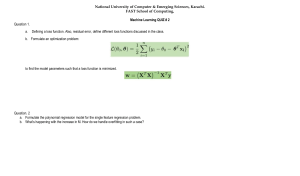
DELHI TECHNOLOGICAL UNIVERSITY ECONOMETRICS (HU-351) PROJECT REPORT Case Study on USD – INR conversion rates using linear and polynomial regression SUBMITTED TO: SUBMITTED BY: Ms. Reenu Ahluwaliya Siddhant Vijay (2K20/CH/065) DELHI TECHNOLOGICAL UNIVERSITY (Formerly Delhi College of Engineering) Bawana Road, Delhi-110042 CANDIDATE’S DECLARATION I, SIDDHANT VIJAY (Roll No. 2K20/CH/065), student of B.Tech. (CHEMICAL ENGINEERING), hereby declare that the project Dissertation titled “Modeling COVID-19 daily cases in Senegal using a generalized Waring regression model” which is submitted by me to the Department of CHEMICAL ENGINEERING, Delhi Technological University, Delhi in partial fulfilment of the requirement for the award of the 5TH semester of the Bachelor of Technology, is original and not copied from any source without proper citation. This work has not previously formed the basis for the award of any Degree, Diploma Associateship, Fellowship or other similar title or recognition. Place: Delhi Date: 19-11-2021 Siddhant Vijay DELHI TECHNOLOGICAL UNIVERSITY (Formerly Delhi College of Engineering) Bawana Road, Delhi-110042 CERTIFICATE I, hereby certify that the Project:Modeling COVID-19 daily cases in Senegal using a generalized Waring regression model, which is submitted by Siddhant Vijay, Roll No – 2K20/CH/065, CHEMICAL ENGINEERING, Delhi Technological University, Delhi in fulfillment of the requirement for the 5TH semester of Bachelor of Technology, is a record of the project work carried out by the students under my supervision. To the best of my knowledge this work has not been submitted in part or full for any Degree or Diploma to this University or elsewhere. Place: Delhi Date: 13-11-2021 Submitted To:Ms. Reenu Ahluwalia DEPARTMENT OF INFORMATION TECHNOLOGY DELHI TECHNOLOGICAL UNIVERSITY (Formerly Delhi College of Engineering) Bawana Road, Delhi-110042 ACKNOWLEDGEMENT I would like to convey my heartfelt thanks to Ms. Reenu Ahluwalia for her ingenious ideas, tremendous help and cooperation. I am extremely grateful to my friends who gave valuable suggestions and guidance for completion of my project. The cooperation and healthy criticism came handy and useful with them. INDEX ✔ Introduction 1. Regression 2. Linear Regression 3. Polynomial Regression 4. Independent and dependant variables 5. Assumptions 6. Hypothesis 7. Cost function 8. Gradient Descent 9. Mean Squared Error 10.R2 Score 11.Overfitting in Regression ✔ Problem Statement ✔ Approach ✔ Dataset ✔ Mechanics of Regression ✔ Performing Linear Regression ✔ Model Slope and Intercept ✔ Performing Polynomial Regression ✔ Graph for Polynomial Regression ✔ Predictions ✔ Regression metrices for model performance ✔ Checking Residual Analysis ✔ Graph for Residual Analysis ✔ Interpretation and conclusions ✔ References INTRODUCTION 1. Regression: Regression is a statistical method used in finance, investing, and other disciplines that attempts to determine the strength and character of the relationship between one dependent variable (usually denoted by Y) and a series of other variables (known as independent variables). Regression helps investment and financial managers to value assets and understand the relationships between variables, such as commodity prices and the stocks of businesses dealing in those commodities. 2. Linear Regression: Linear regression is a linear approach to modelling the relationship between the scalar response and one or more explanatory variables. The case of one explanatory variable is called simple linear regression. For more than one explanatory variable, the process is called multiple linear regression. ● Simple linear regression: Y = a + bX + u Multiple linear regression: Y = a + b 1X1 + b2X2 + b3X3 + … + btXt + ● u Where: ● Y = the variable that you are trying to predict (dependent variable). ● X = the variable that you are using to predict Y (independent variable). ● a = the intercept. ● b = the slope. ● u = the regression residual. 3. Polynomial Regression: o Polynomial Regression is a regression algorithm that models the relationship between a dependent(y) and independent variable(x) as nth degree polynomial. The Polynomial Regression equation is given below: y= b0+b1x1+ b2x12+ b x 3+ bnx1n o It is also called the special case of Multiple Linear Regression in ML. Because we add some polynomial terms to the Multiple Linear regression equation to convert it into Polynomial Regression. o It is a linear model with some modification in order to increase the accuracy. o The dataset used in Polynomial regression for training is of non-linear nature. o It makes use of a linear regression model to fit the complicated and non-linear functions and datasets. o In Polynomial regression, the original features are converted into Polynomial features of required degree (2, 3…, n) and then modeled using a linear model. Why Polynomial Regression? o In the above image, we have taken a dataset which is arranged non-linearly. So, if we try to cover it with a linear model, then we can clearly see that it hardly covers any data point. On the other hand, a curve is suitable to cover most of the data points, which is of the Polynomial model. o Hence, if the datasets are arranged in a non-linear fashion, then we should use the Polynomial Regression model instead of Simple Linear Regression. 4. Independent and dependant variables: i. Independent variable Independent variable is also called an Input variable and is denoted by X. In practical applications, an independent variable is also called a Feature variable or Predictor variable. We can denote it as: Independent or Input variable (X) = Feature variable = Predictor variable ii. Dependent Variable Dependent variable is also called an Output variable and is denoted by y. Dependent variable is also called a Target variable or Response variable. It can be denoted as follows: Dependent or Output variable (Y) = Target variable = Response variable 5. Assumptions: There are 5 key assumptions in Linear Regression Model: i. Linear Relationship between the features and target: According to this assumption there is linear relationship between the features and target. Linear regression captures only linear relationship ii. Little or no Multicollinearity between the features: Multicollinearity is a state of very high inter-correlations or inter associations among the independent variables. It is therefore a type of disturbance in the data if present weakens the statistical power of the regression model. The interpretation of a regression coefficient is that it represents the mean change in the target for each unit change in a feature when you hold all of the other features constant. However, when features are correlated, changes in one feature in turn shifts another feature. The stronger the correlation, the more difficult it is to change one feature without changing another. It becomes difficult for the model to estimate the relationship between each feature and the target independently because the features tend to change in unison. If we have 2 features which are highly correlated, we can drop one feature or combine the 2 features to form a new feature, which can further be used for prediction. iii. Homoscedasticity Assumption: Homoscedasticity describes a situation in which the error term (that is, the “noise” or random disturbance in the relationship between the features and the target) is the same across all values of the independent variables iv. Normal distribution of error terms: The fourth assumption is that the error(residuals) follows a normal distribution. v. Little or No autocorrelation in the residuals: Autocorrelation occurs when the residual errors are dependent on each other. The presence of correlation in error terms drastically reduces model’s accuracy. This usually occurs in time series models where the next instant is dependent on previous instant. 6. Hypothesis: 7. Cost Function: Where m is the number of instances present in the dataset. 8. Gradient Descent: Gradient descent is an iterative algorithm which we will run many times. On each iteration, we apply the following “update rule” (the: = symbol means replace theta with the value computed on the right): Alpha is a parameter called the learning rate. The learning rate gives the engineer some additional control over how large of steps we make. Selecting the right learning rate is critical. If the learning rate is too large, you can overstep the minimum and even diverge. In linear regression, the model targets to get the best-fit regression line to predict the value of y based on the given input value. While training the model, the model calculates the cost function which measures the Root Mean Squared error between the predicted value and true value. The model targets to minimize the cost function. Update Rules: This process can be represented in the plot of Cost functions V/S the independent variables as follows: Imagine that we plot our cost function on the z-axis based on Ө0 on the x axis and Ө1 on the y-axis The process is continued until we reach the very bottom of the curve i.e., when the value becomes minimum. The red arrows show the minimum points in the curve. The movement is done in the direction tangential to the curve with the help of derivative of our cost function. Mathematical derivation of this algorithm is as follows: Ө1: Finally, Derivatives: Let’s consider our hypothesis function: And the Cost function: Since we are monitoring the value of cost function at each step using gradient descent it is easy to see if convergence is achieved or not with the help of suitable value of the learning rate which controls how large the steps are. Here epoch represents the number of steps taken in gradient descent. The most commonly used rates are: 0.001, 0.003, 0.01, 0.03, 0.1, 0.3 • If α is very small, it would take long time to converge and become computationally expensive. • If α is large, it may fail to converge and overshoot the minimum. The above image shows the convergence of gradient descent with a learning rate of 0.01 and 5000 epochs. 9. MEAN SQUARED ERROR: In regression analysis, the plotting is a more natural way to view the overall trend of the whole data. The mean of the distance from each point to the predicted regression model can be calculated and shown as the mean squared error. The squaring is critical to reducing the complexity with negative signs. To minimize MSE, the model could be more accurate, which would mean the model is closer to actual data. One example of a linear regression using this method is the least- squares method—which evaluates the appropriateness of linear regression model to model bivariate dataset, but whose limitation is related to the known distribution of the data. 10. R2 Score: R2 Score is another metric to evaluate performance of a regression model. It is also called coefficient of determination. It gives us an idea of goodness of fit for the linear regression models. It indicates the percentage of variance that is explained by the model. Mathematically, R2 Score = Explained Variation/Total Variation In general, the higher the R2 Score value, the better the model fits the data. Usually, its value ranges from 0 to 1. So, we want its value to be as close to 1. Its value can become negative if our model is wrong. 11. OVERFITTING IN REGRESSION Overfitting is where the model is too complex for the data —it happens when the sample size is too small. If enough predictor variables are used in the regression model, we nearly always get a model that looks significant. While an overfitted model may fit the idiosyncrasies of the data extremely well, it won’t fit additional test samples or the overall population. The model’s p-values, R Squared and regression coefficients can all be misleading. How to Avoid Overfitting In linear modelling (including multiple regression), you should have at least 10-15 observations for each term you are trying to estimate. Any less than that, and you run the risk of overfitting your model. “Terms” include: ● Interaction Effects, ● Polynomial expressions (for modelling curved lines), ● Predictor variables. While this rule of thumb is generally accepted, Green (1991) takes this a step further and suggests that the minimum sample size for any regression should be 50, with an additional 8 observations per term. For example, if you have one interacting variable and three predictor variables, you’ll need around 45-60 items in your sample to avoid overfitting, or 50 + 3(8) = 74 items according to Green. How to Detect and Avoid Overfitting The easiest way to avoid overfitting is to increase your sample size by collecting more data. If you can’t do that, the second option is to reduce the number of predictors in your model —either by combining or eliminating them. Factor Analysis is one method you can use to identify related predictors that might be candidates for combining. Cross-Validation Use cross-validation to detect overfitting: this partitions your data generalizes your model and chooses the model which works best. One form of cross-validation is predicted R-squared. best statistical software will include this statistic, which is calculated by: ● Removing one observation at a time from your data, ● Estimating the regression equation for each iteration, ● Using the regression equation to predict the removed observation. Cross-validation isn’t a magic cure for small data sets though, and sometimes a clear model isn’t identified even with adequate sample size. Shrinkage & Resampling Shrinkage and resampling techniques (like this R-module) can help you to find out how well your model might fit a new sample. Automated Methods Automated stepwise regression shouldn’t be used as an overfitting solution for small data sets. PROBLEM STATEMENT COVID-19 is an acute and severe viral respiratory infection, with high fever, fatigue, cough and shortness of breath as the most common symptoms. The ongoing pandemic has affected more than 184 million people in the world, with severe public health and economic/financial crisis globally, and countries such as Senegal were constrained to the most restrictive preventive measures such as lock downs, closure or limited openings of shops and schools. As of July 9, 2021, the Word Health Organization’s (WHO) statistics reported that death rate of detected infectious individuals with COVID-19 was on average of 2.16 percent (0.1%–9.3%) depending on individuals’ age group and other factors such as health status as well as immunity system among the many factors . However, with the persistent issue of under-reporting, asymptomatic or untested cases, one would expect the actual case fatality rate to be (perhaps much) lower. In Senegal, COVID-19 third wave tends to have a higher peak than the first and second waves, with the number of deaths following the same pattern. A powerful tool used to mitigate the spread and potential impact of the pandemic is mathematical modeling to forecast the long term dynamical spread of the disease, and to help support, using data the effective allocation and mobilization of scarce public health resources. AIM OF THE REGRESSION ANALYSIS We aim to investigate the spread of COVID-19 by fitting a generalized Waring regression model (GWRM) to the number of new daily cases in Senegal, and make short time predictions. The application of this model to COVID-19 is to the best of our knowledge relatively new. Next, we compare the performance of this model with that of the Poisson regression model (PRM) and the negative binomial regression model (NBRM). An extension of the negative binomial distribution that allows three sources of variation to be distinguished is the univariate generalized Waring distribution (UGWD), while the GWRM is a regression model with a UGWD as its underlying distribution. One main advantage of the implementation of the GWRM over other traditional models such as the NBRM is that the GWRM distinguishes individual heterogeneity due to intrinsic (internal) factors, and those due to external factors (e.g., covariates that influence the variability of data but because they cannot be observed or measurable are not included in the model) DATA Our dataset consists of the historical daily new confirmed COVID-19 cases in Senegal. Cases during the first wave of the outbreak were collected from the Senegalese Ministry of Health website (https://www.sante.gouv.sn/), available at this github repository (https://github.com/senegalouvert/COVID-19). The timeline of the observations are from 2nd of March 2020 to the ends of 22nd of October 2020. The daily reported number of new cases and the cumulative number of reported cases are depicted in Fig. 2 while the ratio number of reported cases over number of screening tests is depicted in Fig. 3. We simulate the possibility of near real-time predictions during an actual outbreak to test the predictive power of each of the models by using 90% of the data for estimation, and the remaining 10% for evaluation. The other variables of the data used are the following: • the number of daily screening tests • the number of daily contact COVID-19 cases. A contact case is a COVID-19 case transmission in which the person has been infected by another person with whom he was in contact, whether it is a member of family, a friend, or a neighbor, etc . . . That is to say the person exactly knows the person who contaminated him/her and where he/she was infected. • the daily number of community COVID-19 cases. A community case is a COVID-19 case transmission in which the person does not know where he is infected. • the number of daily COVID-19 imported cases (from foreign countries or from outside the community). • the number of total recovered cases. • the number of total deaths due to COVID-19. • the number of total evacuated cases at foreign countries. • the number of daily critical cases. The descriptive statistics of all variables used in this paper are summarized in Table 1 L. Gning, C. Ndour and J.M. Tchuenche Physica A 597 (2022) 127245 where λxt −τ = exp x′t −τ β . It follows that: ( ) Var(Yt |Ft −τ ) = E(Yt |Ft −τ ) = λxt −τ = exp x′t −τ β . ( ) (2) The conditional mean is then equal to the conditional variance (equidispersion) for this model that constitutes a major shortcoming for its application on count data. 3.2. Negative binomial regression model (NBRM) In this model, we make the following distributional assumption for Yt given Ft −τ : Yt |Ft −τ , λxt −τ ∼ Poisson λxt −τ , ) ( (3) where λxt −τ is a gamma distributed variable: λxt −τ ∼ Gamma(θ, vxt −τ ). (4) Then, using straightforward calculations we obtain (see, for example, [14,19] or [20]): Yt |Ft −τ ∼ NegativeBinomial θ, pxt −τ , ( ) (5) where pxt −τ = 1/(1 + vxt −τ ). Its follows, for the NBRM, that the conditional mean is given by: µxt −τ = E(Yt |Ft −τ ) = E(λxt −τ ) = θvxt −τ . (6) Remark that this model is a Poisson–gamma mixture model and, for the gamma distribution, the parameter θ does not depend on the covariates contrary to vxt −τ . The model parametrized that way is referred as NegbinII model in the literature (see [19]). The variance of the model is now given by: Var(Yt |Ft −τ ) = E(λxt −τ ) + Var(λxt −τ ) = µxt −τ + 1 θ µ2xt −τ (7) The decomposition of this variance is interpreted as follow: the first term of the model variance represents the variability due to randomness inherent in any random phenomenon and the second to differences between the days between 2 March 2020 and 22 October 2020 in our practical application. For more details on the interpretation of the variance decomposition see [14] or [20]. 3.3. Generalized waring regression model (GWRM) The GWMR is an extension of NBRM. Contrary to the last model, it allows to distinguish the variability due to individual differences related to the covariates included in the model, to the one only due to individual differences not related to the model covariates [14,20]. Next, we present a brief background on the genesis of the GWRM and some properties of the model. In this model we assume that Yt given Ft −τ has a univariate generalized Waring distribution: Yt |Ft −τ ∼ UGWD θxt−τ , k, ρ . ( ) (8) This implies that the probability mass function of Yt |Ft −τ is given by [16]: f (y|x) = Γ (θx + ρ )Γ (k + ρ ) (θx )y (k)y 1 , y = 0, 1, 2, . . . , Γ (ρ )Γ (θx + k + ρ ) (θx + k + ρ )y y! (9) Γ (α+r) where θx , k, ρ > 0 and (α )r = Γ (α ) for α > 0 and Γ is the gamma function defined by Γ (x) = x > 0. Then, the conditional mean is given by: E(Yt |Ft −τ ) = µxt −τ = ∫∞ 0 ux−1 exp(−x) dx, for θxt −τ k , (ρ > 1). ρ−1 (10) In addition, linking the conditional mean with the explanatory variables as µxt −τ = exp x′t −τ β , we have: ( θxt −τ = µxt −τ (ρ − 1) . (11) k Furthermore, the variance of the GWRM is given by [14,20]: Var(Yt |Ft −τ ) = µxt −τ + k+1 ρ−2 k + ρ − 1 µxt −τ ) 2 µxt −τ + ρ−2 k , (ρ > 2). (12) The decomposition of this variance is interpreted as follow: • the first term referred as randomness represents the variability inherent to any random phenomenon • the second one called liability represents the variability due to individual differences related to the explanatory variables 4 L. Gning, C. Ndour and J.M. Tchuenche Physica A 597 (2022) 127245 • the third one called proneness is the variability due to individual differences not related to the explanatory variables included in the model Finally, note that the GWRM is a NBRM-beta mixture model and then a limiting case of the NBRM [14]. For further details of this regression model the reader can be refer to [14] or [20]. 3.4. Maximum likelihood estimation Each of the three models considered above, namely the PRM, the NBRM and the GWRM was fitted using the maximum likelihood estimation. That is by maximizing: ℓ1 (β) = ( T ∑ ) yt x′t −τ β − exp(x′t −τ β) − log(yt !) , (13) t =τ +1 ( T ∑ exp(x′t −τ β) ( ) ( )) exp(x′t −τ β) Γ (θ + y t ) − log 1 + + log , θ + exp(x′t −τ β) θ Γ (θ ) + yt ! t =τ +1 ( ) )) ( ( T ∑ (ρ − 1) exp x′t −τ β Γ (θxt−τ + ρ )Γ (ρ + k)Γ (θxt−τ + yt )Γ (k + yt ) , where θxt −τ = . ℓ3 (β, ρ, k) = log Γ (ρ )Γ (θxt−τ )Γ (k)Γ (θxt−τ + k + ρ + yt )yt ! k ℓ2 (β, θ ) = ( yt log ) (14) (15) t =τ +1 The numerical calculations of the maximum likelihood estimators of the model parameters are carried out with the open source software R [21]. Note that, because PRM and NBRM are special cases of the generalized linear model, we estimate their parameters using the Iteratively re-weighted least squares (IRLS) algorithm (for more details see [22]). The PRM case is implemented in the glm function included in the R package stats, while the NBRM case is implemented in the glm.nb included in the package MASS. Finally, we use the R package GWRM developed by [20] to fit the GWRM. The algorithm used in this package is based on non linear minimization, Nelder–Mead optimization and L-BFGS-B methods (see [20]). For adaptation and reproducibility, the source codes of all these packages are available on the Comprehensive R Archive Network (CRAN) repository (http://cran.r-project.org/web/packages/GWRM). 4. Application New daily COVID-19 cases is considered as a discrete count variable over-dispersion tendency. With comparison with the Poisson model, several causes might be responsible of this variability. First, there are external observable factors that could significantly influence the number of daily COVID-19 cases, such as – x1 : the number of daily contact COVID-19 cases. – x2 : the daily number of community COVID-19 cases. – x3 : the number of daily COVID-19 imported cases. Second, there are missing external covariates which would affect the response variable, and the GWRM could provide more information on the data through the quantification of the effect and sources of variability. Finally, the variable number of daily screening COVID-19 tests, has been included as an offset in the models. This means that the ratio number of new daily COVID-19 cases/number of daily screening COVID-19 tests is used as response variable for all models. Choice of τ . The 14-day COVID-19 incubation period from exposure to symptoms onset is denoted by the parameter τ — the median time is about 5 days [23]. In a machine learning point of view, τ can be considered as a hyper-parameter, then, in the following, we use a grid search process to automate the tuning of τ . In Fig. 4, we plot the Root Mean Square Error (RMSE) of the models as functions of τ , and it can be observed that τ = 3 is the optimal value. 4.1. Results and discussion We fit the models from Section 3 to Senegal data. The computed values of the log-likelihood function shown in Table 2 enables us to evaluate the accuracy of the model fit, the Akaike and the Bayesian information criteria. In addition, for each model, using a cross-validation procedure, we calculate in Table 2 the values of RMSE, the Mean Absolute Error (MAE), and the Mean Error (ME). Our results show that the best model out of the three is the Generalized Waring regression model (GWRM). In Table 3, we display the maximum likelihood parameter estimates for the GWRM (the best fitted model), their respective standard errors, and the associated partial Wald tests (statistics and p-values). The estimates of k and ρ are respectively 53.68050 and 12.85094, with standard errors of 46.012067 and 4.668843. Note that the GWRM includes only the daily number of COVID-19 contact cases as a significant covariate at 5% significance level. 5 4. Application New daily COVID-19 cases is considered as a discrete count variable over-dispersion tendency. With comparison with the Poisson model, several causes might be responsible of this variability. First, there are external observable factors that could significantly influence the number of daily COVID-19 cases, such as – x1: the number of daily contact COVID-19 cases. – x2: the daily number of community COVID-19 cases. – x3: the number of daily COVID-19 imported cases. Second, there are missing external covariates which would affect the response variable, and the GWRM could provide more information on the data through the quantification of the effect and sources of variability. Finally, the variable number of daily screening COVID-19 tests, has been included as an offset in the models. This means that the ratio number of new daily COVID-19 cases/number of daily screening COVID-19 tests is used as response variable for all models. Choice of τ . The 14-day COVID-19 incubation period from exposure to symptoms onset is denoted by the parameter τ — the median time is about 5 days [23]. In a machine learning point of view, τ can be considered as a hyper-parameter, then, in the following, we use a grid search process to automate the tuning of τ . In Fig. 4, we plot the Root Mean Square Error (RMSE) of the models as functions of τ , and it can be observed that τ = 3 is the optimal value. 4. Results and Discussion We fit the models from Section 3 to Senegal data. The computed values of the log-likelihood function shown in Table 2 enables us to evaluate the accuracy of the model fit, the Akaike and the Bayesian information criteria. In addition, for each model, using a cross-validation procedure, we calculate in Table 2 the values of RMSE, the Mean Absolute Error (MAE), and the Mean Error (ME). Our results show that the best model out of the three is the Generalized Waring regression model (GWRM). In Table 3, we display the maximum likelihood parameter estimates for the GWRM (the best fitted model), their respective standard errors, and the associated partial Wald tests (statistics and p-values). The estimates of k and ρ are respectively 53.68050 and 12.85094, with standard errors of 46.012067 and 4.668843. Note that the GWRM includes only the daily number of COVID-19 contact cases as a significant covariate at 5% significance level. Now, we focus on how well the GWRM fits the observed data. We investigate this issue by carrying out a visual residual analysis. Indeed, in Figs. 5, 6, and 7, we depict the quantile quantile plots (QQ-plots) with simulated envelope for residuals of type deviance, of type Pearson and of type response (for more details see [24,25]), [20]. For the deviance and Pearson residuals, all the plotted points fall within the boundaries of the envelope, which indicates a good accuracy of the fitted model. However, for the response residuals, there are some points outside the envelope, a sign of a potential lack of accuracy, and hence no evidence against the adequacy of the fitted model. Thus, the GWRM is appropriate to fit the data. We observe that, on average, the proneness represents 50.18 percent of the variability of the model while the liability represents 41.57 percent and the randomness 8.25. This means about 50% of the variation of the model are due to external factors that are unknown because they could be difficult to observe or to measure. Fig. 8 depicts the relationship between the proportion of each component of the variance and the daily new COVID-19 cases. It can be seen that, on average, the randomness and the liability decrease as the number of new daily COVID-19 cases increase, while the proneness increases on average. Fig. 9 shows the proportion of the variance related to randomness, liability and proneness, in terms of the number of screening tests. In general, the variability in the number of new daily COVID-19 cases due to randomness and liability decreases as the number of screening tests increases, whereas the variability due to proneness increases. So, we can deduce that increasing the number of screening tests emphasizes the role of the daily characteristics as a cause of differences between days in relation to the number of new daily COVID-19 cases, whereas randomness and liability are somewhat less relevant. The Figs. 10–12 illustrate the relationships between the model’s covariates and the variance related to randomness, liability and proneness. In general, the variability in the number of daily new COVID-19 infections due to randomness and liability decreases as the number of daily contact or the number of daily community cases increases, whereas the variability due to proneness increases. Thus, an increase of the daily number of cases emphasizes the role of the proneness, whereas randomness and liability become less relevant. In contrast, the variability in the number of new daily infections varies randomly as the number of daily imported cases increases (Fig. 12). However the fraction of proneness is globally more important than the other sources of variation. It is important to note that the imported cases represent 2.24% of the total COVID-19 cases in Senegal during the first stage outbreak with a peak at 27 on June 13, 2020, and, the GWRM does not include this variable as a significant covariate at 5% significance level. 5. Conclusion The rapid spread of the COVID-19 pandemic has triggered substantial economic and social disruptions worldwide. Several mathematical studies have been proposed to investigate and forecast long term dynamics of the disease. We use count regression models such as the generalized Waring regression model to forecast the daily confirmed new COVID-19 cases in Senegal. We fitted the generalized Waring model (GWRM) to the Senegal data (daily number of confirmed new COVID-19 infections). Results from this study reveal that the generalized Waring regression model fits better the data than most of the usual count data such as the negative binomial regression model (NBRM). Therefore the former model can explain specific characteristics of the studied phenomenon. We also analyze the variance decomposition for each day. An advantage of the GWRM is that it accounts for factors that are known to affect the number of daily COVID-19 cases. This study is not exhaustive, and future work can explore several other avenues such as modeling the daily mortality due to Covid-19 using a GWRM, investigating (1) the impact of testing capacity on the number of detected daily cases [26], (2) the effect of various prevention and therapeutic intervention measures on COVID-19 dynamics. One can also try to model the daily number of COVID-19 cases using Generalized Waring auto-regressive process in order to understand better the COVID-19 dynamics. A possible issue with daily confirmed cases data is the potential of weekly seasonality effects, few tests performed over weekends, hence less cases and oscillations in data. In our future study, we pan to investigate – if there are any such issues in the Senegal data and how to correct this for further improvements – whether the generalized Waring regression model could capture the disease multiple wave References . https://stackabuse.com/linear-regression-in-python-with-scikit-learn https://www.statisticssolutions.com/assumptions-of-linear-regression https://en.wikipedia.org/wiki/Coefficient_of_determination https://en.wikipedia.org/wiki/Polynomial_regression https://www.javatpoint.com/machine-learning-polynomial-regression : www.elsevier.com/locate/physa