similar in sequence to the homologous human genes, and thus can be used tostudy human diseases. 1-67 Nurse and Hartwell first treated yeast cells with a chemical mutagen. The mutated population of cells was then grown and observed. Cells that demonstrated defects in cell-cycle regulation (characterized by cell-cycle arrest, larger-than-normal cells, and smaller-than-normal cells) were then isolated. The use of a library of plasmids that each express a normal gene from yeast cells allowed the scientists to identify exactly which gene could be used to “rescue” the mutant, because when the normal gene is expressed again, the cells return to a normal cell cycle. After this big result, the scientistswent on to show that the homologous gene from other organisms could also rescue the mutant phenotype. The most exciting result was obtained with the human version of the cdc2 gene, which demonstrated that there are common principles underlying cell-cycle regulation across a large range of eukaryotic organisms. 1-68 You could use modern technology to discover the sequence of the DNA. If you are right, you would expect to find parts of this sequence that are unmistakably similar to corresponding sequences in other, familiar, living organisms; it would be highly improbable that such similar sequences would have evolved independently. You could, of course, also analyze other features of the chemistry of his cells; forexample, do they contain proteins made of the same set of 20 amino acids? This could all be supporting evidence that this newly discovered species arose from the same common ancestral cells as all other lifeon Earth. ESSENTIAL CELL BIOLOGY, FOURTH EDITION CHAPTER 2: CHEMICAL COMPONENTS OF CELLS Chemical Bonds 2-1 Select the answer that best completes the following statement: Chemical reactionsin living systems occur in an environment, within a narrow range of temperatures. (a) optimal (b) organic (c) extracellular Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (d) aqueous 2-2 Indicate whether the statements below are true or false. If a statement is false,explain why it is false. A. The chemistry of life is carried out and coordinated primarily by the actionof small molecules. B. Carbon-based compounds make up the vast majority of molecules foundin cells. C. The chemical reactions in living systems are loosely regulated, allowing for a wide range of products and more rapid evolution. 2-3 Which subatomic particles contribute to the atomic number for any givenelement? (a) protons (b) protons and neutrons (c) neutrons (d) protons and electrons 2-4 Which subatomic particles contribute to the atomic mass for any given element? (a) protons (b) protons and neutrons (c) neutrons (d) protons and electrons 2-5 Which subatomic particles can vary between isotopes of the same element,without changing the observed chemical properties? (a) electrons (b) protons and neutrons (c) neutrons (d) neutrons and electrons Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 2-6 Figure Q2-6 depicts the structure of carbon. Use the information in the diagram tochoose the correct atomic number and atomic weight, respectively, for an atom ofcarbon. Figure Q2-6 (a) (b) (c) (d) 2-7 6, 12 12, 12 6, 18 12, 6 Carbon 14 is an unstable isotope of carbon that decays very slowly. Compared tothe common, stable carbon 12 isotope, carbon 14 has two additional . (a) (b) (c) (d) 2-8 electrons. neutrons. protons. ions. If the isotope 32S has 16 protons and 16 neutrons, how many protons, neutrons,and electrons will the isotope 35S have, respectively? (a) (b) (c) (d) 16, 20, 15 16, 19, 15 16, 19, 16 16, 19, 17 2-9 A. If 0.5 mole of glucose weighs 90 g, what is the molecular mass of glucose? B. What is the concentration, in grams per liter (g/L), of a 0.25 M solution ofglucose? C. How many molecules are there in 1 mole of glucose? 2-10 Which of the following elements is least abundant in living organisms? (a) sulfur (b) carbon (c) oxygen (d) nitrogen Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 2-11 You explain to a friend what you have learned about Avogadro’s number. Your friend thinks the number is so large that he doubts there is even a mole of livingcells on the Earth. You have recently heard that there are about 50 trillion (5 × 1013) human cells in each adult human body and that each human contains morebacterial cells (in the digestive system) than human cells, so you bet your friend $5 that there is more than a mole of cells on Earth. The human population is approximately 7 billion (7 × 109). What calculation can you show your friend toconvince him you are right? 2-12 Avogadro’s number, calculated from the atomic weight of hydrogen, tells us how many atoms or molecules are in a mole. The resulting base for all calculations of moles and molarity (how many molecules are present when you weigh out a substance or measure from a stock solution) is the following: 1 g of hydrogen atoms = 6 × 1023 hydrogen atoms = 1 mole of hydrogen Sulfur has a molecular weight of 32. How many moles and atoms are there in 120grams of sulfur? (a) 3.75 and 6 × 1023 (b) 32 and 6 × 1023 (c) 1.75 and 1.05 ×1024 (d) 3.75 and 2.25 × 1024 2-13 The first task you are assigned in your summer laboratory job is to prepare a concentrated NaOH stock solution. The molecular weight of NaOH is 40. How many grams of solid NaOH will you need to weigh out to obtain a 500 mL solution that has a concentration of 10 M? (a) 800 g (b) 200 g (c) 400 g (d) 160 g 2-14 You have a concentrated stock solution of 10 M NaOH and want to use it to produce a 150 mL solution of 3 M NaOH. What volume of water and stock solutions will you measure out to make this new solution? (a) 135 mL of water, 15 mL of NaOH stock (b) 115 mL of water, 35 mL of NaOH stock (c) 100 mL of water, 50 mL of NaOH stock (d) 105 mL of water, 45 mL of NaOH stock 2-15 Indicate whether the statements below are true or false. If a statement is false,explain why it is false. A. Electron shells fill discrete regions around the nucleus of the atom andlimit the number of electrons that can occupy a specific orbit. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 B. C. H, C, O, and N are the most common elements in biological molecules because they are the most stable. Some atoms are more stable when they lose one or two electrons, eventhough this means they will have a net positive charge. 2-16 A covalent bond between two atoms is formed as a result of the (a) sharing of electrons. (b) loss of electrons from both atoms. (c) loss of a proton from one atom. (d) transfer of electrons from one atom to the other. 2-17 An ionic bond between two atoms is formed as a result of the (a) sharing of electrons. (b) loss of electrons from both atoms. (c) loss of a proton from one atom. (d) transfer of electrons from one atom to the other. 2-18 For each of the following sentences, fill in the blanks with the best word or phraseselected from the list below. Not all words or phrases will be used; each word or phrase should be used only once. Whereas ionic bonds form a(n) between atoms form a(n) have a characteristic bond more rigid when two electrons are shared in a(n) , covalent bonds . These covalent bonds and become stronger and . Equal sharing of electrons yields a(n) covalent bond. If one atom participating in the bond has a stronger affinity for the electron, this produces a partial negative charge on one atom and a partial positive charge on the other. These covalent bonds should not be confused with the weaker bonds that are critical for the three-dimensional structure of biological molecules andfor interactions between these molecules. charge covalent double bond ionic 2-19 length molecule noncovalent nonpolar polar salt single bond weight Indicate whether the statements below are true or false. If a statement is false,explain why it is false. A. Electrons are constantly moving around the nucleus of the atom, but theycan move only in discrete regions. B. There is no limit to the number of electrons that can occupy the fourthelectron shell. C. Atoms with unfilled outer electron shells are especially stable and aretherefore less reactive. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) . . lOMoARcPSD|30659517 2-20 Table Q2-20 indicates the electrons in the first four atomic electron shells for selected elements. On the basis of the information in the chart and what you knowabout atomic structure, which elements are chemically inert? Table Q2-20 (a) (b) (c) (d) 2-21 carbon, sulfur helium, neon sodium, potassium magnesium, calcium Table Q2-21 indicates the electrons in the first four atomic electron shells for selected elements. On the basis of the information in the chart and what you knowabout atomic structure, which elements will form ions with a net charge of +1 in solution? Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Table Q2-21 (a) (b) (c) (d) 2-22 carbon, sulfur helium, neon sodium, potassium magnesium, calcium Table Q2-22 indicates the electrons in the first four atomic electron shells for selected elements. On the basis of the information in the chart and what you knowabout atomic structure, which elements will form ions with a net charge of +2 in solution? Table Q2-22 (a) (b) (c) (d) 2-23 carbon, sulfur helium, neon sodium, potassium magnesium, calcium Table Q2-23 indicates the electrons in the first four atomic electron shells for selected elements. On the basis of the information in the chart and what you knowabout atomic structure, which elements form stable but reactive diatomic gases? Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Table Q2-23 (a) (b) (c) (d) nitrogen, oxygen helium, neon sodium, potassium magnesium, calcium 2-24 Indicate whether the statements below are true or false. If a statement is false,explain why it is false. A. There are four elements that constitute 99% of all the atoms found in thehuman body. B. Copper, zinc, and manganese are among 11 nonessential trace elementsthat contribute less than 0.1% of all the atoms in the human body. C. Approximately 0.9% of the atoms in the human body come from seven essential elements—Na, Mg, K, Ca, P, S, and Cl—all of which form stable ions in aqueous solution. 2-25 Which of the following factors do not influence the length of a covalent bond? (a) the tendency of atoms to fill the outer electron shells (b) the attractive forces between negatively charged electrons and positivelycharged nuclei (c) the repulsive forces between the positively charged nuclei (d) the minimization of repulsive forces between the two nuclei by the cloudof shared electrons 2-26 Double covalent bonds are both shorter and stronger than single covalent bonds, but they also limit the geometry of the molecule because they . (a) (b) (c) (d) create a new arrangement of electron shells. change the reactivity of the bonded atoms. limit the rotation of the bonded atoms. prevent additional bonds from being formed with the bonded atoms. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 B. 2-27 Polar covalent bonds are formed when the electrons in the bond are not sharedequally between the two nuclei. Which one of these molecules contains polar bonds? (a) molecular oxygen (b) methane (c) propane (d) water 2-28 A. In which scientific unit is the strength of a chemical bond usually expressed? If 0.5 kilocalories of energy are required to break 6 × 1023 bonds of aparticular type, what is the strength of this bond? 2-29 Oxygen, hydrogen, carbon, and nitrogen atoms are enriched in the cells and tissues of living organisms. The covalent bond geometries for these atoms influence their biomolecular structures. Match the elements on the left with thebond geometries illustrated on the right. Some elements assume more than onebond geometry. Figure Q2-29 A. B. C. 2-30 oxygen carbon nitrogen Which combination of answers best completes the following statement: When atoms are held together by , they are typically referredto as . (a) hydrogen bonds, molecules. (b) ionic interactions, salts. (c) ionic interactions, molecules. (d) double bonds, nonpolar. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 2-31 Although covalent bonds are 10–100 times stronger than noncovalent interactions,many biological processes depend upon the number and type of noncovalent interactions between molecules. Which of the noncovalent interactions below willcontribute most to the strong and specific binding of two molecules, such as a pair of proteins? (a) electrostatic attractions (b) hydrogen bonds (c) hydrophobic interactions (d) Van der Waals attractions 2-32 Which of the following expressions accurately describes the calculation of pH? (a) pH = –log10[H+] (b) pH = log10[H+] (c) pH = –log2[H+] (d) pH = –log10[OH–] 2-33 The pH of an aqueous solution is an indication of the concentration of available protons. However, you should not expect to find lone protons in solution; rather, the proton is added to a water molecule to form a(n) ion. (a) hydroxide (b) ammonium (c) chloride (d) hydronium 2-34 The relative strengths of covalent bonds and van der Waals interactions remain thesame when tested in a vacuum or in water. However, this is not true of hydrogen bonds or ionic bonds, whose bond strengths are lowered considerably in the presence of water. Explain these observations. 2-35 Indicate whether the statements below are true or false. If a statement is false,explain why it is false. A. Any covalently bonded H atom can participate in a hydrogen bond if it comes in close proximity with an oxygen atom that forms part of a watermolecule. B. Protons are constantly moving between water molecules, which means there is an overall equilibrium between hydroxyl ions and hydronium ionsin aqueous solutions. C. A strong base is defined as a molecule that can readily remove protonsfrom water. 2-36 Larger molecules have hydrogen-bonding networks that contribute to specific, high-affinity binding. Smaller molecules such as urea can also form these networks. How many hydrogen bonds can urea (Figure Q2-36) form if dissolved in water? Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Figure Q2-36 2-37 (a) (b) (c) (d) 6 5 3 4 A. Sketch three different ways in which three water molecules could be heldtogether by hydrogen-bonding. On a sketch of a single water molecule, indicate the distribution ofpositive and negative charge (using the symbols δ+ and δ–). How many hydrogen bonds can a hydrogen atom in a water moleculeform? How many hydrogen bonds can the oxygen atom in a water molecule form? B. C. 2-38 A. B. C. What is the pH of pure water? What concentration of hydronium ions does a solution of pH 8 contain? Complete the following reaction: CH3COOH + H2O ↔ D. 2-39 . Will the reaction in (C) occur more readily (be driven to the right) if the pH of the solution is high? Aromatic carbon compounds such as benzene are planar and very stable. Doublebond character extends around the entire ring, which is why it is often drawn as ahexagon with a circle inside. This characteristic is caused by electron . (a) (b) (c) (d) 2-40 resonance. pairing. partial charge. stacking. The amino acid histidine is often found in enzymes. Depending on the pH of its environment, sometimes histidine is neutral and at other times it acquires a protonand becomes positively charged. Consider an enzyme with a histidine side chain that is known to have an important role in the function of the enzyme. It is not clear whether this histidine is required in its protonated or its unprotonated state. To answer this question, you measure enzyme activity over a range of pH, with the results shown in Figure Q2-40. Which form of histidine is necessary for the active enzyme? Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Figure Q2-40 2-41 Silicon is an element that, like carbon, has four vacancies in its outer electron shell and therefore has the same bonding chemistry as carbon. Silicon is not foundto any significant degree in the molecules found in living systems, however. Does this difference arise because elemental carbon is more abundant than silicon? What other explanations are there for the preferential selection of carbon over silicon as the basis for the molecules of life? 2-42 Selenium (Se) is an element required in the human body in trace amounts. Selenium is obtained through the diet and levels of selenium found in food depend greatly on the soil where it is grown. Once ingested and absorbed as selenate, it can become incorporated into a small number of polypeptides. These selenoproteins are formed when selenium replaces an element that is found in twoof the twenty “standard” amino acids. Using your knowledge of atomic structure, the periodic table in Figure 2–7, and the structure of amino acids found in Panel 2–5, deduce which two amino acids may be converted to “seleno” amino acids and used to make selenoproteins. Small Molecules in Cells 2-43 Indicate whether the molecules below are inorganic (I) or organic (O). A. glucose B. ethanol C. sodium chloride D. water E. cholesterol F. adenosine G. calcium H. glycine I. oxygen J. iron K. phospholipid Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 2-44 Which of the following monomer building blocks is necessary to assemble selectively permeable boundaries around and inside cells? (a) sugars (b) fatty acids (c) amino acids (d) nucleotides 2-45 The variety and arrangement of chemical groups on monomer subunits contributeto the conformation, reactivity, and surface of the macromolecule into which they become incorporated. What type of chemical group is circled on the nucleotide shown in Figure Q2-45? Figure Q2-45 (a) (b) (c) (d) 2-46 pyrophosphate phosphoryl carbonyl carboxyl The amino acids glutamine and glutamic acid are shown in Figure Q2-46. They differ only in the structure of their side chains (circled). At pH 7, glutamic acid can participate in molecular interactions that are not possible for glutamine. Whattypes of interactions are these? Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Figure Q2-46 (a) (b) (c) (d) ionic bonds hydrogen bonds van der Waals interactions covalent bonds 2-47 Cells require one particular monosaccharide as a starting material to synthesizenucleotide building blocks. Which of the monosaccharides below fills this important role? (a) glucose (b) fructose (c) ribulose (d) ribose 2-48 Oligosaccharides are short sugar polymers that can become covalently linked to proteins and lipids through condensation reactions. These modified proteins and lipids are called glycoproteins and glycolipids, respectively. Within a protein, which of the amino acids (shown in Figure Q2-48) is the most probable target forthis type of modification? Figure Q2-48 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (a) (b) (c) (d) serine glycine phenylalanine methionine 2-49 Which of the following are examples of isomers? (a) glucose and galactose (b) alanine and glycine (c) adenine and guanine (d) glycogen and cellulose 2-50 A. How many carbon atoms does the molecule represented in Figure Q250have? B. How many hydrogen atoms does it have? C. What type of molecule is it? Figure Q2-50 2-51 Most types of molecules in the cell have asymmetric (chiral) carbons. Consequently there is the potential to have two different molecules that look much the same but are mirror images of each other and therefore not equivalent. These special types of isomer are called stereoisomers. Which of the four carbonscircled in Figure Q251 is the asymmetric carbon that determines whether the amino acid (threonine in this case) is a ᴅ or an ʟ stereoisomer? Figure Q2-51 (a) (b) (c) (d) 2-52 1 2 3 4 Indicate whether the statements below are true or false. If a statement is false,explain why it is false. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 A. B. C. 2-53 A disaccharide consists of a sugar covalently linked to another moleculesuch as an amino acid or a nucleotide. The hydroxyl groups on monosaccharides are reaction hot spots and canbe replaced by other functional groups to produce derivatives of the original sugar. The presence of double bonds in the hydrocarbon tail of a fatty acid doesnot greatly influence its structure. On the phospholipid molecule in Figure Q2-53, label each numbered line with thecorrect term selected from the list below. Figure Q2-53 2-54 Many types of cells have stores of lipids in their cytoplasm, usually seen as fatdroplets. What is the lipid most commonly found in these droplets? (a) cholesterol (b) palmitic acid (c) isoprene (d) triacylglycerol 2-55 Choose the answer that best fits the following statement: Cholesterol is an essential component of biological membranes. Although it is much smaller thanthe typical phospholipids and glycolipids in the membrane, it is a(n) molecule, having both hydrophilic and hydrophobic regions. (a) (b) (c) (d) polar oxygen-containing hydrophobic amphipathic Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 2-56 For each of the following sentences, fill in the blanks with the best word or phraseselected from the list below. Not all words or phrases will be used; each word or phrase should be used only once. Proteins are built from amino acids, which each have an amino group and a group attached to the central . There are twenty possible that differ in structure and are generally referred to as “R.” In solutions of neutral pH, amino acids are , carrying both a positive and negative charge. When a protein is made, amino acids are linked together through , which are formed by condensation reactions between the carboxyl end of the last amino acidand the end of the next amino acid to be added to the growing chain. amino α-carbon carbon carboxyl hydroxide ionized length noncovalent peptide bonds polypeptides protein R group side chains 2-57 DNA and RNA are different types of nucleic acid polymer. Which of the following is true of DNA but not true of RNA? (a) It contains uracil. (b) It contains thymine. (c) It is single-stranded. (d) It has 5′-to-3′ directionality. 2-58 Match each term related to the structure of nucleic acids (A–I) with one of thedescriptions provided. A. base B. glycosidic bond C. nucleoside D. nucleotide E. phosphoanhydride bond F. phosphoester bond G. ribose H. phosphodiester bond I. deoxyribose the linkage between two nucleotides the linkage between the 5′ sugar hydroxyl and a phosphate group the nitrogen-containing aromatic ring five-carbon sugar found in DNA sugar unit linked to a base linkage between the sugar and the base linkages between phosphate groups Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 sugar linked to a base and a phosphate five-carbon sugar found in RNA 2-59 A. Write out the sequence of amino acids in the following peptide, using thefull names of the amino acids. Pro-Val-Thr-Gly-Lys-Cys-Glu B. C. 2-60 Write the same sequence with the single-letter code for amino acids. According to the conventional way of writing the sequence of a peptide or a protein, which is the C-terminal amino acid and which is the Nterminalamino acid in the above peptide? The cell is able to harvest energy from various processes in order to generate ATPmolecules. These ATPs represent a form of stored energy that can be used later todrive other important processes. Explain how the cell can convert the chemical energy stored in ATP to generate mechanical energy, for example changing the shape of a protein. Macromolecules in Cells 2-61 Both DNA and RNA are synthesized by covalently linking a nucleoside triphosphate to the previous nucleotide, constantly adding to a growing chain. Inthe case of DNA, the new strand becomes part of a stable helix. The two strandsare complementary in sequence and antiparallel in directionality. What is the principal force that holds these two strands together? (a) ionic interactions (b) hydrogen bonds (c) covalent bonds (d) van der Waals interactions 2-62 Each nucleotide in DNA and RNA has an aromatic base. What is the principal force that keeps the bases in a polymer from interacting with water? (a) hydrophobic interactions (b) hydrogen bonds (c) covalent bonds (d) van der Waals interactions 2-63 Because there are four different monomer building blocks that can be used to assemble RNA polymers, the number of possible sequence combinations that canbe created for an RNA molecule made of 100 nucleotides is . (a) (b) (c) (d) 1004 4100 4 × 100 100/4 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 2-64 There are 20100 different possible sequence combinations for a protein chain with 100 amino acids. In addition to the amino acid sequence of the protein, what otherfactors increase the potential for diversity in these macromolecules? (a) free rotation around single bonds during synthesis (b) noncovalent interactions sampled as protein folds (c) the directionality of amino acids being added (d) the planar nature of the peptide bond 2-65 Indicate whether the statements below are true or false. If a statement is false,explain why it is false. A. “Nonpolar interactions” is simply another way of saying “van der Waals attractions.” B. Condensation reactions occur in the synthesis of all the macromoleculesfound in cells. C. All proteins and RNAs pass through many unstable conformations as theyare folded, finally settling on one single, preferred conformation. 2-66 Macromolecules in the cell can often interact transiently as a result of noncovalentinteractions. These weak interactions also produce stable, highly specific interactions between molecules. Which of the factors below is the most significantin determining whether the interaction will be transient or stable? (a) the size of each molecule (b) the concentration of each molecule (c) the rate of synthesis (d) surface complementarity between molecules 2-67 As a protein is made, the polypeptide is in an extended conformation, with every amino acid exposed to the aqueous environment. Although both polar and chargedside chains can mix readily with water, this is not the case for nonpolar side chains. Explain how hydrophobic interactions may play a role in the early stages of protein folding, and have an influence on the final protein conformation. 2-68 You are trying to make a synthetic copy of a particular protein but accidentally join the amino acids together in exactly the reverse order. One of your classmatessays the two proteins must be identical, and bets you $20 that your synthetic protein will have exactly the same biological activity as the original. After havingread this chapter, you have no hesitation in staking your $20 that it won’t. What particular feature of a polypeptide chain makes you sure your $20 is safe and thatthe project must be done again. 2-69 A protein chain folds into its stable and unique three-dimensional structure, or conformation, by making many noncovalent bonds between different parts of the chain. Such noncovalent bonds are also critical for interactions with other proteinsand cellular molecules. From the list provided, choose the class(es) of amino acidsthat are most important for the interactions detailed below. A. forming ionic bonds with negatively charged DNA Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 B. C. D. E. acidic basic forming hydrogen bonds to aid solubility in water binding to another water-soluble protein localizing an “integral membrane” protein that spans a lipid bilayer tightly packing the hydrophobic interior core of a globular protein nonpolar uncharged polar 2-70 It is now a routine task to determine the exact order in which individual subunitshave been linked together in polynucleotides (DNA) and polypeptides (proteins).However, it remains difficult to determine the arrangement of monomers in a polysaccharide. Explain why this is the case. 2-71 Your lab director requests that you add new growth medium to the mammalian cell cultures before heading home from the lab on a Friday night. Unfortunately, you need to make fresh medium because all the premixed bottles of medium havebeen used. One of the ingredients you know you need to add is a mix of the essential amino acids (those that cannot be made by the cells, but are needed in proteins). On the shelf of dry chemicals you find the amino acids you need, and you mix them into your medium, along with all the other necessary nutrients, andreplace the old medium with your new medium. On Sunday, you come in to the lab just to check on your cells and find that the cells have not grown. You are sureyou made the medium correctly, but on checking you see that somebody wrote a note on the dry mixture of amino acids you used: “Note: this mixture contains only ᴅ-amino acids.” A. What is the meaning of the note and how does it explain the lack of cellgrowth in your culture? B. Are there any organisms that could grow using this mixture? Justify youranswer. 2-72 Eukaryotic cells have their DNA molecules inside their nuclei. However, to package all the DNA into such a small volume requires the cell to use specialized proteins called histones. Histones have amino acid sequences enriched for lysinesand arginines. A. What problem might a cell face in trying to package DNA into a small volume without histones, and how do these special packaging proteinsalleviate the problem? B. Lysine side chains are substrates for enzymes called acetylases. A diagram of an acetylated lysine side chain is shown in Figure Q2-72. How do you think the acetylation of lysines in histone proteins will affect the ability ofa histone to perform its role (refer to your answer in part A)? Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Figure Q2-72 CHAPTER 3: DNA REPLICATION, REPAIR, AND RECOMBINATION 6-1 the The process of DNA parental DNA strands used as of the replication be a to opposing (a) catalyst (b) competito requires that each of produce a duplicate strand. r(c) template (d) copy 6-2 DNA replication is considered semiconservative because of replication, the original consists of the two . (a) after many rounds double helix is still DNA intact. (b) each daughter newstrands parent DNA DNA molecule copied from molecule. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) DNA lOMoARcPSD|30659517 (c) each daughter DNA fromthe parent DNA molecule molecule strand. (d) new and one new DNA strands must be 6-3 The classic experiments conducted demonstrated that consists of one strand copied from a DNA template. by DNA and Meselson Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) Stahl lOMoARcPSD|30659517 replication is mechanism. accomplished by (a) continuous (b) semiconservative (c) dispersive (d) conservative 6-4 Initiator anddisrupt proteins employing a bind to replication hydrogen bonds between the two DNA strands being copied. below does not contribute to origins Which of the the relative ease of strand separation by initiator (a) replication origins are A-T base pairs (b) the reaction (c) they only (d) once opened, other proteins replicationmachinery can separate rich in proteins? occur at room temperature a base pairs at a of bind the few the to DNA E. factors time origin 6-5 If the genome requiresabout 20 itself, of the minutes bacterium to how can the genome bereplicated in of only the 3 fruit fly Drosophila minutes? is smaller than the (a) The genome. (b) rate genome Drosophila Eukaryotic than DNA polymerase synthesizes DNA at coli replicate a Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) E. coli much faster lOMoARcPSD|30659517 prokaryotic DNA polymerase. Page of 25 1 (c) one The nuclear place in the (d) Drosophila DNA replicationthan Meselson isotopes produced A. DNA cell, membrane keeps the which increases contains E. Drosophila the rate of more origins coli DNA concentrated in polymerization. of DNA. and Stahl grew cells in media that contained different of nitrogen (15N and 14N) so that the DNA molecules from these different isotopes could be distinguished by mass. Explain in the how “light” DNA was separated from “heavy” model s for DNA them was ruled dispersive model of Meselsonand Stahl B. Describe the three existing experiments. replication when these studies were begun, and 6 definitively by explain the how one 6- of experiment you described for part A. C. What experimental result eliminated the out replication? 6- Indicate whether 7 a statement is the following false, explain statements are why it is true or false. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) false. If DNA lOMoARcPSD|30659517 A. When DNA is heatingoccurs, being replicated allowing two strands to separate. replication origins B. DNA base pairs. C. Meselson and Stahl forDNA replication. D. DNA process that is initiated are inside a the cell, local typically rich in G-C ruled out the replication is at multiple dispersive model a bidirectional along chromosomes in eukaryotic cells. How many replication forks are is opened? (a) 1 formed when an origin of locations 6- (b) replication 2 8 (c) 3 (d) 4 Answer A. is 6- the the following questions On a DNA growing—the 3′ strand that end, 5′ both ends? Explain end, or about DNA replication. is being synthesized, which end your answer. 9 B. On a DNA template,where is occurring 5′, or relative both? 6- How does the 10 cells compare strand that is being used as the copying to the total number of with the number (a) 1 versus 100 (b) 5 versus 500 replication origin—3′ replication origins of origins in Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) a of the in bacterial human cells? origin, lOMoARcPSD|30659517 6- (c) 10 versus 1000 (d) 1 versus 10,000 Which of 11 for the following DNA replication Page 2 of (a) The replication theconditions. (b) on The the statements correctly to be bidirectional? fork can open or explains what it means 25 DNA replication template machinery can close, depending move in on either direction strand. (c) Replication-fork converges on another (d) The opposite movement replication can switch directions at the 6-12 The chromatin structure complicated than that in eukaryotic observe dreason This is prokaryotic cells. DNA replication occurs much faster in (a) 2× (b) 5× fork fork. replication forks formed directions. in tha t when the prokaryotes. How origin move in cells is much more thought to be much faster is it? Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) the lOMoARcPSD|30659517 (c) 10× (d) 100 × 6-13 DNA polymerase catalyzes nucleotideto a the joining growing of from catalyzing the What prevents this enzyme (a) (Pi) hydrolysis + Pi of pyrophosphate (b) release of PPi from the (c) hybridization of the new strand to the (d) loss as an energy source knowledge explain of of ATP 6-14 Use your synthesized to replication words, must occur in what would be consequences 6-15 of Figure Q6-15 shows a to strand. reverse inorganic reaction? phosphate nucleotide how a why DNA the the 3′–to-5′ (PPi) a DNA 5′-to-3′ template new strand of direction. In DNA is other strand elongation? replication bubble. Figure Q6-15 On A. (use the O). figure, indicate where the origin of replication Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) was located lOMoARcPSD|30659517 Label the B. of the leading-strand template and the fork [R] X and Y, respectively. by arrows the direction (indicated by dark lines) were synthesized. Number D. 3 in the the Okazaki order in right-hand Indicate C. strands E. as which they were synthesized. Indicate where the on the direction Page of 25 of movement which the of template newly made DNA each strand as most recent DNA synthesis Indicate F. arrows. 3 fragments in lagging-strand 1, 2, and S). has occurred (use the replication forks with Use the following information about a series of in vitro DNA replication experiments to answer questions 6-16 through 622. You prepare bacterial cell extracts by lysing the cells andremoving insoluble debris via centrifugation. These extracts provide the proteins required for DNA replication. Your DNA template is a small, double-stranded circular piece of DNA (a plasmid) with a single origin of replication and a single replication termination site. The termination site is on the opposite side of the plasmid from the origin. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 6-16 In addition to the extracts are there any additional materials and you should add system?Explain replication 6-17 to Which of this in to your this in vitro answer. the following statements is vitro replication the plasmid DNA, true with respect strand and one lagging system? (a) There will strand produced be only using one leading this template. (b) of The leading and lagging each newly synthesized DNA strand. (c) The places on DNA replication this strands compose machinery can assemble molecule will be other. at one half multiple plasmid. (d) One daughter shorterthan You protei n examine 6-18 decide to of DNA the use different the replication the role of normalprocess of What part of the mostdirectly affected of bacteria extracts? bacterial lacking DNA if initiation of DNA (b) Okazaki fragment (each having one mutated) proteins replication primase (a) strains machinery individual DNA slightly in order to in the replication. process a would be strain were used to make the synthesis synthesis Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) cell lOMoARcPSD|30659517 (c) leading-strand elongation (d) lagging-strand completion 6-19 What part of directly affected of bacteria polymerase the if DNA a replication strain process lacking the exonuclease activity were used to make the cell extracts? Page 4 of 25 (a) initiation of (b) Okazaki (c) leading-strand elongation (d) lagging-strand completion What part of affected if make the cell would be of most DNA DNA synthesis fragment synthesis the DNA replication process would be most directly a strain of bacteria lacking helicase were used to extracts? 6- (a) 20 initiation of DNA synthesis (b) Okazaki fragment (c) leading-strand elongation (d) lagging-strand completion synthesis Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 What part of affected if bacteria the a lacking DNA replication strain of the process cell single-strand binding would be protein most directly were used to make extracts? 621 (a) initiation of (b) Okazaki fragment (c) leading-strand elongation (d) lagging-strand completion What part of affected if make the cell DNA synthesis synthesis the DNA replication process would be most directly a strain of bacteria lacking DNA ligase were used to extracts? 6- (a) 22 initiation of (b) Okazaki fragment (c) leading-strand elongation (d) lagging-strand completion 6- Which of the 23 a human (a) a DNA synthesis synthesis following statements chromosome is true? about the newly synthesized strand of It was synthesized from a single origin solely by continuousDNA synthesis. (b) It was synthesized from a single origin by mixture of continuous anddiscontinuous DNA synthesis. (c) It was discontinuous synthesized from multiple DNA origins solely by synthesized from multiple origins by synthesis. (d) It was Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) a lOMoARcPSD|30659517 mixture of continuous anddiscontinuous DNA synthesis. You have discovered an “Exo–” mutant form of DNA polymerase in which the 3′-to-5′ exonuclease function has been destroyed but the ability to join nucleotides together is unchanged. Which of the following properties do you expect the mutant polymerase to have? 6- (a) It 24 and the slowly than (c) the to will polymerize in 3′-to-5′ direction. the normal Exo+ It will normal generate of both the 5′-to-3′ direction (b) It will polymerize polymerase. fall off the template Exo+ polymerase. (d) It mismatched base pairs. Page 5 6-25 A molecule yeastcell nucleus more frequently than will be more likely 25 of is but fails think the to arises? that is Choose the most probably bacterial DNA imported replicate with the block to replication protei n responsible for the failure to anexplanation for introduced into into yeast DNA. Where primase (b) helicase a the do or protein complex replicate your bacterial DNA. Give answer. (a) more Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) you below lOMoARcPSD|30659517 (c) DNA polymerase Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (d) initiator 6-26 Most cells in thetelomerase gene is important proteins turned step in the body of enzyme an adult human because off is therefore not cell into cancer which circumvents and lack its expressed. An the cell, growth conversion normal control, why be over of a normal is the resumption of telomerase might necessary again. for the of a replication Page 6 of telomerase ability of 6-27 Which diagram accurately strands at one side a expression. Explain cancer cells represents the to divide over and directionality of DNA fork? 25 6- Indicate whether 28 a statement is the following false, explain statements are why it is A. on Primase both th e is neede to leading d strand and the lagging strand . initiat e true or false. DNA Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) false. If replication lOMoARcPSD|30659517 B. The sliding clamp is strand,where it associated until loaded remains replication once on is complete. each DNA C. own Telomerase is RNA molecule a to DNA use polymerase that as a primer the end of a proofreading function at D. Primase there are no the RNA requires errors in primers used for DNA the carries its lagging strand. that ensures replication. Because all DNA polymerases synthesize DNA in the 5′-to-3′ direction, and the parental strands are antiparallel, DNA replication is accomplished with the use of two whether the 629 mechanisms: continuous and following items relate to discontinuous replication, discontinuous (1) or replication. continuous replication, (3) both modes of Indicate (2) replication. primase single-strand binding protein sliding clamp RNA primers leading strand lagging strand Okazaki fragments DNA helicase DNA ligase 6- The synthesis of 30 direction. However, DNA in living systems scientists synthesize short occurs in the DNA sequences Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) 5′-to-3′ needed lOMoARcPSD|30659517 for their experiments on an instrument A. The chemical proceeds in the synthesis 3′-to-5′ of direction. possible diagram toexplain the Draw a and dedicated to this by this instrument show how this is of is chemically DNA task. process. B. Although possible, it 3′-to-5′ synthesisdoes not occur in living systems. Why DNA not? DNA polymerases are processive, which means that they remain tightly associated with the template strand while moving rapidly and adding nucleotides to the growing daughter strand. Which piece of the replication machinery accounts for this characteristic? 6- (a) 31 helicase (b) sliding clamp (c) single-strand binding (d) primase Page 7 6-32 Use diagram protein of 25 the of components in the list a replication fork in below to Figure Q6-32. 1. DNA polymerase 2. single-strand binding protein Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) label the lOMoARcPSD|30659517 3. Okazaki fragment 4. primase 5. sliding clamp 6. RNA primer 7. DNA helicase Figure Q6-32 6-33 Researchers have isolated a that carries a temperature sensitive variant of thepermissive mutant the cells grow just nonpermissive grown at compared DNA isolated results are strain of the enzyme temperature, the DNA ligase. At as wild-type temperature, all of the cells 2 hours. DNA from mutant cells is mutant well as the in the culture the nonpermissive with the temperature for from cells the grown at E. coli cells. At tube die 30 permissive minutes temperature. The shown in size by Figure Q6-33, where DNA molecules have been separated means of Explain electrophoresis the permissive; NP, nonpermissive). distinct band with the sample a size appearanc of epairs (bp) a in (P, within of Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) 200 by base lOMoARcPSD|30659517 collected at the nonpermissive temperature. Figure Page 8 of 25 6-34 Indicate false. If a whether the statement is explain it why is 1. The repair polymerase is synthesized strands to 1. There is and ensure a lays corresponding DNA fragments following false, statements are the enzyme that proofreads the accuracy of DNA replication. the RNA primers on sequence the to it is synthesizing DNA 6-35 Which of the following proofreadingduring DNA the that degrades behind it. seal sugar–phosphate the lagging 1. The repair polymerase does not clamp, because only true or false. single enzyme down the 1. DNA ligase is required between all the DNA Q6-33 newly backbone strand. require the over very aid of short stretches. statements about sequence Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) the sliding lOMoARcPSD|30659517 replication is false? (a) DNA The the exonuclease activity polymerase. (b) The direction. is in exonuclease activity a cleaves different domain DNA the in (c) The DNA proofreading activity strandelongation. occurs concomitantly (d) If an incorrect beforeremoval. added, it 6-36 The binds to base is sliding clamp complex DNA polymerase. This of DNA helps the the polymerase synthesize is “unpaired” DNA template must happen each time made on the lagging a new Okazaki does the expedite this process? The DNA duplex consists of covalent polymers wrapped around two each long it is strand. How cell many times over their entire length. The strands for separation replicatio causes n the the replication to be the torsional strands and much longer stretches clamp only strand, fragment How the with the dissociating. While the the leading loading 5′-to-3′ of without once o n 6-37 encircles of occurs other of the “overwound” in DNA front of fork. does the cell DNA duplex relieve during stress created Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) along lOMoARcPSD|30659517 replication? (a) will Nothing needs to be separated be done because the two strands the backbone afte r replication is complete. (b) break the covalent bonds of Topoisomerase s allowing unwinding the local of DNA (c) Helicase replication is DNA (d) replac e ahead of the replication unwinds the complete. DNA and remov e repair enzymes incorrectly fork. rewinds torsiona l it stress as after they paired bases. 6-38 Telomeres chromosomes. serve as Which of caps at the ends of the following is not regarding the (a) The lagging-strand replicatedby true replication telomeres linear of telomeric are not DNA completely sequences? polymerase. (b) Telomeres are made of (c) Additional repeated templatestrand. (d) The leading aprimer repeating sequences strand doubles for sequences. are added to the back the on itself lagging to strand. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) form lOMoARcPSD|30659517 Page 9 of 25 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 DNA Repair Sickle-cell anemia disease. Individuals 6-39 disorder by with have misshapen change in is this an example red (sickle-shaped) the of an blood cells inherited caused a sequence of the thechange? (a) chromosome loss (b) base-pair (c) gene duplication (d) base-pair 6-40 Even function,it in gene. What is the nature of change insertion though still the β-globin DNA polymerase has a introduces errors proofreading what degree newly synthesized strand at does the mismatch arising from DNA a repair system rate of 1 per 107nucleotides. decrease the error rate replication? (a) 2-fold (b) 5-fold (c) 10-fold Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) To lOMoARcPSD|30659517 (d) 100-fold Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 6-41 to Which of repair the shown in Figure below represents the correct way in DNA Figure Q6-41? Q6-41 6-42 A backbone. only of the choices mismatch mismatched base pair causes If this were the a indication mutatio n higher. of an would be Explain replication, the the overall rate backbone caused by DNA indicate which why. Page 10 6-43 a Beside the distortion in the mismatche base pair, what d additiona lstrand of error in much distortion 25 mark is need to s ther e be on the template DNA eukaryotic to repaired? (a) a nick in (b) a chemical modification of strand the new strand Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (c) a nick in the new strand (d) a sequence gap in the 6-44 A pregnant achemical. litter deformed, new strand mouse is exposed Many of the to mice but interbred when they are high in levels of her with each ar e all other , their offspring are normal. Which two explain these results? of (a) In were mice, somatic the deformed mutated. the following cells statements could but not germ cells (b) The mouse’s original (c) In were the deformed mutated . (d) The toxic chemical 6-45 The repair of nucleotidesin were mutated. mice, germ cells but affects development but mismatched base pairs or a requires known a multistep sequence of events in this (a) DNA isidentified damage by an existing new germ cells not somatic is not damaged DNA cells mutagenic. strand process. Which choice below describes the recognized, the newly synthesized strand nick in the backbone, strand is removed by a segment by DNA process? is repair proteins, the gap is thestrand is sealed filled of the polymerase, and Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 by DNA ligase. (b) it DNA and repair polymerase simultaneously polymerizes the correc sequenc moves along tthe etemplate. DNA ligase seals (c) DNA isidentified existing new the damage by an is the recognized, the (d) A nick proteinsswitch out in gap is the DNA is wrong the insert the correct bases ahead of behin d it repaired strand. the nick in the backbone, a strand is removed by an exonuclease, and base and nicks in remove s repaired as it newly synthesized strand segment by DNA recognized, base, and DNA of the ligase. DNA repair ligase seals the 6-46 Human pigmentosum beings with the have seriou s inherited disease problem scancer with with lesions repeate d on their skin and often develop exposur ebeing to sunlight. recognized in DNA damage of these individuals? (a) chemical (b) X-ray irradiation (c) mismatched bases (d) ultraviolet What typ the ecells of xeroderma damage damage irradiation nick. damage Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) skin is not lOMoARcPSD|30659517 6-47 You are theenzyme examining phosphofructokinase noticethat the sequence tha t in the Page 11 the DNA in skinks and coding actually enzyme of 25 similar in vary quite a the bit. sequences that Komodo dragons. directs is the sequence code for of but that the surrounding most likely explanation for (a) Coding sequences are repaired more efficiently. (b) Coding sequences are replicated more accurately. (c) Coding sequences thechromosomes to are packaged protect more tightly in damage. (d) Mutations deleterious to in the organism coding sequences than mutations amino acids very two organisms What is the them from DNA You in are noncoding more likely to sequences this? be sequences . In somatic cells, if a base is mismatched in one new daughter strand during DNA replication, and is not repaired, what fraction of the DNA duplexes will have a permanent change in the DNA sequence after the second round of DNA replication? 6- (a) 1/2 4 8 (b) 1/4 (c) 1/8 (d) 1/16 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Sometimes, replication correct mutation. and not 6- mutation 4 rounds 9 (b) TUAT (c) TGAT (d) TAAT chemical damage to DNA can occur just before DNA begins, not giving the repair system enough time to the error before the DNA is duplicated. This gives rise to If the cytosine in the sequence TCAT is deaminated repaired, which of the following is the point you would observe after this segment has undergone two of DNA replication? (a) TTAT During DNA replication in a bacterium, a C is accidentally incorporated instead of an A into one newly synthesized DNA strand. Imagine that this error was not corrected and that it has no effect on the ability of the progeny to grow and reproduce. A. After this original bacterium has divided once, what its 6- proportion of 5 would you expect to contain the mutation? 0 progeny B. What proportion tocontain the after three more rounds of its mutation of progeny DNA would you replication and expect cell division? 6- Sometimes, chemical damage to DNA can occur just before DNA 5 replication begins, not giving the repair system enough time to 1 correct the error before the DNA is duplicated. This gives rise to mutation. If the adenosine in the sequence TCAT is depurinated and not repaired, which of the following is the point mutation you would observe after this segment has undergone two rounds of DNA replication? (a) TCGT (b) TAT (c) TCT Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 TGTT (d) Page 12 of 25 6-52 Which of the accuratestatement following statements is about thymine not an dimers? (a) to Thymine stall. dimers can cause the DNA replication machinery (b) on Thymine opposite dimers DNA are covalent links thymidines exposure to sunlight causes thymine recognize the thymine dimers DNA 6-53 Indicate false. If a whether the statement is following false, statements are explain it is false. 1. Ionizing radiation and oxidative between strands. (c) to Prolonged form. (d) Repair proteins adistortion in dimers as backbone. why damage can true or cause DNA double-strand breaks. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1. After backbone damaged are DNA has been repaired, nicks in the phosphate maintained as a way to identify the repaired. is caused by mechanism that ensures strand that 1. Depurination of DNA is ultraviolet irradiation. 2. Nonhomologous end DNA double- strand breaks fidelity to was a rare event that joining is a are repaired the original with a DNA high degree that of sequence. 6-54 the Several same members kind of when they were unusually the most likely of the cancer young. Which one explanation for this phenomenon? theindividuals with have same family were diagnosed It the of the following is possible cancer . cancer-causing an gene that ancestor’s inherited amutation a mutation in a gene required for (c) inherited a mismatch repair. mutation in a gene required for inherited a thesynthesis of mutation purine in a gene required for somatic suffered cells. (b) inherited synthesis. (d) is that a in (a) with nucleotides. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) DNA lOMoARcPSD|30659517 6-55 You have made a thataredefective collection of in various mutant aspects fruit of to each mutant for hypersensitivity DNA repair. You test three DNA-damaging agents: cytosine), acid sunlight, nitrous and formic (which causes depurination). summarizedin “yes” indicates normal fly, where a than a blanks indicate normal Page 25 Table acid 13 of its (which causes deamination of The results Table Q6-55, that the and flies mutant are is more sensitive sensitivity. Q6-55 1. polymerase? Which mutant is most likely to theDNA repair 1. What aspect of other mutants? 6-56 is nucleic caused The a repair is most likely to deamination of cytosine naturally occurring acid by base, and damage be so due defective be generates does not to in affected a represent in the uracil base. This a Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) DNA lesion lOMoARcPSD|30659517 chemicals “foreign” or and irradiation. Why is why is it important for cells andremove to found in the duplex? DNA this base recognized have a mechanism uracil when it to is as recognize Homologous Recombination 6-57 Select the option that Nonhomologous joining end is is best completes process by which a statement : to a similar thecomplementary stretch (b) after repairing mismatches. (c) to (d) after filling in maintainthe DNA sequence. 6-58 of Nonhomologous end the following? any nearest recovery double-stranded DNA . (a) (a) the strand following end a joined the the of sequence chromosome. on double-stranded DNA nucleotides, helping of to the available any lost integrity of (b) the interruption of (c) loss of (d) translocations entirelydifferent joining can result in lost nucleotides on all a but which damaged DNA gene expression nucleotides at the site of repair of DNA fragments chromosome to an end. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 6-59 Homologous recombination which organisms use a “backup” genetic for is an copy of the DNA as double-strand breaks without information. Which of homologous recombination to the important a mechanism template loss following is in to of fix not necessary occur? (a) 3′ DNA strand overhangs (b) 5′ DNA strand overhangs (c) a long stretch Page 14 of 25 (d) nucleases of sequence similarity 6-60 In addition to the repair of strandbreaks, homologous recombination is DNA a mechanism segments of for generating parental diversity chromosomes. During (a) DNA replication (b) DNA repair (c) meiosis (d) transposition genetic which process double- by does swapping Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) swapping occur? lOMoARcPSD|30659517 Recombination has occurred chromosomesegments shown in 6-61 Q6-61. The b, are genes A and B, used as markers the maternal alignment and , and paternal between and the on the Figure recessive chromosomes, alleles a and respectively. After have b homologous recombination, B, aand the specific arrangements of A, changed. Figure Q6-61 Which of from the the choices below correctly replication products of the (a) AB and aB (b) ab and Ab (c) AB and Ab (d) aB and Ab 6- The 62 to events listed below are occur properly: A. Holliday junction B. strand invasion C. DNA synthesis D. DNA ligation all indicates maternal necessary for cut ligated and the gene combination chromosome? homologous Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) recombination lOMoARcPSD|30659517 E. double-strand F. nucleases Page 15 Which of of break create uneven strands 25 the following events during is the correct homologous order of recombination? (a) E, B, F, D, C, A (b) B, E, F, D, C, A (c) C, E, F, B, D, A (d) E, F, B, C, D, A 6-63 Homologous recombination strandbreaks (DSBs) in is a chromosome. DSBs arise from DNA harmfulchemicals or radiation specialized (for cell produce s the gametes (spermcells intentionally cause DSBs so at least one homologous chronot example, division initiated damage by X-rays). that and by caused double- by During meiosis, the eggs) for sexual reproduction, as to stimulate crossover homologousrecombination. If not there is occurrence within each pair of crossing-over segregate Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) of lOMoARcPSD|30659517 mosomes during meiosis, those noncrossover chromosomes will properly. Figure Q6-43 1. Consider the copy of Chromosome 3 your mother. Is it identical mother to (her the Chromosome 3 that tha t you received sh e received from from her materna chromosome) lshe received from or identica l to the Chromosome 3 her chromosome), or neither? Explain. father (her 1. Starting with paternal the representation in maternal and paternal mother,draw two chromosomes possible chromosomes you 1. What does this indicate may Figure Q6-43 of the found in your have received from about your resemblance to double-stranded your mother. your grandfather and grandmother? 6-64 Indicate false. If a whether the statement is explain it why is haploid, and therefore thechromosome to use as template for statements are true or false. 1. Homologous recombination they are following false, cannot occur in have no a prokaryotic cells, because extra copy of repair. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1. The first stretch from step in repair requires of base pairs 5′ the 1. The 3′ can end overhang be used as a primer Page 16 of of to the site the remove of homologous DNA duplex, the repair DNA polymerase. used to repair the broken a the break. which 25 chromosome inherited 17 nuclease each strand at “invades” for 1. The DNA template homologous Page of a from the strand is the other parent. 25 ANSWERS 6-1 (c) 6-2 (b) Choice (c) is the are false. Although a correct replication statement, is called correct answer. choice (d) is Choices (a) and it reason that DNA is not the Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 semiconservative. 6-3 (b) 6-4 (b) 6-5 Choice (d) is the of and correct answer. Bacteria has many. Choice (a) genome is is incorrect because genome. Choice (b) eukaryotic is Drosophila Drosophila bigger than the E. coli incorrect,because polymerases are not faster than prokaryotic A. The DNA samples centrifugetubes containing 6-6 cesium the light chloride. heavy and DNA have one products collected the polymerases. were placed into After high-speed centrifugation were separated by 1. The three models origin replication, for 2 density. were conservative, semiconservative, and dispersive. conservative model suggested parental a mechanism by which the strands stayed together after replication and the duplex was made entirely of newly synthesized DNA. The semiconservative model two produced during proposedthat the replication DNA duplexes days, original daughte r were hybrid Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) The lOMoARcPSD|30659517 molecules, each having one oneof the of the newly synthesized strands. newDNA The dispersive model predicted each contained all along segments of duplexes strands the molecule. The density-gradient parental conservative model was strands and that the parental and daughter ruled out by the experiments. 1. The dispersive DNA model was ruled out duplexes stranded and then comparing DNA. If the dispersive have had model had an the been by using heat to densities of individual strand s was the correct denature the the single- should , intermediat density. eheavy strands light strands However, and were observed, thesemiconservative DNA 6-7 A. replication this not case; only which convincingly supported model for replication. False. The two strands to occur, but this is accomplished by the binding theorigin of replication. 1. False. DNA replication pairs, which are held together for C-G by base origins only do need to of initiator are typically separate proteins rich at in A-T of two hydrogen bonds (instead pairs), making it easier to atthese sites. separate the strands Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) for base three lOMoARcPSD|30659517 1. True. 2. True. Page 18 6-8 (b) 6-9 to A. the nucleic of The 3′ end. DNA 3′-OH end of a acid 1. Both, as a replication. 6-10 25 polymerase can add nucleotides only chain. result of the bidirectional nature of chromosomal (d) 6-11 (d) 6-12 (c) 6-13 (a) 6-14 There would be several to-5′ strand elongation. detrimental consequences Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) to 3′– lOMoARcPSD|30659517 One of replication those most directly involves synthesis of the lagging are degraded the DNA , segments group. The remaining will incoming nucleotid eprovided will by have linked to the strand . After the have 5′ ends with 3′-OH group. release Without of processes RNA of DNA primers a single phosphat e the energy athe PPi from the 5′ end, nolonger be energetically the process of elongation would favorable. 6-15 See Figure Figure A6-15. A6-15 You will probably add exogenous nucleoside triphosphates to serve as the building blocks needed to make new strands of DNA. Although these monomers will be present in the extracts, they 6will be present at lower concentrations than are normally found 1 inside the cell. They may also be subject to hydrolysis, and 6 the nucleoside diphosphates that are the products of this hydrolysis are not usable substrates for DNA replication. For both of these reasons, it is important to add excess nucleotides to the reaction mixture for efficient DNA replication to occur. 6- (b) Leading and lagging strands are 1 from the replication origin, and are joined 7 the two replication forks meet at the synthesized bidirectionally together by DNA ligase where termination site. Choice (a) is Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 not correct, because this answer implies that the replication fork is not bidirectional and that replication continues around the plasmid until the process makes it back to the origin of replication. Choice (c) is Page 19 of 25 incorrect because the origin is a specialized sequence where initiator proteins bind and open the DNA so that the DNA replication machinery can assemble. Choice (d) is incorrect because the daughter DNA molecules will be same size as the original plasmid (and each other). 6- Choice (a) is the best answer because DNA synthesis cannot 1 begin without the initial primers. Choice (b) is a good answer 8 because lagging-strand synthesis requires continual use of RNA primers for discontinuous replication to occur. 61 (d) 9 6- (a) Because replication 0 helicase of at helicase unwinds both strands the time of the two depends2 upon initiation. DNA template strands, the of activity Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 62 (b) 1 62 (d) 2 (d) Each newly synthesized strand in a daughter duplex was synthesized by a mixture of continuous and discontinuous DNA synthesis from multiple origins. Consider a single replication origin. The fork 6- moving in one direction synthesizes a daughter strand continuously 2 as part of leading-strand synthesis; the fork moving in the 3 opposite direction synthesizes a portion of the same daughter strand discontinuously as part of lagging-strand synthesis. 62 (d) 4 Choice (d) is the correct answer. DNA from all organisms is 6- chemically identical except for the sequence of nucleotides. The 2 proteins listed in choices (a) to (c) can act on any DNA 5 regardless of its sequence. In contrast, the initiator proteins recognize specific DNA sequences at the origins of replication. These sequences differ between bacteria and yeast. 6- In the absence of telomerase, the life-span of a cell and 2 its progeny cells is limited. With each round of DNA replication, 6 the length of telomeric DNA will shrink, until finally all the telomeric DNA has disappeared. Without telomeres capping the chromosome ends, the ends might be treated like breaks arising from DNA damage, or crucial genetic information might be lost. Cells whose DNA lacks telomeres will stop dividing or die. However, if Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 telomerase indefinitely despite is provided to cells, they may be because their telomeres will remain repeated rounds of DNA replication. able to divide a constant length 62 (d) 7 62 A. True. 8 Page 20 of 25 1. False. Although the leading strand, sliding clamp is only loaded once on the lagging strand needs to thepolymerase unload reaches the the RNA primer reloadit previous new segment primer has from the where a the clamp once and then been synthesized. 1. True. 2. False. Primase need one does not have a because the RNA primers the DNA. The primers are are removed, and a aproofreading function DNA polymerase that fills in gaps. the remaining proofreading function, not a nor permanent part does have Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) does it of lOMoARcPSD|30659517 6-29 primase 3 2 single-strand binding 3 sliding clamp 3 RNA protein primers 1 leading strand 2 lagging strand 2 Okazaki fragments 3 DNA helicase 2 DNA ligase 6-30 the A. The actual chemical same regardless of whether direction. reaction going in the 5′-to-3′ The most important or distinction between synthesized in the these two 3′- options to-5′ direction, rather tha n 5′ 3′ of will th e the en the in DNA synthesis in the 3′-to-5′ is that elongating if DNA is is strand, d end, have a nucleoside 1. DNA synthesis the last from 3′ triphosphate. to nucleotide added is mispaired lastnucleotide growing nucleotide strand is coming a 5′ and nucleoside does not is on allow proofreading. removed, the the monophosphate and in Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) the If lOMoARcPSD|30659517 only no has a hydroxyl favorable hydrolysis 6-31 (b) Page 21 reaction 6-32 to of 25 See Figure A6-32. 3′ group on the drive the addition end. Thus, there is of new nucleotides. Figure A6-32 DNA ligase has an important role in DNA replication. After Okazaki 6- fragments are synthesized, they must be ligated (covalently connected) to 3 each other so that they finally form one continuous strand. At the 3 nonpermissive temperature this does not happen, and although theremay be a range of fragments, the notable band at 200 base pairs is the typical size of an individual Okazaki fragment. 6- A. False. The 3 vacant after the 4 RNA primers repair polymerase is are used to in the spaces left degraded. B. False. This is different enzymes. a two-step First, a nuclease polymeras e the th e removes fills in fill RNA process primers. that Then, the Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) requires repair two lOMoARcPSD|30659517 complementary C. True. D. True. DNA sequence. 63 (b) 5 The cell employs an additional protein in order to make the 6constant reloading of the sliding clamp on the lagging strand much 3 more efficient. The protein, called the clamp loader, harnesses energy 6 from ATP hydrolysis to lock a sliding clamp complex around the DNA for every successive round of DNA synthesis. 63 (b) 7 63 (d) 8 63 (b) 9 64 (d) 0 6- (a) 4 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 1 Page 22 of 25 The distortion in the DNA backbone is insufficient information for the mismatch repair system to identify which base is incorrect and which was originally part of the chromosome when replication began. Without additional marks that identify the difference between the 6- newly synthesized strand and the template strand, the repair would 42 be corrected only 50% of the time by random chance. The error rate (and therefore the mutation rate) would still be less than in a system that lacked the mismatch repair enzymes (1 mistake per 107 base pairs), but greater than the error rate in a system that accurately identifies the newly synthesized strand (1 mistake per 109 base pairs). 6- (c) 43 Choices (a) these results in 644 was the or (d) because original are a mouse’s correct. mutation germ cells already carrying. Neither mutations in the germ cells of utero would have had no effect on theymight have led to their offspring. 6- Choice (b) cannot the fetuses would have no (a) (d) 46 6- (d) 47 6- effect on can choice (c), because the fetuses while in their development, but mutant mice among 45 6- account she (b) 48 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) for lOMoARcPSD|30659517 6- (a) 49 A. will One-half, create two or 50%. DNA replication new DNA molecules, mismatchedC-T one of base which will pair. now So a one daughter cell completely norma l of DNA that division will molecule with round of DNA molecule; the other cell themutation to correct a the original bacterium carry a carry nucleotide. or 25%. At cell division, the carrying the from the bacterium on one cell will have the mispaired B. Onequarter, replication and 6- pass in the next mismatched C-T will produce and 50 normal the T DNA and molecule one mutan So tpair. DNA at molecule with this stage, one a fully mutant C-G out the four of the original bacteriu m rise of mutant. containing base is Subsequent cell divisions of to mutant these mutant bacteria, bacteri a will give whereas bacteria . other bacteria normalThe will give proportion rise to containing 25%. mutation will therefore of 6- progeny undamaged strand the progeny remainat the (c) 51 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) only lOMoARcPSD|30659517 6- (b) 52 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Page 23 of 6-53 A. True. 1. False. It replication 25 is as believed a that means of easy identification of aresealed by ligase shortly DNA the nicks are the newly synthesized after replication 1. False. Depurination occurs constantly hydrolysis of deoxyribose the sugar. involves nucleases 6-54 bond linking the DNA base to DNA a loss of genetic becauseare degraded can be Choice (c) is affectedindividuals the in correct of in early-onset mismatch repair choice (a) is genes. Mutations arising incorrect. would A defect in probabl be y spontaneous the repair double-strand sequence, always before they history mutations through cells change in nonhomologousjoining strand but completed. our can during DNA is in 1. False. Homologous recombination any generated breaks but end information by ligated back without the ends together. answer. some families In with fact, a colon cancer have been found to carry in somatic cells are inherited, DNA synthesis or nucleotide not Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) biosynthesis so lOMoARcPSD|30659517 lethal, so choices (b) and (d) are incorrect. 6-55 to A. be Mr Self-destruct is more likely than defective in the DNA the repair polymerase repair of becaus e Mr all thre e thre kinds of e of kinds DNA damage. The in later steps, including are damage for requiremen t similar the DNA the the Self-destruct is other mutants defective repair pathways in for all a repair polymerase. 1. The other mutants are damage. Thus, the mutations are likely to thefirst stage of specific be recognitio and excision of n and Mole are likely to for a particular in genes required repair—the the damaged type of for bases. Dracula be defective in thymidinedimers; the recognition or excision Faust is likely to G be defective in the mismatched base pairs; recognition or excision of and or Marguerite excision be defective in recognition it it recognized as it be is likely to of the U- of abasic sites. 6-56 Uracil is amutational formed with a an RNA base and lesion because, as from the guanine deamination of in the is is cytosine, will Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) paired lOMoARcPSD|30659517 context hydrogen of the DNA bonds, similar thymine, forms and is thus a three hydrogen bonds with cytosine. The backbone, allowing duplex. to mismatch the Uracil pairs by poor partner causes a for distortion repair machinery Becauseuracil to recognize pairs preferably the uracil as with adenine deamination of (its partner cytosine to in double-stranded uracil is transition highly mutagenic. If unrepaired, it of a C-G base pair to a T-A 6-57 (c) 6-58 (a) 6-59 (b) of 25 Page 24 6-60 6-61 a forming two guanine, which of DNA the lesion. can RNA), the result in base pair. (c) (d) 6-62 (d) Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) the lOMoARcPSD|30659517 A. Neither. mother is hybrid of The a copy of Chromosome 3 you received from your the ones she herfather. received from her mother and See Figure A6-43. The chromosome in which a correct answers include any chromosome 6-63 B. portion and th e matches the information from the paternal matches the information from the maternal extensive crossing-over, you resemble there were no crossing-over, you one other. remainder chromosome . Figure C. A6-43 As a result of grandmother and a your grandfather. If much stronger 6- resemblance to A. False. Homologous recombination than the also occurs in prokaryotic both your might have cells, and 64 typically occurs very shortly replicated duplexes after DNA replication, are in when the newly is not close proximity. B. True. C. True. D. False. Although a process it is called homologous recombination, that depends as a on the proximity of parental this homologs. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) When used lOMoARcPSD|30659517 mechanism for DNA repair, homologous recombination chromatids as a in an undamaged, newly replicated uses (homologous) the sister DNA helix template. Page 25 of 25 ESSENTIAL CELL BIOLOGY, FOURTH EDITION CHAPTER 7: FROM DNA TO PROTEIN © 2014 GARLAND SCIENCE PUBLISHING 7-1 For each of the following sentences, fill in the blanks with the best word or phrase selected from the list below. Not all words or phrases will be used; use each word or phrase only once. The instructions specified by the DNA will ultimately specify the sequence of proteins. This process involves DNA, made up of different nucleotides, which gets into RNA, which is then into proteins, made up of different amino acids. In eukaryotic cells, DNA gets made into RNA in the ,while proteins are produced from RNA in the . The segment of DNA called a portionthat is copied into RNA; this process is catalyzed by RNA is the . 4 gene proteasome 20 Golgi replisome 109 kinase sugar-phosphate 128 nuclear pore transcribed cytoplasm nucleus transferase exported polymerase translated 7-2 Use the numbers in the choices below to indicate where in the schematic diagram of a Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 eukaryotic cell (Figure Q7-2) those processes take place. Figure Q7-2 1. transcription 2. translation 3. RNA splicing Page 1 of 29 4. polyadenylation 5. RNA capping From DNA to RNA 7-3 Consider two genes that are next to each other on a chromosome, as arranged in FigureQ7-3. Figure Q7-3 Which of Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 the following statements is true? (a) The two genes must be transcribed into RNA using the same strand of DNA. (b) If gene A is transcribed in a cell, gene B cannot be transcribed. (c) Gene A and gene B can be transcribed at different rates, producing differentamounts of RNA within the same cell. (d) If gene A is transcribed in a cell, gene B must be transcribed. 7-4 RNA in cells differs from DNA in that . (a) it contains the base uracil, which pairs with cytosine. (b) it is single-stranded and cannot form base pairs. (c) it is single-stranded and can fold up into a variety of structures. (d) the sugar ribose contains fewer oxygen atoms than does deoxyribose. 7-5 Transcription is similar to DNA replication in that . (a) an RNA transcript is synthesized discontinuously and the pieces are then joinedtogether. (b) it uses the same enzyme as that used to synthesize RNA primers during DNA replication. (c) the newly synthesized RNA remains paired to the template DNA. (d) nucleotide polymerization occurs only in the 5′-to-3′ direction. Figure Q7-6 is to be used with Questions 7-6, 7-7, and 7-8. These three questions can be used separately or together. Figure Q7-6 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Page 2 of 29 Figure Q7-6 shows a ribose sugar. RNA bases are added to the part of the ribose sugar pointed to by arrow . (a) 3. 7-6 (b) 4. (c) 5. (d) 6. Figure Q7-6 shows a ribose sugar. The part of the ribose sugar that is different from the deoxyribose sugar used in DNA is pointed to by arrow . (a) 1. 7-7 (b) 4. (c) 5. (d) 6. Figure Q7-6 shows a ribose sugar. The part of the ribose sugar where a new ribonucleotide will attach in an RNA molecule is pointed to by arrow . (a) 1. 7-8 (b) 3. (c) 4. (d) 5. 7-9 For each of the following sentences, fill in the blanks with the best word or phrase selected from the list below. Not all words or phrases will be used; use each word or phrase only once. For a cell’s genetic material to be used, the information is first copied from the DNA into the nucleotide sequence of RNA in a process called . Various kinds of RNA are produced, each with different functions. molecules code for proteins, Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 molecules act as adaptors for protein synthesis, molecules are integral components of the ribosome, and important in the splicing of RNA transcripts. molecules are incorporation rRNA translation mRNA snRNA transmembrane pRNA transcription tRNA proteins 7-10 Match the following structures with their names. Page 3 of 29 Figure Q7-10 Which of the following statements is false? (a) A new RNA molecule can begin to be synthesized from a gene before theprevious RNA molecule’s synthesis is completed. (b) If two genes are to be expressed in a cell, these two genes can be transcribed 7-11 withdifferent efficiencies. (c) RNA polymerase is responsible for both unwinding the DNA helix and catalyzing the formation of the phosphodiester bonds between nucleotides. (d) Unlike DNA, RNA uses a uracil base and a deoxyribose sugar. 7-12 Unlike DNA, which typically forms a helical structure, different molecules of RNA can fold into a Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 variety of three-dimensional shapes. This is largely because . (a) RNA contains uracil and uses ribose as the sugar. (b) RNA bases cannot form hydrogen bonds with each other. (c) RNA nucleotides use a different chemical linkage between nucleotides comparedto DNA. (d) RNA is single-stranded. Which of the following molecules of RNA would you predict to be the most likely to fold into a specific structure as a result of intramolecular base-pairing? (a) 5′-CCCUAAAAAAAAAAAAAAAAUUUUUUUUUUUUUUUUAGGG-3′ 7-13 (b) 5′-UGUGUGUGUGUGUGUGUGUGUGUGUGUGUGUGUGUGUGUG-3′ (c) 5′-AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA-3′ (d) 5′-GGAAAAGGAGAUGGGCAAGGGGAAAAGGAGAUGGGCAAGG-3′ Which one of the following is the main reason that a typical eukaryotic gene is able to respond to a far greater variety of regulatory signals than a typical prokaryotic gene or operon? 7-14 (a) Eukaryotes have three types of RNA polymerase. (b) Eukaryotic RNA polymerases require general transcription factors. Page 4 of 29 (c) The transcription of a eukaryotic gene can be influenced by proteins that bind farfrom the promoter. (d) Prokaryotic genes are packaged into nucleosomes. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 7-15 7-16 Match the following types of RNA with the main polymerase that transcribes them. List three ways in which the process of eukaryotic transcription differs from the process of bacterial transcription. 7-17 For each of the following sentences, fill in the blanks with the best word or phrase selected from the list below. Not all words or phrases will be used; each word or phrase should be used only once. In eukaryotic cells, general transcription factors are required for the activity of all promoters transcribed by RNA polymerase II. The assembly of the general transcriptionfactors begins with the binding of the factor to DNA, causing a marked local distortion in the DNA. This factor binds at the DNA sequence called the box, which is typically located 25 nucleotides upstream from the transcription start site. Once RNA polymerase II has been brought to the promoter DNA,it must be released to begin making transcripts. This release process is facilitated by the addition of phosphate groups to the tail of RNA polymerase by the factor . It must be remembered that the general transcription factors and RNA polymerase are not sufficient to initiate transcription in the cell and are affected by proteins bound thousands of nucleotides away from the promoter. Proteins that link the distantly bound transcription regulators to RNA polymerase and the general transcription Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 factors include the large complex of proteins called the . The packing of DNA into chromatin also affects transcriptional initiation, and histone is an enzyme that can render the DNA less accessible to thegeneral transcription factors. activator lac TFIIACAP ligase TFIID deacetylase Mediator TFIIH enhancer TATA 7-18 You have a piece of DNA that includes the following sequence: 5′-ATAGGCATTCGATCCGGATAGCAT-3′ 3′-TATCCGTAAGCTAGGCCTATCGTA-5′ Page 5 of 29 Which of the following RNA molecules could be transcribed from this piece of DNA? (a) 5′UAUCCGUAAGCUAGGCCUAUGCUA-3′ (b) 5′-AUAGGCAUUCGAUCCGGAUAGCAU-3′ (c) 5′-UACGAUAGGCCUAGCUUACGGAUA-3′ (d) none of the above 7-19 The following segment of DNA is from a transcribed region of a chromosome. You know that RNA polymerase moves from left to right along this piece of DNA, that the promoter for this gene is to Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 the left of the DNA shown, and that this entire region of DNA is made into RNA. 5′-GGCATGGCAATATTGTAGTA-3′ 3′-CCGTACCGTTATAACATCAT-5′ Given this information, a student claims that the RNA produced from this DNA is: 3′-GGCATGGCAATATTGTAGTA-5′ Give two reasons why this answer is incorrect. 7-20 You have a segment of DNA that contains the following sequence: 5′-GGACTAGACAATAGGGACCTAGAGATTCCGAAA-3′ 3′-CCTGATCTGTTATCCCTGGATCTCTAAGGCTTT-5′ You know that the RNA transcribed from this segment contains the following sequence: 5′-GGACUAGACAAUAGGGACCUAGAGAUUCCGAAA–3′ Which of the following choices best describes how transcription occurs? (a) the top strand is the template strand; RNA polymerase moves along this strandfrom 5′ to 3′ (b) the top strand is the template strand; RNA polymerase moves along this strandfrom 3′ to 5′ (c) the bottom strand is the template strand; RNA polymerase moves along this strandfrom 5′ to 3′ (d) the bottom strand is the template strand; RNA polymerase moves along this strandfrom 3′ to 5′ 7-21 Imagine that an RNA polymerase is transcribing a segment of DNA that contains the following sequence: 5′-AGTCTAGGCACTGA-3′ Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 3′-TCAGATCCGTGACT-5′ Page 6 of 29 A. If the polymerase is transcribing from this segment of DNA from left to right, which strand (top or bottom) is the template? B. What will be the sequence of that RNA (be sure to label the 5′ and 3′ ends of yourRNA molecule)? The sigma subunit of bacterial RNA polymerase . (a) contains the catalytic activity of the polymerase. 7-22 (b) remains part of the polymerase throughout transcription. (c) recognizes promoter sites in the DNA. (d) recognizes transcription termination sites in the DNA. 7-23 Which of the following might decrease the transcription of only one specific gene in a bacterial cell? (a) a decrease in the amount of sigma factor (b) a decrease in the amount of RNA polymerase (c) a mutation that introduced a stop codon into the DNA that precedes the gene’scoding sequence (d) a mutation that introduced extensive sequence changes into the DNA that Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 precedes the gene’s transcription start site There are several reasons why the primase used to make the RNA primer for DNA replication is not suitable for gene transcription. Which of the statements below is not one of those reasons? (a) Primase initiates RNA synthesis on a single-stranded DNA template. 7-24 (b) Primase can initiate RNA synthesis without the need for a base-paired primer. (c)Primase synthesizes only RNAs of about 5–20 nucleotides in length. (d) The RNA synthesized by primase remains base-paired to the DNA template. You have a bacterial strain with a mutation that removes the transcription termination signal from the Abd operon. Which of the following statements describes the most likely effect of this mutation on Abd transcription? (a) The Abd RNA will not be produced in the mutant strain. (b) The Abd RNA from the mutant strain will be longer than normal. 7-25 (c) Sigma factor will not dissociate from RNA polymerase when the Abd operon is being transcribed in the mutant strain. (d) RNA polymerase will move in a backward fashion at the Abd operon in the mutant strain. Transcription in bacteria differs from transcription in a eukaryotic cell because . (a) RNA polymerase (along with its sigma subunit) can initiate transcription on its own. 7-26 (b) RNA polymerase (along with its sigma subunit) requires the general transcription factors to assemble at the promoter before polymerase can begin transcription. (c) the sigma subunit must associate with the appropriate type of RNA polymerase to produce mRNAs. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Page 7 of 29 (d) RNA polymerase must be phosphorylated at its C-terminal tail for transcription to proceed. Which of the following does not occur before a eukaryotic mRNA is exported from the nucleus? (a) The ribosome binds to the mRNA. 7-27 (b) The mRNA is polyadenylated at its 3′ end. (c) 7-methylguanosine is added in a 5′-to-5′ linkage to the mRNA. (d) RNA polymerase dissociates. Total nucleic acids are extracted from a culture of yeast cells and are then mixed with resin beads to which the polynucleotide 5′-TTTTTTTTTTTTTTTTTTTTTTTTT-3′ has been covalently attached. After a short incubation, the beads are then extracted from the mixture. When to beads, which you analyze the cellular nucleic acids that have stuck the of the following is most abundant? 7-28 (a) DNA (b) tRNA (c) rRNA (d) mRNA 7-29 7-30 Name three covalent modifications that can be made to an RNA molecule in eukaryotic cells before the RNA molecule becomes a mature mRNA. Which of the following statements about RNA splicing is false? (a) Conventional introns are not found in bacterial genes. (b) For a gene to function properly, every exon must be removed from the primarytranscript in the same fashion on every mRNA molecule produced from the same Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 gene. (c) Small RNA molecules in the nucleus perform the splicing reactions necessary for the removal of introns. (d) Splicing occurs after the 5′ cap has been added to the end of the primarytranscript. 7-31 7-32 The length of a particular gene in human DNA, measured from the start site for transcription to the end of the protein-coding region, is 10,000 nucleotides, whereas the length of the mRNA produced from this gene is 4000 nucleotides. What is the most likely reason for this difference? Why is the old dogma “one gene—one protein” not always true for eukaryotic genes? Genes in eukaryotic cells often have intronic sequences coded for within the DNA. These sequences are ultimately not translated into proteins. Why? (a) Intronic sequences are removed from RNA molecules by the spliceosome, 7-33 whichworks in the nucleus. (b) Introns are not transcribed by RNA polymerase. (c) Introns are removed by catalytic RNAs in the cytoplasm. Page 8 of 29 (d) The ribosome will skip over intron sequences when translating RNA into protein. 7-34 snRNAs . (a) are translated into snRNPs. (b) are important for producing mature mRNA transcripts in bacteria. (c) are removed by the spliceosome during RNA splicing. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (d) can bind to specific sequences at intron–exon boundaries through complementarybase-pairing. 7-35 Is this statement true or false? Explain your answer. “Since introns do not contain protein-coding information, they do not have to be removed precisely (meaning, a nucleotide here and there should not matter) from the primary transcript during RNA splicing.” 7-36 You have discovered a gene (Figure Q7-36A) that is alternatively spliced to produce several forms of mRNA in various cell types, three of which are shown in Figure Q7-36B. The lines connecting the exons that are included in the mRNA indicate the splicing. From your experiments, you know that protein translation begins in exon 1. For all forms of the mRNA, the encoded protein sequence is the same in the regions of the mRNA that correspond to exons 1 and 10. Exons 2 and 3 are alternative exons used in different mRNA, as are exons 7 and 8. Which of the following statements about exons 2 and 3 is the most accurate? Explain your answer. Figure Q7-36 (a) Exons 2 and 3 must have the same number of nucleotides. (b) Exons 2 and 3 must contain an integral number of codons (that is, the number of nucleotides divided by 3 must be an integer). (c) Exons 2 and 3 must contain a number of nucleotides that when divided by 3, leaves the same remainder (that is, 0, 1, or 2). (d) Exons 2 and 3 must have different numbers of nucleotides. Page 9 of 29 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 From RNA to Protein 7-37 Which of the following statements about the genetic code is correct? (a) All codons specify more than one amino acid. (b) The genetic code is redundant. (c) All amino acids are specified by more than one codon. (d) All codons specify an amino acid. NOTE: The following codon table is to be used for Problems Q7-38 to Q7-49. 7-38 The piece of RNA below includes the region that codes for the binding site for the initiator tRNA needed in translation. 5′-GUUUCCCGUAUACAUGCGUGCCGGGGGC-3′ Which amino acid will be on the tRNA that is the first to bind to the A site of the ribosome? (a) methionine (b) arginine (c) cysteine (d) valine 7-39 The following DNA sequence includes the beginning of a sequence coding for a protein. What would be the result of a mutation that changed the C marked by an asterisk to an A? 5′-AGGCTATGAATGGACACTGCGAGCCC… Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 * 7-40 Which amino acid would you expect a tRNA with the anticodon 5′-CUU-3′ to carry? (a) lysine Page 10 of 29 (b) glutamic acid (d) leucine (d) phenylalanine Which of the following pairs of codons might you expect to be read by the same tRNA as a result of wobble? (a) CUU and UUU 7-41 (b) GAU and GAA (c) CAC and CAU (d) AAU and AGU 7-42 Below is a segment of RNA from the middle of an mRNA. 5′-UAGUCUAGGCACUGA-3′ If you were told that this segment of RNA was part of the coding region of an mRNA for a large protein, give the amino acid sequence for the protein that is encoded by this segment of mRNA. Write your answer using the one-letter amino acid code. 7-43 Below is the sequence from the 3′ end of an mRNA. 5′-CCGUUACCAGGCCUCAUUAUUGGUAACGGAAAAAAAAAAAAAA-3′ If you were told that this sequence contains the stop codon for the protein encoded by this mRNA, what is the anticodon on the tRNA in the P site of the ribosome when release factor binds to the A site? Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (a) 5′-CCA-3′ (b) 5′-CCG-3′ (c) 5′-UGG-3′ (d) 5′-UUA-3′ 7-44 One strand of a section of DNA isolated from the bacterium E. coli reads: 5′-GTAGCCTACCCATAGG-3′ A. Suppose that an mRNA is transcribed from this DNA using the complementary strand as a template. What will be the sequence of the mRNA in this region (make sure you label the 5′ and 3′ ends of the mRNA)? B. How many different peptides could potentially be made from this sequence ofRNA, assuming that translation initiates upstream of this sequence? C. What are these peptides? (Give your answer using the one-letter amino acidcode.) 7-45 A strain of yeast translates mRNA into protein inaccurately. Individual molecules of a particular protein isolated from this yeast have variations in the first 11 amino acids compared with the sequence of the same protein isolated from normal yeast cells, as Page 11 of 29 listed in Figure Q7-45. What is the most likely cause of this variation in protein sequence? Figure Q7-45 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (a) a mutation in the DNA coding for the protein (b) a mutation in the anticodon of the isoleucine-tRNA (tRNAIle) (c) a mutation in the isoleucyl-tRNA synthetase that decreases its ability todistinguish between different amino acids (d) a mutation in the isoleucyl-tRNA synthetase that decreases its ability todistinguish between different tRNA molecules A mutation in the tRNA for the amino acid lysine results in the anticodon sequence 5′- UAU-3′ (instead of 5′-UUU-3′). Which of the following aberrations in protein synthesis might this tRNA cause? 7-46 (a) read-through of stop codons (b) substitution of lysine for isoleucine (c) substitution of lysine for tyrosine (d) substitution of lysine for phenylalanine After treating cells with a mutagen, you isolate two mutants. One carries alanine and the other carries methionine at a site in the protein that normally contains valine. After treating these two mutants again with mutagen, you isolate mutants from each that 7-47 now carry threonine at the site of the original valine (see Figure Q7-47). Assuming that all mutations caused by the mutagen are due to single nucleotide changes, deduce the codonsthat are used for valine, alanine, methionine, and threonine at the affected site. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Figure Q7-47 What do you predict would happen if you created a tRNA with an anticodon of 5′-CAA- 7-48 3′ that is charged with methionine, and added this modified tRNA to a cell-free translation system that has all the normal components required for translating RNAs? Page 12 of 29 (a) methionine would be incorporated into proteins at some positions where glutamine should be (b) methionine would be incorporated into proteins at some positions where leucineshould be (c) methionine would be incorporated into proteins at some positions where valineshould be (d) translation would no longer be able to initiate 7-49 7-50 In a diploid organism, the DNA encoding one of the tRNAs for the amino acid tyrosine is mutated such that the sequence of the anticodon is now 5′-CTA-3′ instead of 5′-GTA-3′. What kind of aberration in protein synthesis will this tRNA cause? Explain your answer. The ribosome Which of the is important for catalyzing the formation of peptide bonds. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 following statements is true? (a) The number of rRNA molecules that make up a ribosome greatly exceeds thenumber of protein molecules found in the ribosome. (b) The large subunit of the ribosome is important for binding to the mRNA. (c) The catalytic site for peptide bond formation is formed primarily from an rRNA. (d) Once the large and small subunits of the ribosome assemble, they will not separate from each other until degraded by the proteasome. Which of the following statements is true? (a) Ribosomes are large RNA structures composed solely of rRNA. 7-51 (b) Ribosomes are synthesized entirely in the cytoplasm. (c) rRNA contains the catalytic activity that joins amino acids together. (d) A ribosome binds one tRNA at a time. 7-52 Figure Q7-52A shows the stage in translation when an incoming aminoacyl-tRNA has bound to the A site on the ribosome. Using the components shown in Figure Q7-52A as a guide, show on Figures Q7-52B and Q7-52C what happens in the next two stages to complete the addition of the new amino acid to the growing polypeptide chain. Page 13 of 29 Figure Q7-52 7-53 A poison added to an in vitro translation mixture containing mRNA molecules with the sequence 5′-AUGAAAAAAAAAAAAUAA-3′ has the following effect: the only product made is a Met-Lys dipeptide that remains attached to the ribosome. What is the most likely way in which the poison acts to inhibit protein synthesis? (a) It inhibits peptidyl transferase activity. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (b) It inhibits movement of the small subunit relative to the large subunit. (c) It inhibitsrelease factor. (d) It mimics release factor. In eukaryotes, but not in prokaryotes, ribosomes find the start site of translation by . 7-54 (a) binding directly to a ribosome-binding site preceding the initiation codon. (b)scanning along the mRNA from the 5′ end. (c) recognizing an AUG codon as the start of translation. (d) binding an initiator tRNA. Which of the following statements about prokaryotic mRNA molecules is false? (a) A single prokaryotic mRNA molecule can be translated into several proteins. (b) Ribosomes must bind to the 5′ cap before initiating translation. (c) mRNAs are not polyadenylated. 7-55 (d) Ribosomes can start translating an mRNA molecule before transcription iscomplete. Page 14 of 29 7-56 Figure Q7-56 shows an mRNA molecule. Figure Q7-56 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 A. Match the labels given in the list below with the label lines in Figure Q7-56. (a) ribosome-binding site (b) initiator codon (c) stop codon (d) untranslated 3′ region (e) untranslated 5′ region (f) protein-coding region B. Is the mRNA shown prokaryotic or eukaryotic? Explain your answer. 7-57 You have discovered a protein that inhibits translation. When you add this inhibitor to a mixture capable of translating human mRNA and centrifuge the mixture to separate polyribosomes and single ribosomes, you obtain the results shown in Figure Q7-57. Which of the following interpretations is consistent with these observations? Figure Q7-57 (a) The protein binds to the small ribosomal subunit and increases the rate of initiation of translation. Page 15 of 29 (b) The protein binds to sequences in the 5′ region of the mRNA and inhibits the rate of initiation of translation. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (c) The protein binds to the large ribosomal subunit and slows down elongation of thepolypeptide chain. (d) The protein binds to sequences in the 3′ region of the mRNA and preventstermination of translation. The concentration of a particular protein, X, in a normal human cell rises gradually from a low point, immediately after cell division, to a high point, just before cell division, and then drops sharply. The level of its mRNA in the cell remains fairly constant throughout this time. Protein X is required for cell growth and survival, but the drop in its level just before cell division is essential for division to proceed. You have isolated a line of human cells that grow in size in culture but cannot divide, and on analyzing these mutants, you find that following levels of X mRNA in the mutant cells are normal. Which of the 7-58 mutations in the gene for X could explain these results? (a) the introduction of a stop codon that truncates protein X at the fourth amino acid (b) achange of the first ATG codon to CCA (c) the deletion of a sequence that encodes sites at which ubiquitin can be attached tothe protein (d) a change at a splice site that prevents splicing of the RNA 7-59 For each of the following sentences, fill in the blanks with the best word or phrase selected from the list below. Not all words or phrases will be used; use each word or phrase only once. Once an mRNA is produced, its message can be decoded on ribosomes. The ribosome is composed of two subunits: the subunit, which catalyzes the formation of the peptide bonds that link the amino acids together into a polypeptide chain, and the subunit, which matches the tRNAs to the codons of the mRNA. During the chain elongation process of translating an mRNA into protein, the growing polypeptide chain attached to a tRNA is bound to the site of the ribosome. An incoming aminoacyl-tRNA carrying the next amino acid in the chain will bind to the site by forming base pairs with the exposed codon in the mRNA. The enzyme catalyzes the formation of a new peptide bond between the growing polypeptide chain and the newly arriving amino acid. The end of a protein-coding message is signaled by the presence of a stop codon, which binds the called release factor. Eventually, most proteins will be degraded by a large complex of proteolytic enzymes called the . Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 A medium protein central P RNA DNA peptidyl transferase smallE polymerase T large proteasome ubiquitin Page 16 of 29 7-60 Which of the following methods is not used by cells to regulate the amount of a protein inthe cell? (a) Genes can be transcribed into mRNA with different efficiencies. (b) Many ribosomes can bind to a single mRNA molecule. (c) Proteins can be tagged with ubiquitin, marking them for degradation. (d) Nuclear pore complexes can regulate the speed at which newly synthesizedproteins are exported from the nucleus into the cytoplasm. 7-61 Which of the following statements about the proteasome is false? (a) Ubiquitin is a small protein that is covalently attached to proteins to mark themfor delivery to the proteasome. (b) Proteases reside in the central cylinder of a proteasome. (c) Misfolded proteins are delivered to the proteasome, where they are sequestered from the cytoplasm and can attempt to refold. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (d) The protein stoppers that surround the central cylinder of the proteasome use theenergy from ATP hydrolysis to move proteins into the proteasome inner chamber. RNA and the Origins of Life 7-62 Which of the following molecules is thought to have arisen first during evolution? (a) protein (b) DNA (c) RNA (d) all came to be at the same time 7-63 According to current thinking, the minimum requirement for life to have originated onEarth was the formation of a . (a) molecule that could provide a template for the production of a complementarymolecule. (b) double-stranded DNA helix. (c) molecule that could direct protein synthesis. (d) molecule that could catalyze its own replication. 7-64 Ribozymes catalyze which of the following reactions? (a) DNA synthesis (b) transcription (c) RNA splicing (d) protein hydrolysis Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 7-65 You are studying a disease that is caused by a virus, but when you purify the virusparticles and analyze them you find they contain no trace of DNA. Which of the following molecules are likely to contain the genetic information of the virus? (a) high-energy phosphate groups (b) RNA (c) lipids (d) carbohydrates Page 17 of 29 7-66 Give a reason why DNA makes a better material than RNA for the storage of genetic information, and explain your answer. How We Know: Cracking the Genetic Code 7-67 When using a repeating trinucleotide sequence (such as 5′-AAC-3′) in a cell-free translation system, you will obtain: (a) three different types of peptides, each made up of a single amino acid (b) peptides made up of three different amino acids in random order (c) peptides made up of three different amino acids, each alternating with each otherin a repetitive fashion Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 (d) polyasparagine, as the codon for asparagine is AAC 7-68 You have discovered an alien life-form that surprisingly uses DNA as its genetic material, makes RNA from DNA, and reads the information from RNA to make protein using ribosomes and tRNAs, which read triplet codons. Because it is your job to decipherthe genetic code for this alien, you synthesize some artificial RNA molecules and examine the protein products produced from these RNA molecules in a cell-free translation system using purified alien tRNAs and ribosomes. You obtain the results shown in Table Q7-68. Table Q7-68 From this information, which of the following peptides can be produced from poly UAUC? (a) Ile-Phe-Val-Tyr (b) Tyr-Ser-Phe-Ala (c) Ile-Lys-His-Tyr (d) Cys-Pro-Lys-Ala 7-69 An extraterrestrial organism (ET) is discovered whose basic cell biology seems pretty much the same as that of terrestrial organisms except that it uses a different genetic code Page 18 of 29 to translate RNA into protein. You set out to break the code by translation experiments using RNAs of known sequence and cell-free extracts of ET cells to supply the necessary protein- Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 synthesizing machinery. In experiments using the RNAs below, the following results were obtained when the 20 possible amino acids were added either singly or in different combinations of two or three: RNA 1: 5′-GCGCGCGCGCGCGCGCGCGCGCGCGCGC-3′ RNA 2: 5′-GCCGCCGCCGCCGCCGCCGCCGCCGCCGCC-3′ Using RNA 1, a polypeptide was produced only if alanine and valine were added to the reaction mixture. Using RNA 2, a polypeptide was produced only if leucine and serine and cysteine were added to the reaction mixture. Assuming that protein synthesis can start anywhere on the template, that the ET genetic code is nonoverlapping and linear, and that the many each codon is the same length (like terrestrial triplet code), how nucleotides does an ET codon contain? (a) 2 (b) 3 (c) 4 (d) 5 (e) 6 7-70 NASA has discovered an alien life-form. You are called in to help NASA scientists to deduce the genetic code for this alien. Surprisingly, this alien life-form shares many similarities with life on Earth: this alien uses DNA as its genetic material, makes RNA from DNA, and reads the information from RNA to make protein using ribosomes and tRNAs. Even more amazing, this alien uses the same 20 amino acids, like the organisms found on Earth, and also codes for each amino acid by a triplet codon. However, the scientists at NASA have found that the genetic code used by the alien life-form differs from that used by life on Earth. NASA scientists drew this conclusion after creating a cell-free protein-synthesis system from alien cells and adding an mRNA made entirely of uracil (poly U). They found that poly U directs the synthesis of a peptide containing only glycine. NASA scientists have synthesized a poly AU mRNA and observe that it codes for a polypeptide of alternating serine and proline amino acids. From these experiments, can you determine which codons code for serine and proline? Explain. Bonus question. Can you propose a mechanism for how the alien’s physiology is altered so that it uses a different genetic code from life on Earth, despite all the similarities? Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Page 19 of 29 ANSWERS The instructions specified by the DNA will ultimately specify the sequence of proteins. This process involves DNA, made up of 4 different nucleotides, which gets transcribed into RNA, which is then translated into proteins, made up of 20 different amino acids. In 7-1 eukaryotic cells, DNA gets made into RNA in the nucleus, while proteins are produced from RNA in the cytoplasm. The segment of DNA called a gene is the portion that is copied into RNA; this process is catalyzed by RNA polymerase. 7-2 See Figure A7-2. Figure A7-2 7-3 (c) Choice (c) is correct. Choice (a) is untrue because although RNA contains uracil, uracil pairs with adenine, not cytosine. Choice (b) is false because RNA can form base pairs with a complementary 7 RNA or DNA sequence. Choice (d) is false because ribose contains one more oxygen atom than deoxyribose. - 4 7-5 Choice (d) is correct. Choice (a) is incorrect because an RNA transcript is made by a single polymerase molecule that proceeds from the start site to the termination site without falling off. The enzyme used to make primers during DNA synthesis is indeed an RNA polymerase, but it is a special enzyme, primase, and not the enzyme that is used for transcription, which is why choice (b) is incorrect. Choice (c) is false. 7-6 (a) 7-7 (c) Page 20 of 29 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 7-8 (c) 7-9 For a cell’s genetic material to be used, the information is first copied from the DNA into the nucleotide sequence of RNA in a process called transcription. Various kinds of RNA are produced, each with different functions. mRNA molecules code for proteins, tRNA molecules act as adaptors for protein synthesis, rRNA molecules are integral components of the ribosome, and snRNA molecules are important in the splicing of RNA transcripts. 7-10 A—4; B—2; C—1; D—3 7-11 Choice (d) is false. RNA nucleotides contain the sugar ribose. Choice (d) why RNA is correct. Choice 7 (a) is true, but is not the main reason different molecules can form different three-dimensional structures (although ribose does increase potential hydrogenbonding potentials 1 compared to deoxyribose). Choices (b) and (c) are untrue. 2 Choice (a) is correct. Choices (b) and (c) do not have any opportunity for intramolecular basepairing and thus a specific structure is unlikely. Although there is some opportunity for intramolecular base-pairing in choice (d), choice (a) has much more intrastrand complementarity 7 and is a better choice. 1 3 7-14 (c) 7-15 A—1; B—3; C—3; D—2; E – 2 7-16 Any three of the following are acceptable. 1. Bacterial cells contain a single RNA polymerase, whereas eukaryotic cells have three. 2. Bacterial RNA polymerase can initiate transcription without the help of additional proteins, whereas eukaryotic RNA polymerases need general transcription factors. 3. Ineukaryotic cells, transcription regulators can influence transcriptional initiation thousands of nucleotides away from the promoter, whereas bacterial regulatory sequences are very close to the promoter. 4. Eukaryotic transcription is affected by chromatin structure and nucleosomes, Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 whereas bacteria lack nucleosomes. In eukaryotic cells, general transcription factors are required for the activity of all promoters transcribed by RNA polymerase II. The assembly of the general transcription factors begins with the binding of the factor TFIID to DNA, causing a marked local distortion in the DNA. This factor binds at the DNA sequence called the TATA box, which is typically located 25 nucleotides upstream from the transcription start site. Once RNA polymerase II has been brought to the promoter DNA, 7 it must be released - to begin making transcripts. This release process is facilitated by the addition of phosphate groups 1 to the tail of RNA polymerase by the factor TFIIH. It must be remembered that the 7 Page 21 of 29 general transcription factors and RNA polymerase are not sufficient to initiate transcription in the cell and are affected by proteins bound thousands of nucleotides away from the promoter. Proteins that link the distantly bound transcription regulators to RNA polymerase and the general transcription factors include the large complex of proteins called the Mediator. The packing of DNA into chromatin also affects transcriptional initiation, and histone deacetylase is an enzyme that can render the DNA less accessible tothe general transcription factors. 7-18 Choice (b) is correct. The molecules listed in choices (a) and (c) have incorrect polarity. First, the RNA molecule should have uracil instead of thymine bases. Second, the polarity of the molecule is incorrectly labeled. The correct RNA molecule produced, using the bottom strand of the DNA duplex as a template, would be: 7-19 5′-GGCAUGGCAAUAUUGUAGUA-3′ 7-20 (d) The bottom strand can hybridize with the RNA molecule and thus is the template strand. The polymerase moves along the DNA in a 3′-to-5′ direction, because the RNA nucleotides are joined in a 5′-to-3′ polarity. A. The bottom strand. 7 B. 5′-AGUCUAGGCACUGA-3′ 2 1 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 7-22 (c) (d) Such changes would probably destroy the function of the promoter, making RNA polymerase unable to bind to it. Decreasing the amount of sigma factor or RNA polymerase [choices (a) or (b)] would affect the transcription of most of the genes in the cell, not just one specific gene. 7 codon before the coding sequence [choice (c)] would have no effect on Introducing a stop - gene, because the transcription machinery does not recognize translational transcription of the 2 stops. 3 7-24 7-25 7-26 Choice (b) is true for both primase and RNA polymerase, so it does not describe why primase cannot be used for gene transcription. (b) Without the termination signal, the polymerase will not halt and release from the DNA template at the normal location when transcribing the Abd operon. Most probably, the polymerase will continue to transcribe RNA until it reaches a sequence in the DNA that can serve as a termination sequence, either from the next downstream operon or in the intervening sequence between the Abd operon and the next operon. Dissociation of sigma factor occurs once an approximately 10-nucleotide length of RNA has been synthesized by RNA polymerase and should not be affected by the lack of a termination signal [choice (c)]. Choice (a) is correct. Eukaryotic cells, but not bacteria, require general transcription factors [choice (b)]. There is only a single type of RNA polymerase in bacterial cells Page 22 of 29 [choice (c)]. The general transcription factor TFIIH phosphorylates the C-terminal tail of RNA polymerase in eukaryotic cells but not in bacteria [choice (d)]. 7-27 7-28 (a) Ribosomes are in the cytosol and will bind to the mRNA once it has been exported from the nucleus. (d) mRNA is the only type of RNA that is polyadenylated, and its poly-A tail would be able to basepair with the strands of poly T on the beads and thus stick to them. DNA would not be found in the sample, because the poly-A tail is not encoded in the DNA and long runs of T are rare in DNA. 1. A poly-A tail is added. 2. A 5′ cap is added. 7-29 3. Introns can be spliced out. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 7-30 7-31 7-32 (b) The primary transcript of a gene can sometimes be spliced differently so that different exons can be stitched together to produce distinct proteins in a process called alternative splicing. The gene contains one or more introns. The transcripts from some genes can be spliced in more than one way to give mRNAs containing different sequences, thus encoding different proteins. A single eukaryotic gene may therefore encode more than one protein. 7-33 (a) 7-34 (d) snRNAs are part of the snRNPs, which include proteins and RNA molecules. The proteins within the snRNPs are encoded by their own genes and not translated from snRNPs, which is why choice (a) is incorrect. Bacteria do not have introns, which is why choice (b) is incorrect. 7-35 False. Although it is true that the sequences within the introns are mostly dispensable, the introns must still be removed precisely because an error of one or two nucleotides would shift the reading frame of the resulting mRNA molecule and change the protein it encodes. 7-36 Choice (c) is the only answer that must be true for exons 2 and 3. Although choices (a), (b), and (d) could be true, they do not have to be. Because the protein sequence is the same in segments of the mRNA corresponding to exons 1 and 10, the choice of either exon 2 or exon 3 would not alter the reading frame. To maintain the normal reading frame, whatever it is, the alternative exons must have a number of nucleotides that when divided by 3 (the number of nucleotides in a codon) give the same remainder. Page 23 of 29 7-37 (b) Most amino acids can be specified by more than one codon. Each codon specifies only one amino acid [choice (a)]. Tryptophan and methionine are encoded by only one codon [choice (c)]. Some codons specify translational stop signals [choice (d)]. 7-38 Choice (b) is correct. The initiator methionine is underlined on the RNA molecule below. 5′-GUUUCCCGUAUACAUGCGUGCCGGGGGC-3′ The first tRNA to bind at the A site is the second codon of the protein, because the initiator tRNA is already bound to the P site when translation begins. The codon that follows the binding site for the initiator tRNA is CGU, which codes for arginine. 7-39 The change creates a stop codon (TGA, or UGA in the mRNA) very near the beginning of the (the of the coding Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 protein-coding sequence and in the correct reading frame beginning sequence is indicated by the ATG). Thus, translation would terminate after only four amino acids had been joined together, and the complete protein would not be made. 7-40 (a) As is conventional for nucleotide sequences, the anticodon is given reading from 5′ to 3′. The complementary base-pairing occurs between antiparallel nucleic acid sequences, and the codon recognized by this anticodon will therefore be 5′-AAG-3′. 7-41 Choice (c) is the correct answer. These two codons differ only in the third position and also encode the same amino acid, which is the definition of wobble. Although the codons GAU and GAA [choice (b)] also differ only in the third position, they are unlikely in normal circumstances to be read by the same tRNA, because they encode different amino acids. 7-42 SLGT is the correct answer. (Reading frame two is the only reading frame that does not contain a stop codon.) 7-43 (a) The stop codon (UAA) is underlined in the mRNA sequence below; this is the only stop codon on this piece of mRNA. The codon (UGG) preceding the stop codon will be binding to a tRNA in the P site of the ribosome when release factor binds to the A site. The anticodon of the tRNA will bind to the codon UGG and will be CCA. 5′-CCGUUACCAGGCCUCAUUAUUGGUAACGGAAAAAAAAAAAAAA-3′ A. 5′-GUAGCCUACCCAUAGG-3′ B. Two. (There are three potential reading frames for each RNA. In this case, theyare 7-44 GUA GCC UAC CCA UAG …UAG CCU ACC CAU AGG … AGC CUA CCC AUA GG? … The center one cannot be used in this case, because UAG is a stop codon.) Page 24 of 29 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 C. VAYP SLPIG Note: PTHR will not be a peptide because it is preceded by a stop codon. 7-45 (c) A mutation in the isoleucyl-tRNA synthetase that decreases its ability to distinguish between amino acids would allow an assortment of amino acids to be attached to the tRNAIle. These assorted aminoacyl-tRNAs would then base-pair with the isoleucine codon and cause a variety of substitutions at positions normally occupied by isoleucine. A mutation in the gene encoding the protein would cause only a single variant protein to be made [choice (a)]. A mutation in the anticodon loop of tRNAIle [choice (b)] or a mutation in the isoleucyl-tRNA synthetase that decreases its ability to distinguish between different tRNA molecules [choice (d)] would cause the substitution ofisoleucine for some other amino acid (which is the opposite of what is observed). (b) The mutant tRNALys will be able to pair with the codon 5-AUA-3, which codes for ′′ 7-46 isoleucine. 7-47 Given that there are only single nucleotide changes, the only codons consistent with the changes are GUG for valine, GCG for alanine, AUG for methionine, and ACG for threonine. 7-48 (b) The 5′-CAA-3′ anticodon binds to the 5′-UUG-3′ codon, which normally codes for leucine. 7-49 If the DNA sequence specifying the anticodon is changed from 5′-GTA-3′ to 5′-CTA-3′, this tRNA will now pair with the 5′-UAG-3′ codon (instead of 5′ -UAC-3′). The UAG codon normally serves as a stop codon. Thus, this change will result in the amino acid tyrosine being incorrectly incorporated where there is a stop codon, resulting in the addition of amino acids at the end of proteins that normally would come to a stop because of the UAG codon in the mRNA. (Note that the tyrosine codons will NOT cause premature termination of translation, as tyrosine should continue to be incorporated into proteins, as there are additional tyrosine-tRNA genes in the cell that will provide a normal supply of tyrosine-tRNAs.) 7-50 Choice (c) is correct. A ribosome is built from many more proteins than rRNA molecules, although the ribosome is about two-thirds RNA and one-third protein by weight. Thus, (a) is incorrect. The small subunit binds to mRNA, so (b) is incorrect. Choice (d) is incorrect, as the assembly and disassembly of the small subunit with the large subunit occurs every time a protein is produced from an mRNA. When release factor binds to an mRNA, the ribosome will release the mRNA and dissociate into its two subunits, to be recycled during another round of protein synthesis. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 7-51 Choice (c) is correct. Ribosomes contain proteins as well as rRNA [choice (a)]. rRNA is synthesized in the nucleus, and ribosomes are partly assembled in the nucleus [choice (b)]. A ribosome must be able to bind two tRNAs at any one time [choice (d)]. 7-52 See Figure A7-52. Page 25 of 29 Figure A7-52 7-53 Choice (b) is correct. Choice (a) would prevent all peptide bond formation. Choice (c) would have no effect on translation until the stop codon was reached. Choice (d) would be likely to result in a mixture of polypeptides of various lengths; a poison mimicking a release factor could conceivably cause only Met-Lys to be made, but this dipeptide would not remain bound to the ribosome. 7-54 Choice (b) is correct. Choice (a) is true only for prokaryotes. Choices (c) and (d) are true for both prokaryotes and eukaryotes. (b) Bacterial mRNAs do not have 5′ caps. Instead, ribosome-binding sites upstream of the start 7-55 codon tell the ribosome where to begin searching for the start of translation. A. (a)—3; (b)—2; (c)—4; (d)—6;(e)—1; (f)—5 B. The mRNA is prokaryotic. It contains coding regions for more than one protein, 7-56 as shown by the multiple initiation codons, each preceded by a ribosome-binding site. It contains an unmodified 5′ end, as shown by the three phosphate groups, and an unmodified 3′ end, as shown by the absence of a poly-A tail. 7-57 (b) The results in Figure Q7-57 show a marked decrease in the number of polyribosomes formed relative to normal. Polyribosomes form because the initiation of translation is fairly rapid: ribosomes can bind successively to the free 5′ end of an mRNA molecule and start translation before the first ribosome has had a chance to finish translating the message. Therefore, inhibition of the rate of initiation will tend to decrease the number of Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Page 26 of 29 ribosomes in the polyribosome, and in the extreme case there will be only one ribosome per mRNA. Conversely, increasing the rate of initiation or slowing the rate of elongation would result in an increased number of ribosomes per polyribosome (up to a maximum point), making choices (a) and (c) false. Choice (d) is incorrect, because preventing termination would prevent release of the ribosomes at the end of the coding sequence and would be expected to “freeze” the assembled polyribosomes, so that the ratio of polyribosomes to ribosomes would be much as normal. 7-58 (c) The decrease in the level of protein X in the normal cell is most probably due to protein degradation, because levels of mRNA remain constant. The inability of the mutant cell to divide could be due to a mutation that inhibits protein degradation. This would be achieved by the removal of sites for attachment of ubiquitin, which targets proteins for destruction. Choices (a), (b), and (d) would probably not produce the results mutant cells described, because without the production of a functional protein X,the could not grow in size. 7-59 Once an mRNA is produced, its message can be decoded on ribosomes. The ribosome is composed of two subunits: the large subunit, which catalyzes the formation of the peptide bonds that link the amino acids together into a polypeptide chain, and the small subunit, which matches the tRNAs to the codons of the mRNA. During the chain elongation process of translating an mRNA into protein, the growing polypeptide chain attached to a tRNA is bound to the P site of the ribosome. An incoming aminoacyl-tRNA carrying the next amino acid in the chain will bind to the A site by forming base pairs with the exposed codon in the mRNA. The peptidyl transferase enzyme catalyzes the formation of a new peptide bond between the growing polypeptide chain and the newly arriving amino acid. The end of a protein-coding message is signaled by the presence of a stop codon, which binds the protein called release factor. Eventually, most proteins will be degraded by a large complex of proteolytic enzymes called the proteasome. 7-60 (d) Proteins are synthesized in the cytoplasm and therefore newly synthesized proteins would not be exported from the nucleus into the cytoplasm. 7-61 (c) Once proteins are sent to the proteasome, proteases degrade them. Chaperone proteins provide a place for misfolded proteins to attempt to refold. 7-62 (c) Because RNA is known to catalyze reactions within the cell, because the components of RNA are thought to be more readily formed in the conditions on primitive Earth, and because RNA can contain genetic information, it is the most likely of the three molecules to have arisen first in evolution. 7-63 Choice (d) is correct. Choice (a) is incorrect in that, although this may have been a step in self- Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 replication, it would not by itself be sufficient. Choices (b) and (c) are incorrect, as these stages in the evolution of the cell must have succeeded the formation of the first self-replicating molecules. 7-64 (c) Page 27 of 29 7-65 (b) 7-66 Three possible answers are: 1. The deoxyribose sugar of DNA makes the molecule much less susceptible than RNA to breakage, because of the lack of the hydroxyl group on carbon 2 of the ribose sugar. 2. DNA is double-stranded and therefore the complementary strand provides a template from which damage can be repaired accurately. 3. The use of “T” in DNA instead of “U” (as in RNA) protects against the effect of deamination, a common form of damage. Deamination of T produces an aberrantbase (methyl C), whereas deamination of U generates C, a normal base. The presence of an abnormal base eases the cell’s job of recognizing the damaged strand. 7-67 (a) An mRNA composed of a trinucleotide repeat of AAC can be “read” in three different frames: AAC, ACA, and CAA. Thus, this mRNA will yield polyasparagine (codon = AAC), Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 polythreonine (codon = ACA), and polyglutamine (codon = CAA). 7-68 (d) All other answers are not possible, because poly UAUC cannot code for Tyr. Tyr must be encoded by AUA, because both poly AUA and poly UA lead to the synthesis of Tyr (see Table A7-68). Table A7-68 7-69 (d) An organism having codons with an even number of nucleotides (such as 2, 4, or 6) could read 5′-GCGCGCGCGC-3′ (RNA 1) in either of two ways, namely “GC GC GC GC …” or “CG CG CG CG …” Either of the two amino acids alone could have supported protein synthesis, so you would not need them in combination [thus eliminating choices (a), (c), and (e)]. An organism having three bases per codon could read the sequence 5′-GCCGCCGCCGCCGCC-3′ (RNA 2) in one of three ways, namely “GCC GCC GCC GCC …,” “CCG CCG CCG CCG …,” or “CGC CGC CGC Page 28 of 29 CGC …,” and so again, any one of the three amino acids could have supported synthesis of a polypeptide, and you would not need to add all three amino acids to produce a polypeptide chain, thus eliminating choice (b). Only a five-nucleotide code gives you two different consecutive codons for RNA 1 and three different consecutive codons for RNA 2. 7-70 No, you cannot definitively determine the codons that code for serine or proline, because it could be either UAU or AUA. Bonus. The alien aminoacyl-tRNA synthetases could adapt a different amino acid to each tRNA, thus matching an amino acid with a different codon compared with the codons used by life on Earth. Page 29 of 29 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 INTRACELLULAR COMPARTMENTSAND TRANSPORT Membrane-Enclosed Organelles 15.1 Name the membrane-bounded compartments in a eucaryotic cell where each of thefunctions listed below takes place. A. Photosynthesis B. Transcription C. Oxidative phosphorylation D. Modification of secreted proteins E. Steroid hormone synthesis F. Degradation of worn-out organelles G. New membrane synthesis H. Breakdown of lipids and toxic molecules 1 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 15.2 Label the structures of the cell indicated by the lines on the figure below: Figure Q15-2 A. B. C. D. E. F. G. H. I. J. 15.3 nucleus free ribosomes rough endoplasmic reticulum Golgi apparatus cytosol endosome plasma membrane lysosome mitochondrion peroxisome You discover a fungus that contains a strange star-shaped organelle not found in anyother eucaryotic cell you have seen. On further investigation you find the following 1. the organelle possesses a small genome in its interior. 2. the organelle is surrounded by two membranes. 3. vesicles do not pinch off the organelle membrane. 4. the interior of the organelle contains proteins similar to those of many bacteria. 5. the interior of the organelle contains ribosomes. How might this organelle have arisen? Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Protein Sorting 15.4 For each of the following sentences, fill in the blanks with the best word or phrase selected from the list below. Not all words or phrases will be used; each word or phraseshould be used only once. Plasma membrane proteins are inserted into the membrane in the . The address information for protein sorting in a eucaryotic cell is contained in the of the proteins. Proteins enter the nucleus in their form. Proteins that remain in the cytosol do not contain a . Proteins are transported into the Golgi apparatus via . The proteins transported into the endoplasmic reticulum by are in their form. amino acid sequence endoplasmic reticulum folded 15.5 Golgi apparatus plasma membrane protein translocators sorting signal transport vesicles unfolded What would happen in each of the following cases? Assume in each case that the proteininvolved is a soluble protein, not a membrane protein. A. You add a signal sequence (for the ER) to the amino-terminal end of a normally cytosolic protein. B. You change the hydrophobic amino acids in an ER signal sequence into charged amino acids. C. You change the hydrophobic amino acids in an ER signal sequence into other, hydrophobic, amino acids. D. You move the amino-terminal ER signal sequence to the carboxyl-terminal end ofthe protein. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 15.6 You are trying to identify the peroxisome-targeting sequence in the thiolase enzyme fromyeast. The thiolase enzyme normally resides in the peroxisome and therefore must contain amino acid sequences that are used to target the enzyme for import into the peroxisome. To identify the targeting sequences, you create a set of hybrid genes that encode fusion proteins containing part of the thiolase protein fused to another protein, histidinol dehydrogenase (HDH). HDH is a cytosolic enzyme required for the synthesis of the amino acid histidine and cannot function if it is localized in the peroxisome. You genetically engineer a series of yeast cells to express these fusion proteins instead of their own versions of these enzymes. If the fusion proteins are imported into the peroxisome, the HDH portion of the protein cannot function and the yeast cells cannot grow on media lacking histidine. You obtain the following results: Figure Q15-6 What region of the thiolase protein contains the peroxisomal targeting sequence? Explainyour answer. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 15.7 What is the role of the nuclear localization sequence in a nuclear protein? (a) It is bound by cytoplasmic proteins that direct the nuclear protein to thenuclear pore. (b) It is a hydrophobic sequence that enables the protein to enter thenuclear membranes. (c) It aids protein unfolding in order for the protein to thread through nuclear pores. (d) It prevents the protein diffusing out of the nucleus via nuclear pores. 15.8 A gene regulatory protein, A, contains a typical nuclear localization signal but surprisingly is usually found in the cytosol of cells. When the cell is exposed to hormones, protein A moves from the cytosol into the nucleus where it turns on genes involved in cell division. When you purify protein A from cells that have not been treated with hormones, you find that protein B is always complexed with it. To determine the function of protein B, you engineer cells lacking the gene for protein B. You compare normal and defective cells by using differential centrifugation to separate the nuclear fraction from the cytoplasmic fraction and then separate the proteins in thesefractions by gel electrophoresis. You identify the presence of protein A and protein B bylooking for their characteristic bands on the gel. The gel you run is shown below: Figure Q15-8 On the basis of these results, what is the function of protein B? Explain your conclusionand propose a mechanism for how protein B works. 15.9 Which of the following statements about import of proteins into mitochondria are TRUE? (a) The signal sequences on mitochondrial proteins are usually carboxyl terminal. (b) The first stage of import of a mitochondrial protein is across the outermembrane into the intermembrane space. (c) Most mitochondrial proteins are not imported from the cytosol but aresynthesized inside the mitochondria. (d) Mitochondrial proteins are translocated across the inner and outermembranes simultaneously. (e) Mitochondrial proteins cross the membrane in their native, folded state. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 15.10 Proteins destined to enter the endoplasmic reticulum (a) are transported across the membrane after their synthesis is complete. (b) are synthesized on free ribosomes in the cytosol. (c) begin to cross the membrane while still being synthesized. (d) cross the membrane in a folded state. (e) all remain within the endoplasmic reticulum. 15.11 After isolating the rough endoplasmic reticulum from the rest of the cytoplasm, you purify the RNAs attached to it. Which of the following proteins do you expect the RNA from the rough endoplasmic reticulum to encode? (a) Soluble secreted proteins (b) ER membrane proteins (c) Mitochondrial membrane proteins (d) Plasma membrane proteins (e) Ribosomal proteins 15.12 Briefly describe the mechanism by which the presence of an internal stop-transfer sequence in a protein causes the protein to become embedded in the lipid bilayer as a transmembrane protein with a single membrane-spanning region. Assume that the protein has an amino terminal signal sequence and just one internal hydrophobic stop-transfer sequence. 15.13 Using genetic engineering techniques, you have created a set of proteins that contain two (and only two) conflicting signal sequences that specify different compartments. Predict which signal would win out for the following combinations. Explain youranswers. A. Signals for import into the nucleus and import into the ER. B. Signals for export from the nucleus and import into the mitochondria. C. Signals for import into mitochondria and retention in the ER. 15.14 A protein traverses the plasma membrane three times in the orientation shown below (N = amino terminus, C = carboxyl terminus; the hydrophobic membranespanning regions are shown as open boxes). This protein is known to have a signal sequence thatis cleaved by signal peptidase in the ER. Figure Q15-14 Sketch the ER membrane and the arrangement of the newly synthesized protein chainafter it has completed its entry into the ER membrane but before any action of signal Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 peptidase. Be sure to label the cytosol, the ER lumen, the signal sequence, and the aminoand carboxyl termini of the protein in your diagram. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 15.15 The figure below shows the orientation of a multipass transmembrane protein after it hascompleted its entry into the ER membrane (part A) and after it gets delivered to the plasma membrane (part B). This protein has an amino-terminal signal sequence (depicted as the dark grey membrane spanning box), which is cleaved off in the endoplasmic reticulum by signal peptidase. The other membrane-spanning domains in the protein are depicted as open boxes. Given that any hydrophobic membranespanningdomain can act as either a start-transfer or a stop-transfer region, draw the final consequences of the actions described below on the orientation of the protein in the plasma membrane. Be sure to indicate on your drawing the extracellular space, the cytosolic face, and the plasma membrane, as well as the amino- and carboxyl-termini of the protein. Figure Q15-15 Deleting the first signal sequence. Changing the hydrophobic amino acids in the first, cleaved, sequence tocharged amino acids. C. Changing the hydrophobic residues in every other transmembrane sequenceto charged residues, starting with the first, cleaved, signal sequence. 15.16 Examine the multipass transmembrane protein shown in Figure Q15-16. What would you predict would be the effect of converting the first hydrophobic transmembrane segment to a hydrophilic segment? Sketch the arrangement of the modified protein in theER membrane. A. B. Figure Q15-16 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Vesicular Transport 15.17 For each of the following sentences, fill in the blanks with the best word or phrase selected from the list below. Not all words or phrases will be used; each word or phraseshould be used only once. Proteins are transported out of a cell via the or pathway. Fluids and macromolecules are transported into the cell via the pathway. All proteins being transported out of the cell pass through the and the . Transport vesicles link organelles of the system. The formation of in the endoplasmic reticulum stabilizes protein structure. carbohydrate disulfide bonds endocytic endomembrane endoplasmic reticulum endosome exocytic Golgi apparatus hydrogen bonds ionic bonds lysosome protein secretory 15.18 Name two functions of the protein coat of vesicles that bud from membranousorganelles used in vesicular transport. 15.19 An individual transport vesicle (a) contains only one type of protein in its lumen. (b) will fuse with only one type of membrane. (c) is endocytic if it is traveling toward the plasma membrane. (d) is enclosed by a membrane with the same lipid and protein composition asthe membrane of the donor organelle. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 15.20 In class we have discussed how v-SNAREs and t-SNARES mediate the recognition ofa vesicle with its target membrane so that a vesicle displaying a particular type of vSNARE will only fuse with a target membrane containing a complementary type of tSNARE. It is also known that in some cases, v-SNAREs and t-SNAREs may also mediate fusion of identical membranes. In yeast cells, right before the formation of a new cell, vesicles derived from the vacuole will come together and fuse to form a new vacuole destined for the new cell. Unlike the situation we’ve discussed in class, the vacuolar vesicles contain both v-SNAREs and t-SNAREs. Your friend is trying to understand the role of these SNAREs in the formation of the new vacuole and wants toconsult with you regarding the interpretation of his data. Your friend has designed an ingenious assay for the fusion of vacuolar vesicles utilizing alkaline phosphatase. The protein alkaline phosphatase is made in a “pro” form that must be cleaved in order for the protein to be active. Your friend has designed two different strains of yeast: strain A produces the “pro” form of alkaline phosphatase (pro-Pase), while strain B produces the protease that can cleave pro-Pase into the active form(Pase). Neither strain has the active form of the alkaline phosphatase, but when vacuolarvesicles from the strains A and B are mixed, fusion of vesicles generates active alkaline phosphates, whose activity can be measured and quantified. Figure Q15-20A Your friend has taken each of these yeast strains and further engineered them so that they express only the v-SNAREs, the t-SNAREs, both (the normal situation), or neither SNARE. He then isolates vacuolar vesicles from all strains and tests the ability of each variant form of strain A to fuse with each variant form of strain B, using the alkaline phosphatase assay. The data are shown in the graph depicted in Figure Q15-20B. On this graph, the SNARE present on the vesicle of the particular yeast strain is indicated as“v” (for the presence of the v-SNARE) and “t” (for the presence of the t-SNARE). Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Figure Q15-20 B What does his data say about the requirements for v-SNAREs and t-SNAREs in the vacuolar vesicles? Be sure to comment on whether it is important to have a specific typeof SNARE (that is, v- or t-SNARE) on each vesicle. Secretory Pathway 15.21 N-linked oligosaccharides on secreted glycoproteins are attached to (a) nitrogen atoms in the polypeptide backbone. (b) the serine or threonine in the sequence Asn-X-Ser/Thr. (c) the amino terminus of the protein. (d) the asparagine in the sequence Asn-X-Ser/Thr. (e) the aspartic acid in the sequence Asp-X-Ser/Thr. 15.22 Name two types of protein modification that can occur in the ER but not in the cytosol. 15.23 If you were to remove the ER-retention signal from a protein that normally resides inthe ER lumen, where do you expect the protein will ultimately end up? Be sure to explain your reasoning, for full credit. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 15.24 Match the set of labels below with the numbered label lines on Figure 15-24. Figure Q15-24 A. B. C. D. E. Cisterna Golgi stack Secretory vesicle trans Golgi network cis Golgi network Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 15.25 A plasma membrane protein carries an oligosaccharide containing mannose (Man), galactose (Gal), sialic acid (SA), and N-acetylglucosamine (GlcNAc). These sugars are added to the protein as it proceeds through the secretory pathway. First, a core oligosaccharide containing Man and GlcNAc is added, followed by Gal, Man, SA, and GlcNAc in a particular order. Each addition is catalyzed by a different transferase acting at a different stage as the protein proceeds through the secretory pathway. You have isolated mutants defective for each of the transferases, purified the membrane protein from each of the mutants, and identified which sugars are present in each mutant protein. The results are summarized in Table Q15-25. Table Q15-25 Cell lacking: A. Oligosaccharide protein transferase Sugars present in the purified protein SA GlcNAc Ma Gal – – – n – B. Galactose transferase C. SA transferase D. GlcNAc transferase + – – + + + + – – – + less than in normal cells From these results, match each of the transferases (A, B, C, D) to its subcellular locationselected from the list below. (Assume that each location contains only one enzyme.) 1. 2. 3. 4. Central Golgi cisternae cis Golgi network ER trans Golgi network 15.26 For each of the following sentences, choose one of the options enclosed insquare brackets to make a correct statement. New plasma membrane reaches the plasma membrane by the [regulated/constitutive] exocytosis pathway. New plasma membrane proteins reach the plasma membrane by the [regulated/constitutive] exocytosis pathway. Insulin is secreted from pancreatic cells by the [regulated/constitutive] exocytosis pathway. The interior of the trans Golgi network is [acidic/alkaline]. Proteins that are constitutively secreted [aggregate/do not aggregate] in the trans Golgi network. 15.27 In a cell capable of regulated secretion, what are the three main classes of proteinsthat must be separated before they leave the trans Golgi network? Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Endocytic Pathways 15.28 Name three possible fates for an endocytosed molecule that has reached the endosome. 15.29 For each of the following sentences, fill in the blanks with the best word or phrase selected from the list below. Not all words or phrases will be used; each word or phraseshould be used only once. Eucaryotic cells are continually taking up materials from the extracellularspace by the process of endocytosis. One type of endocytosis is , which involves utilizing proteins to form small vesicles containing fluids and molecules. After these vesicles pinch off from the plasma membrane, they will fuse with the , where the uptaken materials are sorted. A second type of endocytosis is , which is used to takeup large vesicles that can contain microorganisms and cellular debris. Macrophages are especially suited for this process, as they extend (sheetlike projections of their plasma membrane)to surround the invading microorganisms. chaperone Golgi apparatus cholesterol clathrin endosome mycobacterium phagocytosis pinocytosis pseudopod s rough ER SNARE transcytosis Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 15.30 Fibroblast cells from patients W, X, Y, and Z, who each have a different inherited defect, all contain “inclusion bodies,” which are lysosomes filled with undigested material. You wish to identify the cellular basis of these defects. The possibilities are: 1. 2. 3. a defect in one of the lysosomal hydrolases. a defect in the phosphotransferase that is required for mannose-6-phosphate tagging of the lysosomal hydrolases. a defect in the mannose-6-phosphate receptor, which binds mannose-6phosphate tagged lysosomal proteins in the trans Golgi network and deliversthem to lysosomes. You find that when some of these mutant fibroblasts are incubated in media in which normal cells have been grown, the inclusion bodies disappear. This leads you to suspect that lysosomal hydrolases are being secreted by the constitutive exocytic pathway in normal cells and are being taken up by the mutant cells. (It is known that some mannose-6-phosphate receptor molecules are found in the plasma membrane and can take up and deliver lysosomal proteins via the endocytic pathway.) You incubate cells from each patient with media from normal cells and media from each of the other mutant cell cultures, and get the following results. Cell Line Normal W X Y Z From normal cells From cultures of W cells Media From cultures of X cells + – + + + + – + + + + – – – – From cultures of Y cells From cultures of Z cells + – – – + + – – + – + indicates that the cells appear normal; – indicates that the cells still have inclusionbodies. For each patient (W, X, Y, Z) indicate which of the defects (1, 2, 3) they are most likelyto have. 15.31 How is it that the low pH of lysosomes protects the rest of the cell fromlysosomal enzymes in case the lysosome breaks? Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 How We Know: Tracking Protein and Vesicle Transport 15.32 You have created a GFP fusion to a protein that is normally secreted from yeast cells. Since you have learned about the use of temperature-sensitive mutations in yeast to study protein and vesicle transport, you obtain a collection of three mutant yeast strains, each one defective in some aspect of the protein secretory process. Being a good scientist, youof course, also obtain a wild-type control strain. You decide to examine the fate of your GFP fusion protein in these various yeast strains and engineer the mutant strains to express your GFP fusion protein. However, in your excitement to do the experiment, yourealize that you did not label any of the mutant yeast strains and no longer know which strain is defective in what process. You end up numbering your strains with the numbers1 through 4, and then you carry out the experiment anyway, obtaining the following results (note that the black dots represent your GFP fusion protein): Figure Q15-32 Name the process defective in each of these strains. Remember that one of these strainsis your wild-type control. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 Answers 15.1 A B. C. D. E. F. G. H. 15.2 Photosynthesis = chloroplast Transcription = nucleus Oxidative phosphorylation = mitochondrion Modification of secreted proteins = Golgi apparatus and roughendoplasmic reticulum (ER) Steroid hormone synthesis = smooth ER Degradation of worn-out organelles = lysosome New membrane synthesis = ER Breakdown of lipids and toxic molecules = peroxisome See Figure A15-2. Figure A15-2 15.3 A genome, a double membrane, ribosomes, and proteins similar to those found in bacteria are evidence for an organelle having evolved from an engulfed bacterium. 15.4 Plasma membrane proteins are inserted into the membrane in the endoplasmic reticulum. The address information for protein sorting in a eucaryotic cell is contained in the amino acid sequence of the proteins. Proteins enter the nucleus in their folded form. Proteins that remain in the cytosol do not contain a sorting signal. Proteins are transported into the Golgi apparatus via transport vesicles. The proteins transported intothe endoplasmic reticulum by protein translocators are in their unfolded form. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 15.5 A. B. C. D. 15.6 The protein will now be transported into the ER lumen. The altered signal sequence will not be recognized and the protein will remainin the cytosol. The protein will still be delivered into the ER. It is the distribution of hydrophobic amino acids that is important, not the actual sequence. The protein will not enter the ER. Because the carboxyl terminus of the proteinis the last part to be made, the ribosomes synthesizing this protein will not be recognized by the SRP and carried to the ER. The peroxisomal targeting sequence lies between amino acids number 100 and number 125. Any fusion protein containing this sequence can be targeted for import into the peroxisome (because the yeast cannot grow on media lacking histidine) while the fusion proteins lacking this region do not target the fusion protein for import into the peroxisome (because the yeast do grow on media lacking histidine). The most importantpieces of data are the fusion proteins containing amino acids 100–200 of the thiolase protein fused to HDH and the fusion protein containing amino acids 1–125 of the thiolase protein fused to HDH. Both of these fusion proteins do not allow growth on media lacking histidine and can be used to define the minimal region necessary for targeting thiolase for import into the peroxisome. (Note that although these experiments show that amino acids 100–125 are necessary, these experiments do not show that this region is sufficient for peroxisomal targeting. Itis possible that amino acids 100–125 is sufficient, or, it could be that this region collaborates with redundant signals between amino acids 1–100 or 125–200.) 15-7 (a) 15-8 The data on the gel shows that protein A is always found in the nucleus in the absenceof protein B. Therefore, any mechanism that is proposed must explain this result. On possible answer is that protein B binds protein A and masks the nuclear localization signal. In the presence of hormone, protein B interacts with the hormone, which changesits conformation so that it can no longer bind protein A. When protein B no longer bindsto protein A, the nuclear localization signal on protein A is now exposed and protein A can enter the nucleus. Therefore, in the absence of protein B, the nuclear localization signal on protein A is always exposed and protein A resides in the nucleus. Another possible answer is that protein B binds protein A and sequesters it by keeping protein A in some subcellular compartment, away from the nucleus. In the presence of hormone, protein B interacts with the hormone, changing its conformation so that it can Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 no longer bind to protein A. When protein B is not present, protein A can enter thenucleus in the presence or absence of hormone. 15-9 (d) 15-10 (c) Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 15.11 (a), (b), and (d) The rough ER consists of ER membranes and polyribosomes that are in the process of translating and translocating proteins into the ER membrane and lumen. Thus all proteins that end up in the lysosome, Golgi apparatus, or plasma membrane, or are secreted, will be encoded by the RNAs associated with the rough ER. Mitochondrial and ribosomal proteins are translated on free cytosolic ribosomes. 15.12 The amino-terminal signal sequence initiates translocation and the protein chain starts tothread through the translocation channel. When the stop-transfer sequence enters the translocation channel, the channel discharges both the signal sequence and the stop- transfer sequence sideways into the lipid bilayer. The signal sequence is then cleaved, sothat the protein remains held in the membrane by the hydrophobic stoptransfer sequence. 15.13 A. B. C. 15.14 The protein would enter the ER. The signal for a protein to enter the ER is recognized as the protein is being synthesized and will end up either in the ERor on the ER membrane. Proteins destined for the nucleus get recognized by cytosolic nuclear transport proteins once they are fully synthesized and fully folded. The protein would enter in the mitochondria. In order for a nuclear export signalto work, the protein would have to end up in the nucleus first and thus would need a nuclear import signal for the nuclear export signal to get utilized. The protein would enter the mitochondria. In order to be retained in the ER, theprotein needs to enter the ER. Since there is no signal for ER import, the ER retention signal would not function. The N-terminal signal sequence initiates translocation of the protein across the ER membrane. The signal sequence will be cleaved off by signal peptidase, leaving the amino-terminus of the protein in the luminal side of the ER membrane. Upon fusion tothe plasma membrane, the amino terminus of the protein will reside in the extracellularspace. Figure A15-14 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 15.15 A. Deleting the first signal sequence completely would convert the next membrane- spanning domain into an internal start-transfer signal and wouldinvert the orientation of the protein (see Figure A15-15A). B. Changing the hydrophobic amino acids to charged amino acids destroys the ability of the sequence both to act as a signal sequence and to become a membrane-spanning sequence. Therefore, the adjacent membrane spanning domain will now become an internal start-transfer sequence and the protein willbe inverted, as seen above in part A. The mutated signal sequence would not getcleaved off, since it would remain on the cytoplasmic side of the membrane andsignal peptidase is found only inside the ER (see Figure A1515B). C. Mutating every other membrane spanning region so that they are now charged (and thus cannot span the membrane) would decrease the number oftransmembrane regions and increase the size of the internal loops between membrane-spanning regions (see Figure 15-15C). Figure A15-15 15.16 As shown in Figure A15-16, elimination of the first transmembrane segment (by making it hydrophilic) would be expected to reverse the orientation of the protein in the membrane. What originally was the second transmembrane segment (a stoptransfer signal), would now be read as a start-transfer signal and would have the opposite orientation in the membrane—as would all the remaining transmembrane segments. Although the N-terminus would still be in the ER lumen, all the rest of the external parts of the protein would swap positions so that what was in the cytosol would now bein the ER lumen, and vice versa. Figure A15-16 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 15.17 Proteins are transported out of a cell via the secretory or exocytic pathway. Fluid andmacromolecules are transported into the cell via the endocytic pathway. All proteins being transported out of the cell pass through the endoplasmic reticulum and the Golgi apparatus. Transport vesicles link organelles of the endomembrane system. The formation of disulfide bonds in the endoplasmic reticulum stabilizes protein structure. 15.18 1. The proteins in the coat help shape the membrane into a bud. 2. The proteins in the coat can also select cargoes for transport. 15.19 Choice (b) is the correct answer. An individual vesicle may contain more than one type of protein in its lumen (choice (a)), all of which will contain the same sorting signal (or will lack specific sorting signals). Endocytic vesicles (choice (c)) generally move away from the plasma membrane. The vesicle membrane will not necessarily contain the samelipid and protein composition as the donor organelle, since the vesicle is formed from a selected subset of the organelle membrane from which it budded (choice (d)). 15.20 In order to get maximal levels of vacuolar vesicle fusion, vesicles from each strain must carry both v-SNAREs and t-SNARES. Experiment 1, which represents the normal scenario, is the only experiment where 100% alkaline phosphatase activity is measured. However, as long as complementary SNAREs are present on the vesicles, some vesicle fusion does occur (see experiments 3, 4, 6, 7, 8, 9). If both vesicles are missing either v- SNAREs (experiment 2) or t-SNAREs (experiment 5) or both SNAREs (experiment 10 and 11), the level of fusion is very low. It does not matter whether a tor v-SNARE is onthe vesicle of a particular strain, as long as the vesicle from the other strain contains a complementary SNARE (compare experiments 3 and 4, 6 and 7, and 8 and 9). 15-21 (d) 15.22 15.23 1. Proteins in the ER can undergo disulfide bond formation. (This does not occur in the cytosol because of its reducing environment.) 2. Proteins in the ER can undergo glycosylation. (Glycosylating enzymes are notfound in the cytosol.) (Signal-sequence cleavage is also an acceptable answer, although not really whatthis question is referring to.) The protein would end up in the extracellular space. Normally, the protein would go from the ER to the Golgi apparatus, get captured because of its ER-retrieval signal, andreturn to the ER. However, without the ER-retrieval signal, the protein would evade capture, ultimately leave the Golgi via the default pathway, and become secreted into the extracellular space. The protein would not be retained anywhere else along the secretory pathway, as it presumably has no signals to promote such localization since itnormally resides in the ER lumen. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 15-24 A—3; B—1; C—5; D—4; E—2 Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 15.25 A—3 (oligosaccharide protein transferase = ER) B—1 (galactose transferase = central Golgi cisternae) C —4 (SA transferase = trans Golgi network) D—2 (GlcNAc transferase = cis Golgi network Proteins are modified in a stepwise fashion in the Golgi apparatus, with early steps takingplace in the cis Golgi, intermediate steps taking place in the central Golgi cisternae, and late steps occurring in the trans Golgi network. If each enzyme produces the substrate for the next step, then a mutant lacking the enzyme that catalyzes the addition of the first sugar will be missing all of the sugars, a mutant lacking the enzyme that catalyzes the addition of the second sugar will contain the first sugar but will lack the other three, and so on. By this logic, mannose and GlcNAc must be the first sugars added, additional GlcNAc the second, galactose the third, and SA the last. Hence, the oligosaccharide protein transferase must be in the ER, the GlcNAc transferase in the cis Golgi, the galactose transferase in the central Golgi, and the SA transferase in the trans Golgi. 15.26 New plasma membrane reaches the plasma membrane by the constitutive exocytosis pathway. New plasma membrane proteins reach the plasma membrane bythe constitutive exocytosis pathway. Insulin is secreted from pancreatic cells by the regulated exocytosis pathway. The interior of the trans Golgi network is acidic. Proteins that are constitutively secreted do not aggregate in the trans Golgi network. 15.27 The three main classes of proteins that must be sorted before they leave the trans Golgi network in a cell capable of regulated secretion are (1) those destined for lysosomes, (2)those destined for secretory vesicles, and (3) those destined for immediate delivery to the cell surface. 15.28 1. 2. 3. 15.29 Eucaryotic cells are continually taking up materials from the extracellular space by the process of endocytosis. One type of endocytosis is pinocytosis, which involves utilizing clathrin proteins to form small vesicles containing fluids and molecules. After these vesicles pinch off from the plasma membrane, they will fuse with the endosome, where the uptaken materials are sorted. A second type of endocytosis is phagocytosis, which is used to take up large vesicles that can contain microorganisms and cellular debris. Macrophages are especially suited for this process, as they extend pseudopods (sheetlike recycled to the original membrane destroyed in the lysosome transcytosed across the cell to a different membrane. projections of their plasma membrane) to surround the invading microorganisms. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com) lOMoARcPSD|30659517 15.30 W—3 (defect in mannose-6-phosphate receptor) X—2 (defect in phosphotransferase) Y—1; Z—1 (defect in lysosomal hydrolases); these will be defects in two differentlysosomal acid hydrolases. A cell that has no mannose-6-phosphate receptor will be able to make all the lysosomal hydrolases properly, but will not be able to send them to the lysosome and will also not be able to scavenge hydrolases from the external media. Hence, this cell line cannot be rescued by culture media that has had lysosomal hydrolases secreted into it and thus willnot be rescued by any of the media tested here. A cell line that has no phosphotransferasewill be able to scavenge hydrolases from the external medium, but since all of the cell’s own hydrolases will lack the mannose-6-phosphate tag, it will be rescued only by media from a cell line that is able to make all of the hydrolases. Cell lines missing one hydrolase will be rescued by media from any cell line that is able to secrete that hydrolase in a mannose-6-phosphate tagged form; in addition, media from cultures of cells missing a hydrolase will rescue any cell line with another type of defect. 15.31 The lysosomal enzymes are all acid hydrolases, which have optimal activity at the low pH (about 5.0) found in the interior of lysosomes. If a lysosome were to break, the acid hydrolases would find themselves at pH 7.2, the pH of the cytosol, and would therefore do little damage to cellular constituents. 15.32 Strain A has protein accumulating in the ER, which means that this cell has a mutation that blocks transport from the ER to the Golgi apparatus. Strain B has secreted protein, and therefore is your wild-type control. Strain C has protein accumulating in the Golgi apparatus, and thus has a mutation that blocks exit of proteins from the Golgi apparatus.Strain D has protein accumulating in the cis-Golgi network, and thus has a mutationthat blocks the travel of proteins through the Golgi cisternae. Downloaded by S? Hi?p (nguyensyhoanghiep2003@gmail.com)