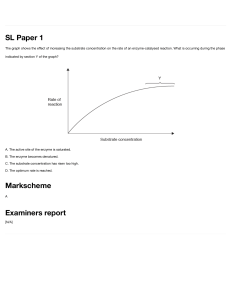
Enzymes: Characteristics, Classification, and Mechanisms
advertisement
ENZYMES ➢ Holoenzyme – biochemically active conjugated enzyme ➢ Coenzyme – a small organic molecule that acts as a cofactor in a conjugated enzyme ➢ Inorganic Ions – include Zn2+, Mg2+, Mn2+, and Fe2+. An important source of this is dietary minerals Greek words – “en” and “zyme” which means in and yeast, respectively GENERAL CHARACTERISTICS - - Most remarkable and highly specialized protein and has a high degree of specificity for their substances “central” to every biochemical process Acts as catalysts for biochemical reactions lowers the activation energy for a chemical reaction less energy is needed to convert reactant molecules to products Most are globular proteins Not all enzymes are proteins – some are ribonucleic acids (1980s) SUBSTRATE - is the substance upon which the enzyme “acts.” NOMENCLATURE - Named based on function, the catalyzed reaction, and substrate identity as focal points. 1) Suffix -ase identifies a substance as an enzyme. Suffixes -in are used to name digestive enzymes. 2) Prefix indicates the type of catalyzed reaction – (hydrolase (hydrolysis), and oxidase (oxidation)) 3) identity of the substrate is often noted in addition to the type of reaction (glucose oxidase, pyruvate carboxylase) General Structure Classes 1. Simple Enzymes – composed only of proteins 2. Conjugated Enzymes – has protein and nonprotein part ➢ Apoenzyme – protein part ➢ Cofactor – non-protein part. Generally, either a small organic molecule or an inorganic ion APOENZYME + COFACTOR = HOLOENZYME Why do apoenzymes need cofactors? Cofactors provide additional chemically reactive functional groups besides those present in amino acid side chains of apoenzymes. Classes Based on Type of Reactions 1. Oxireductase - catalyzes an oxidationreduction reaction (linked process) - requires a coenzyme that is oxidized or reduced as the substrate is reduced or oxidized. Example: Lactate dehydrogenase is an oxidoreductase that removes hydrogen atoms from a molecule. 2. Transferase - catalyzes the transfer of a functional group from one molecule to another Major subtypes: 1) Transaminase – catalyzes the transfer of an amino group from one molecule to another. 2) Kinases – play a major role in metabolic energy-production reactions that catalyze the transfer of a phosphate group from ATP to give ADP and phosphorylated product 3. Hydrolase - catalyzes a hydrolysis reaction (center to the process of digestion) in which the addition of a water molecule to a bond causes the bond to break. • Carbohydrase breaking of glycosidic bonds in oligo- and polysaccharides • Proteases – the breaking of peptide linkages in proteins • Lipases – the breaking of ester linkages in triacylglycerols 4. Lyase - catalyzes the addition of a group to a double bond or the removal of a group to form a double bond in a manner that does not involve hydrolysis or oxidation. • Dehydrase - removal of the components of water from a double bond • Hydratase - effects the addition of the components of water to a double bond. 5. Isomerase - catalyzes a substrate's isomerization (rearrangement of atoms) in a reaction, converting it into a molecule isomeric with itself. 6. Ligase - catalyzes the bonding together of two molecules into one with the participation of ATP. 2 Mechanisms: 1) Lock and Key Theory – the active site is rigid, thus providing a rigid, pre-shaped template fitting with the size and shape of the substrate molecule. 2) Induced Fit Theory or Koshland theory – the active site is flexible. The substrate during its binding induces conformational changes in the active site to attain the final catalytic shape and form. Models of Enzyme Action 2 Important Concepts: 1) Active site 2) Enzyme-Substrate Complex Enzyme Specificity – determined by the active site - Some accommodate only one compound while others can accommodate a family of closely related compounds. Absolute Specificity – enzyme will catalyze a particular reaction for only one substrate Active Site – a relatively small part of an enzyme’s structure that is involved in catalysis. - Usually the “crevice–like” location Enzyme-Substrate Complex – intermediate reaction species formed when a substrate binds to the active site - Most restrictive of all specificities, NOT COMMON. Example: Urease Stereochemical Specificity – enzyme can distinguish between isomers. Chirality is inherent at active site since amino acids are chiral. - Example: L-amino acid oxidase Group Specificity – involves structurally similar compounds with the same functional groups - Example: Carboxypeptidase Linkage Specificity – involves a particular type of bond irrespective of the structural features in the vicinity of the bond. - MOST GENERAL Example: Phosphatases Enzyme Activity - Measure of the rate at which an enzyme converts substrate to in a biochemical reaction. Factors Affecting Enzyme Activity 1. Temperature – reaction rate increases with temperature until the point at which the protein is denatured (above 50 degrees Celsius) and activity drops sharply. - Optimum temperature is around 37 degrees Celsius 2. pH – maximum enzymatic activity is possible only within a narrow pH range (5-9); outside this range, the protein is denatured - Optimum pH is 7.0-7.5 3. Concentration of Substrate – reaction rate increases with substrate concentration until full saturation occurs 4. Concentration of Enzyme – reaction rate increases with increasing enzyme concentration, assuming enzyme concentration is lower than that of substrate. Enzyme Inhibition ➢ Negative regulator – decreases enzyme activity (noncompetitive inhibitor) Regulation of Enzyme Activity - Minimize the process of energy-wasting Several “turn off” and “turn on” mechanisms to regulate Regulation Mechanism 1. Feedback Control – associated with allosteric enzymes - process in which activation/inhibition of the first reaction sequence is controlled by the product of the reaction sequence 2. Proteolytic Enzymes and Zymogens Proteolytic enzymes – catalyzes the breaking of peptide bonds that maintain the primary structure of protein. - Allosteric Enzymes – has two or more protein chains (quaternary structure) and two kinds of the active site (substrate & regulator) Regulator – molecules that regulatory sites ➢ Positive regulator – enzyme activity bind on increases Generated in an inactive form but converts back to active form if needed. Zymogens – inactive form of proteolytic enzyme (proenzyme) - Names of zymogens can be recognized by the suffix -ogen or the prefix pre- or pro-. Table 1.1 Main Classes and Subclasses of Enzymes Main Classes 3. Covalent Modification – a process where enzyme activity is altered by covalently modifying the structure of an enzyme thru attachment of a chemical group or removal of a chemical group from a particular amino acid within enzyme structure - Most encountered type are ➢ Phosphorylation – addition of phosphate group to an enzyme ➢ Dephosphorylation – removal of phosphate group Selected Subclasses Type of Reaction Catalyzed oxidases oxidation of a substrate reductases reduction of a substrate introduction of double bond Oxidoreductase (oxidation) s by formal dehydrogenase removal of s two H atoms from substrate, the H being accepted by a coenzyme transaminases transfer of an amino group between substrate kinases transfer of a phosphate group between substrates lipases hydrolysis of ester linkages in lipids proteases hydrolysis of amide linkages in proteins nucleases hydrolysis of sugar– phosphate ester bonds Transferases Hydrolases in nucleic acids carbohydrases hydrolysis of glycosidic bonds in carbohydrate s phosphatases hydrolysis of phosphate– ester bonds dehydratases removal of H2O from a substrate deaminases removal of NH3 from a substrate hydratases addition of H2O to a substrate decarboxylase s removal of CO2 from a substrate racemases conversion of D isomer to L isomer, or vice versa mutases transfer of a functional group from one position to another in the same molecule synthases formation of new bond between two substrates, with participation of ATP carboxylases formation of new bond between a Lyases Isomerases Ligases substrate and CO2, with participation of ATP