Genetic Engineering of Anopheles gambiae Habilitation Thesis
advertisement
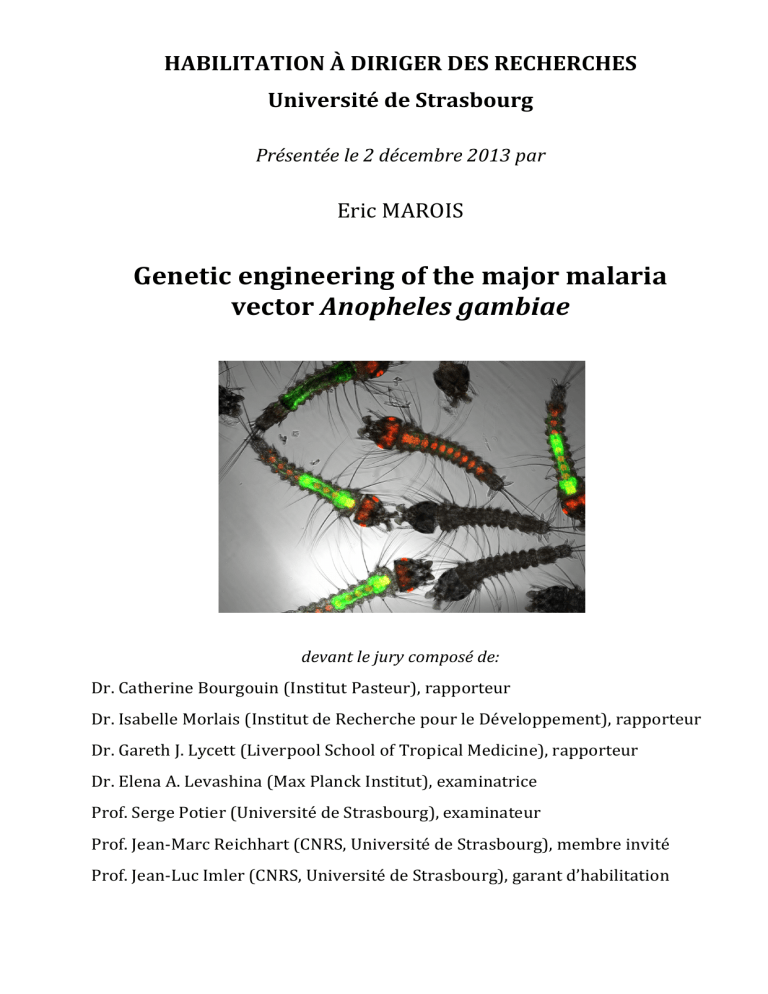
HABILITATION À DIRIGER DES RECHERCHES Université de Strasbourg Présentée le 2 décembre 2013 par Eric MAROIS Genetic engineering of the major malaria vector Anopheles gambiae devant le jury composé de: Dr. Catherine Bourgouin (Institut Pasteur), rapporteur Dr. Isabelle Morlais (Institut de Recherche pour le Développement), rapporteur Dr. Gareth J. Lycett (Liverpool School of Tropical Medicine), rapporteur Dr. Elena A. Levashina (Max Planck Institut), examinatrice Prof. Serge Potier (Université de Strasbourg), examinateur Prof. Jean-­‐Marc Reichhart (CNRS, Université de Strasbourg), membre invité Prof. Jean-­‐Luc Imler (CNRS, Université de Strasbourg), garant d’habilitation TABLE OF CONTENTS Remerciements . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 2 Curriculum vitae . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 3 I. Foreword . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 6 II. Overview of the Research Project . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 8 III. Past work . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 11 III.1. Ph.D studies: The tale of the TALEs . . . . . . . . . . . . . . . . . . . . . . . . . . 11 III.2. Post-­‐Doctoral studies: A good dive in Drosophila biology . . . . 17 III.3. From flies to mosquitoes . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 20 IV. Current Research and Projects . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 23 IV.1. Getting transgenesis to work in A. gambiae. . . . . . . . . . . . . . . . . . . 23 IV.2. Mosquito genome engineering —and what for? . . . . . . . . . . . . . . 27 IV.3. Conclusions and perspectives . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 31 References . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 33 ANR project . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 35 Article 1 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 59 Article 2 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 71 Article 3 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 83 Article 4 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 93 Article 5 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 105 Article 6 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 121 Article 7 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 127 Article 8 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 137 1 Remerciements L’écriture de cette synthèse de mon activité scientifique m’a fait revivre des instants de nostalgie pour des lieux, et surtout pour des personnes, grâce à qui les travaux présentés ici se sont déroulés dans des conditions exaltantes. Grâce à Jacques Tempé, un de mes professeurs de Phytopathologie à l’Institut National Agronomique Paris-­‐Grignon, j’ai pu accéder à des laboratoires de carrure internationale. Naturellement, rien n’eût été possible sans les directeurs de laboratoires qui m’ont successivement accueilli : Noel Keen, Jeff Dangl, Ulla Bonas, Suzanne Eaton et Elena Levashina, dont les personnalités m’ont impressionné et beaucoup inspiré. Mes mentors directs préférés, qui chaque jour m’ont fait sacrifier dans la joie de longues heures sur l’autel du progrès de la connaissance, furent Susanne Kjemtrup et Guido van den Ackerveken. Le sourire que je ne peux m’empêcher d’esquisser en pensant à eux est le signe de l’amitié que leurs personnalités attachantes ont su m’inspirer. De la même manière, dans les constellations de collègues côtoyés au cours de ces années, Carol Boyd, David Slaymaker, Boris Szurek, Laurent Noël, Ombeline Rossier, Elisabeth Huguet, Michèle Pierre, Kai Wengelnik, Patricia Monteiro, Jean-­‐Benoît Morel, Ralf Koebnik, Jens Boch, Daniela Büttner, Carola Kretschmer, Angelika Landgraf, Ali Mahmoud… sont des étoiles qui illuminent mes souvenirs. Le rayonnement d’autres étoiles, amis, colocataires… côtoyés à l’extérieur du laboratoire, a profondément enrichi mon parcours: Yvonne Monsauret, Julia Bryan, Clay Hensley, Shannon Hogan et les membres de la Table Française de Chapel Hill ; Magali Solé, Katja Sommer, Anne-­‐Gaëlle Bébin, Olivier Renaud, Soazig Le Lay… Ma famille compte aussi beaucoup dans mon équilibre, à commencer par Sophie et nos quatre enfants Viki, Johann, Sylvine et Naëlle, et mes parents Robert et Liliane qui soignent les terres de mes racines dans la Nièvre et dans l’Orne pour ma tranquillité d’esprit, ainsi que mes beaux-­‐parents Alain et Danièle dont l’implication auprès des enfants a un impact positif évident sur ma carrière ! Mes projets scientifiques actuels ne seraient rien sans le soutien de tous les membres plus ou moins actuels du laboratoire « Réponse Immunitaire et Développement chez les Insectes », et particulièrement Elena Levashina, Stéphanie Blandin, Jean-­‐Marc Reichhart, Jean-­‐Luc Imler (que je remercie également d’avoir bien voulu être mon garant d’HDR), Jules Hoffmann, Dominique Ferrandon, Hidehiro Fukuyama, Christine Kappler, Julien Soichot, Olivier Terenzi, Gloria Volohonsky, Andrea Smidler, Ferdinand Nanfack et Nathalie Schallon. Que toutes les personnes citées ci-­‐dessus, ainsi que les autres membres des laboratoires par lesquels je suis passé, trouvent ici l’expression de ma profonde reconnaissance pour leur aide, leur travail et leur influence sur mon parcours. 2 Curriculum vitae CURRICULUM VITAE Date of birth: May 18, 1973. I am married and have 4 children. EDUCATION 1995-­‐1996: Institut National Agronomique de Paris-­‐Grignon (INA-­‐PG), France. Graduation from the general course, Diplôme d’Agronomie Approfondie Graduation from the specialized course (Plant Pathology), Diplôme d’Etudes Approfondies (DEA) 1993-­‐1995: First and second year at INA-­‐PG. Diplôme d’Agronomie Générale 1991-­‐1993: Mathématiques Supérieures et Spéciales –Biologie 1991: Graduation from high school, Baccalauréat D exam (grade: very good). INTERNSHIPS AND EXPERIENCE Since October 2007 : INSERM staff researcher in the Anopheles laboratory of CNRS UPR9022, IBMC Strasbourg: mosquito transgenesis and molecular mechanisms of the Plasmodium/Anopheles interaction. ANR project leader for the period Nov. 2011-­‐Oct. 2014. April 2006-­‐ September 2007: Post-­‐doctoral studies as a fellow of the Fondation Recherche Médicale (FRM) in the group of Elena Levashina (Réponse Immunitaire et Développement chez les Insectes , CNRS, Institut de Biologie Moléculaire et Cellulaire, Strasbourg): Role of nutrient transport proteins in the Plasmodium/Anopheles interaction. June 2002-­‐February 2006 : Post-­‐doctoral studies as an Alexander-­‐von-­‐Humboldt fellow in Dr. Suzanne Eaton’s laboratory, Max-­‐Planck Institute of Cell Biology and Genetics, Dresden, Germany : RNAi and dominant negative screen to identify factors involved in the establishment of morphogen gradients in Drosophila. 12 April 2002 : Ph.D. defense (European Ph.D.) at INA-­‐PG, Paris : Characterization of pepper genes induced by the Xanthomonas type III effector AvrBs3. Grade : très honorable avec félicitations du jury. June 1998-­‐May 2002: Ph.D. studies, Dr. Ulla Bonas’ laboratory, Martin-­‐Luther Universität, Halle, Germany: cDNA-­‐AFLP screen for pepper genes induced in response to the Xanthomonas type III effector AvrBs3. October 1996-­‐May 1998: Alternative National Service performed as a researcher, laboratory of Dr. Jeff Dangl, Biology Department, University of North Carolina at Chapel Hill, USA: Study of the function of the type III effectors avrRpm1 and avrB of Pseudomonas 3 Curriculum vitae syringae. March-­‐August 1996: Diploma (DEA) work, Dr. Ulla Bonas’ laboratory, Institut des Sciences Végétales, CNRS Gif-­‐sur-­‐Yvette, France: Recognition of the avirulence protein AvrBs3 of Xanthomonas campestris pv. vesicatoria occurs inside pepper cells carrying the resistance gene Bs3. April-­‐September 1995: Student internship in Dr. Noel Keen’s laboratory, Department of Plant Pathology, University of California, Riverside, USA: Isolation of the primary signal for systemic acquired resistance in cucumber. HPLC purification of phloem compounds. March 1995: research internship, Dr. Ulla Bonas' laboratory, Institut des Sciences Végétales, CNRS Gif-­‐sur-­‐Yvette, France: contribution to the map-­‐based cloning of the Bs3 resistance gene from pepper. Summer 1994: Summer student in Jean-­‐Pierre Boutin’s laboratory, Laboratoire du Métabolisme, INRA of Versailles, France: Role of sucrose synthase in the sink strength of the pea seed. LANGUAGES French, English, German and Spanish: fluent (oral and written). Italian, Chinese: bases. PUBLICATIONS Smidler AL, Terenzi O, Soichot J, Levashina EA, Marois E. (2013) Targeted Mutagenesis in the Malaria Mosquito Using TALE Nucleases. PLoS One 15;8(8):e74511. Marois E, Scali C, Soichot J, Kappler C, Levashina EA, Catteruccia F. (2012) High-­‐throughput sorting of mosquito larvae for laboratory studies and for future vector control interventions. Malar J. 11:302 Marois E. (2011) The multifaceted mosquito anti-­‐Plasmodium response. Current Opinion in Microbiology 14:429–435 Rono M, Whitten M, Oulad-­‐Abdelghani M, Levashina E, Marois, E. (2010) The Major Yolk Protein Vitellogenin Interferes with the Anti-­‐Plasmodium Response in the Malaria Mosquito Anopheles gambiae. PLoS Biology, 8(7):e1000434. Kay S., Hahn S., Marois E., Wieduwild R, Bonas U. (2009) Detailed analysis of the DNA recognition motifs of the Xanthomonas type III effectors AvrBs3 and AvrBs3ΔRep16. Plant Journal 59:859-­‐871 Blandin S.A., Marois E., Levashina E.A. (2008) Antimalarial responses in Anopheles 4 Curriculum vitae gambiae: from a complement-­‐like protein to a complement-­‐like pathway. Cell Host Microbe 3(6):364-­‐74 Kay S., Hahn S., Marois E., Hause G., Bonas U. (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 26:648-­‐651 Marois E., Eaton S. (2007) pFRiPE, a vector for temporally and spatially controlled RNAi in Drosophila. In Methods in Molecular Biology, Humana Press, J. Horabin ed. Marois E., Mahmoud A., Eaton S. (2006) The endocytic pathway and formation of the Wingless morphogen gradient. Development 133(2):307-17 Classen AK, Anderson KI, Marois E, Eaton S. (2005) Hexagonal packing of Drosophila wing epithelial cells by the planar cell polarity pathway. Developmental Cell 9(6):805-­‐17 Panáková D., Sprong H., Marois E., Thiele C., Eaton S. (2005) Lipoprotein particles carry lipid-­‐linked proteins and are required for long-­‐range Hedgehog and Wingless signalling. Nature 43 :558-­‐65 Marois E., Van den Ackerveken G., Bonas U. (2002) The Xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Mol. Plant Microbe Interact. 15:637-­‐46 Szurek B., Marois E., Bonas U., Van den Ackerveken G. (2001) Eukaryotic features of the Xanthomonas type III effector AvrBs3: protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. Plant J. 5:523-­‐34. Marois E., Nimchuk Z., Kjemtrup S., Leister R.T., Katagiri F., Dangl J.L. (2000) Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell, 101(4):353-­‐63. Bonas U., Van den Ackerveken G., Büttner D., Hahn K., Marois E., Nennstiel D., Noël L., Rossier O., Szurek B. (2000) How the bacterial plant pathogen Xanthomonas campestris pv. vesicatoria conquers the host. Mol. Plant Microbiol 1(1):73-­‐76 Van den Ackerveken G., Marois E., Bonas U. (1996) Recognition of the Bacterial Avirulence protein AvrBs3 Occurs inside the Host Plant Cell. Cell, 87 1307-­‐1316. Smith-­‐Becker J., Marois E., Huguet E. J., Midland S.L., Sims J. J., Keen N.T. (1998) Accumulation of salicylic acid and 4-­‐hydroxybenzoic acid in phloem fluids of cucumber during systemic acquired resistance is preceded by a transient increase in phenylalanine ammonia-­‐lyase activity in petioles and stems. Plant Physiol. 116(1):231-­‐8. from extracurricular activities: Marois, E. (1990) Hybridation réussie entre Eudia pavonia and Saturnia pyri. Imago #45 Marois, E. (1993). Les Lépidoptères diurnes de la friche de La Beue. Nature-­‐Nièvre #1 5 Foreword I. FOREWORD My interest in Biological Sciences arose early: as a small child, I was passionately collecting butterflies and moths and rearing their caterpillars from egg to adulthood. When I was five, I was hit by a car that broke my left leg. The cast prevented me from roaming around our house to search for caterpillars. Upon some begging on my side, my mother would then take me in the wheelbarrow to tour the neighboring gardens, so that I could spot some Swallowtail (Papilio machaon) caterpillars in the carrot fields and collect them for my rearings. The shapes, color patterns and behavior of caterpillars and adult Lepidoptera were an endless source of fascination to me. Some of the most exciting experiments I did around the age of 12 were to hold a female Emperor moth (Saturnia pavonia), or Oak Eggar (Lasiocampa quercus) captive, catch the many males flying towards us as they were attracted by the female’s pheromones, and sample their coloration and pattern diversity. As a teenager, I began to be interested in the genetic basis of a moth’s patterns and became fascinated by the possibility of hybridizing different species to see what characteristics would be inherited from each parent. Using an Emperor moth female to attract males, I managed to cross Emperor moth males to a Great Peacock (Saturnia pyri) female newly hatched from my rearings. The rearing of the F1 hybrids to adulthood (picture below) was one of the most exciting science experiments I carried out, and its description the subject of my very first scientific paper (at age 17) in the –now defunct-­‐ French journal Imago. This publishing experience was also my first conflict with a journal’s editor, as the Imago editor altered a sentence in my paper without submitting the change to my validation. This led to the wrong statement that I had injected ecdysone into female pupae to make them eclose. In reality, the eclosion of female pupae was merely delayed compared to that of the males (in interspecific Lepidoptera hybrids, an ecdysone deficiency sometimes causes failed eclosion in females, which can be circumvented by hormone injection). I demanded a corrigendum in the journal, but my requests were ignored, which led me to discontinue my subscription. In parallel to my caterpillar cultures, I particularly enjoyed building inventories of the butterfly and moth species present in given habitats around my hometown, and thus became aware of the value of the insect and plant communities to gauge the ecological health of a given area. This activity led me to publish a Lepidoptera inventory in the bulletin of the local nature protection association. In 1991, when I left my parents’ home in the countryside near Nevers to study in the large and distant city of Lyon, my collection of Lepidoptera had reached about 2000 specimens; I gradually discontinued collecting and donated this collection in 2007 to the Museum “Jardin des Sciences” in Dijon. In the course of my studies, I always took great pleasure in Biology classes, which for me were a hobby rather than studying. At the Institut National d’Agronomie, from which I graduated with a diploma in Agricultural Sciences, I finally became exposed to molecular biology. The loading of DNA samples onto agarose gels was so abstract and distant from my previous contacts with Genetics that I did not immediately recognize that this was to become my daily bread. With time however, I developed a particular euphoria for molecular cloning, and to this day I enjoy looking for the most elegant solution to assemble bits and pieces of DNA into the desired plasmid. Throughout my subsequent career progress, I 6 Foreword retained this taste for molecular engineering and kept devoting time to set up elegant tools and protocols. However, I also felt that the best reward for good engineering was not the established tool or method itself, but the answering to an exciting scientific question through the use of that method. A good balance between molecular engineering work and the advancement of knowledge is still what I am looking for today. With the additional view that some of these tools and knowledge may one day serve humans in the fight against one of their oldest scourges, malaria… Saturnia pavonia male and female, 10 S. pyri specimens and 20 hybrids Another frame from my butterfly collection, just as a tribute to the beauty of the world of Insects. 7 Overview II. OVERVIEW OF THE RESEARCH PROJECT Genetic engineering of the major malaria vector Anopheles gambiae Malaria today remains one of the most deadly human diseases with AIDS and tuberculosis. 225 million people are infected every year in 108 countries, half the human population is at risk, the recorded annual death toll is 600 to 800 000, the majority of victims being infants in sub-­‐Saharan Africa (WHO, malaria report 2010 and 2011). The true annual number of deaths may approach 1.5 million (Murray et al., 2012). Global warming is expected to promote the spread of malaria, and other mosquito-­‐borne illnesses, from tropical regions to higher latitudes. Five different species of Plasmodium are causal agents of malaria, the most deadly being P. falciparum. About 20 species of mosquitoes in the genus Anopheles are known vectors of Plasmodium, transmitting the parasite during their blood meals on humans. In sub-­‐Saharan Africa, Anopheles gambiae is the major malaria vector. Within the Institut de Biologie Moléculaire et Cellulaire (IBMC) in Strasbourg, CNRS unit UPR9022 is investigating insect models of innate imunity. Hosted by this unit, our INSERM unit U963, created under the leadership of Elena Levashina and currently headed by Stéphanie Blandin, is studying Plasmodium transmission by A. gambiae, more specifically the immune response that mosquitoes mount against the parasite. A better understanding of mosquito resistance to the parasite is anticipated to inspire novel control strategies against the disease. Malaria mosquitoes are routinely reared in many laboratories, including ours at the IBMC Strasbourg, to study the biology of the interaction between Plasmodium parasites and their mosquito vectors. Anopheles gambiae mosquitoes are easy to rear and have a short generation time (about 2 weeks from egg to egg), making them amenable to genetic studies and manipulation. I joined the laboratory in 2006 after post-­‐doctoral studies on Drosophila development, and started exploring the interference relationships between immunity and blood meal-­‐induced reproductive proteins in A. gambiae. We discovered that Lipophorin and Vitellogenin, two proteins required to transport nutrients from the blood meal to developing eggs, reduce the efficiency of the mosquito anti-­‐parasitic response. In addition, boosting mosquito antiparasitic immunity by artificially activating the NF-­‐κB pathway reduced mosquito reproduction. We could show that this trade-­‐off between immunity and reproduction is, at least in part, mediated by a reduction in vitellogenin expression (Article 5 presented in this work). In the course of these studies, I became aware of the paucity and poor efficiency of tools for the genetic manipulation of mosquitoes, especially compared to its Dipteran cousin Drosophila. Routine and efficient transgenesis in Drosophila has played a major role in reaching today’s high degree of understanding of the fruit fly’s biology. In contrast, few Genetics tools have been developed for mosquitoes. Transgenesis in many mosquito species, especially the major malaria vector A. gambiae, has proved to be technically much more challenging than in the fruit fly. Today, only a handful of laboratories are able to transform A. gambiae. Transgenic lines are obtained at a low rate; probably only a few dozens transgenic A. gambiae lines have been obtained worldwide to date and fewer than 15 have been published, compared to tens of thousands in Drosophila. Still, the ability 8 Overview to routinely insert synthetic transgenes or mutant versions of mosquito genes by transgenesis would greatly facilitate the further unraveling of mosquito biological processes, especially those involved in the resistance/susceptibility to Plasmodium parasites. From this realization, I decided to focus on the development of tools for mosquito transgenesis. After six years of continued optimization and innovation in the procedure, A. gambiae transgenesis is now routine in our laboratory. The tools we developed place us in a unique position in the community of mosquito research to further develop ambitious Genetics strategies for the engineering of the mosquito genome. In the frame of a young researcher ANR grant that I obtained starting in November 2011, a post-­‐doctoral fellow, two master students, a technician and I have successfully established targeted gene knockout in A. gambiae (Article 5); and are working on the design of synthetic transcription factors to manipulate gene expression in transgenic A. gambiae. Besides using genetically modified mosquitoes for basic studies, a potential future application of mosquito gene engineering technologies is vector control. Historically, the destruction of mosquito vector populations and/or their habitat, and the protection of humans against vector bites with insecticide-­‐impregnated bednets have been the most effective means to combat malaria transmission. DDT and other insecticides have played, and are still playing, an important role in reducing malaria, with unquantified adverse effects on human health and the environment. Over time, mosquito populations have shown a great ability to develop resistances against insecticides, rendering these ineffective. Genetically modified mosquitoes are considered as a serious alternative to achieve the same purpose with a more specific action, being directed only against the correct mosquito vector species, without collateral damage on human health and the ecosystem. Fighting malaria with transgenic mosquitoes remains a controversial topic, but field releases of transgenic Aedes aegypti (a major dengue fever vector mosquito) have already been conducted in Malaysia and the Cayman islands by British company Oxitec (Harris et al., 2012; Lacroix et al., 2012). I am interested in examining the possibilities that transgenic technologies could offer in the fight against malaria. One promising strategy is to harness the power of the vector’s immune system to more efficiently combat the parasite. Mosquitoes could be made refractory to Plasmodium in the laboratory and released in the field, under the provision that no adverse long-­‐term effects of the transgenes would be identified in thorough laboratory and semi-­‐field experiments. Similar strategies avoiding the release of transgenic mosquitoes can be designed. Under the leadership of Elena Levashina and Stéphanie Blandin, our laboratory has been (and is) actively working on identifying natural mosquito antiparasitic genes. In parallel, we established protocols that exploit sexual chromosome-­‐linked transgenes for sorting non-­‐transgenic mosquito populations by flow cytometry. Our results (Article 7) show that we can sort large all-­‐male or all-­‐female non-­‐transgenic populations. If natural malaria refractoriness genes could be identified and fixed in laboratory mosquito populations, we could propose to generate all-­‐ male, non-­transgenic populations carrying these alleles, and release them in the field to increase the frequency of refractory alleles in nature (genetic control). This may be regarded as population gene therapy to cure mosquitoes of malaria. 9 Overview Current trial releases of transgenic mosquitoes to fight dengue fever were conducted without much supervision from public research. In the future, the public might demand that independent research examines both the potential benefits and potential associated short and long-­‐term risks of biotech approaches in the fight against infectious diseases. Our ambition is double: (i) provide the means to perform complete functional analysis of any gene of interest (gene elimination by knock-­‐out, allelic substitutions by gene exchange, transgenic gene silencing, tissue and time-­‐specific over-­‐expression…) to identify candidate genes that could be used in biotechnological approaches to fight malaria. (ii) develop tools and assays for the analysis of genetically modified insects to help determine their utility and potential risks in field applications. Transgenic mosquito larvae expressing Yellow Fluorescent Protein in their nervous system. 10 Past Work III. PAST WORK III.1. Ph.D studies: The tale of the TALEs In 1996 I obtained my Diplôme d’Etudes Approfondies (DEA) from the Institut National Agronomique Paris-­‐Grignon, with a focus on Phytopathology. I was extremely lucky to perform the associated six-­‐month lab training in the lab of Prof. Ulla Bonas at the Institut des Sciences Végétales of CNRS in Gif-­‐sur-­‐Yvette. Ulla’s laboratory was investigating the interaction of Xanthomonas campestris phytopathogenic bacteria with their host plants, pepper and tomato. Some plants can resist an infection with certain bacterial strains, and respond to the bacterial presence with a rapid form of programmed cell death, termed the hypersensitive response (HR). The HR is a cell suicide accompanied by the production of toxic compounds (such as nitric oxide and reactive oxygen species) that circumvent bacterial growth. This response depends on the presence of a plant resistance gene, encoding a component of plant innate immunity, which specifically recognizes the presence of a cognate bacterial protein that betrays the bacterial invasion. These bacterial genes, preventing bacterial success in resistant plants, are thus termed avirulence genes. This gene-­‐for-­‐gene interaction was then the topic of intense study in plant biology, which gradually led to a better understanding of plant innate immunity. Many pairs of plant resistance/bacterial avirulence genes were being studied across the world. In most cases, it was clear that avr activity depended on the integrity of the bacterial type III secretion system, which was just being characterized as a protein injection needle in animal pathogenic bacteria such as Yersinia, Shigella or E. coli. The possibility that Avr proteins might be injected by this apparatus into host plant cells became our working hypothesis. My project was to investigate whether Xanthomonas was injecting our favorite avirulence protein, AvrBs3, into the cells of its host pepper plants. To see if AvrBs3 activity could be observed if the protein was expressed directly in plant cells, I was offered the task of designing a transient expression protocol for plant leaves. One of the options I explored was the use of Agrobacterium tumefaciens bacteria as a transfection agent to deliver the DNA encoding the avirulence protein into the cells of a leaf infiltrated with an Agrobacterium suspension. Genes of interest to be expressed in plant cells are cloned within the T-­‐DNA borders (derived from the Agrobacterium Ti plasmid) in a shuttle plasmid. After transferring this plasmid from E. coli to Agrobacterium (by conjugation or electroporation), an Agrobacterium culture induced in a specific medium is syringe-­‐infiltrated into the leaf mesophyll. T-­‐DNA transfer from Agrobacterium to plant cells occurs and the synthetic genes are expressed transiently in the plant. This procedure allowed us to show that the Xanthomonas AvrBs3 was indeed recognized by its cognate plant resistance gene Bs3 directly within plant cells, rather than indirectly by a hypothetical product of its activity within the bacteria. This was consistent with the fact that avirulence depended on type III secretion. We published these results in Cell in 1996 (abstract below): 11 Past Work Cell. 1996 Dec 27;87(7):1307-16. Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Van den Ackerveken G, Marois E, Bonas U. Institut des Sciences Végétales, CNRS, Gif-sur-Yvette, France. Abstract The molecular mechanism by which bacterial avirulence genes mediate recognition by resistant host plants has been enigmatic for more than a decade. In this paper we provide evidence that the Xanthomonas campestris pv. vesicatoria avirulence protein AvrBs3 is recognized inside the plant cell. Transient expression of avrBs3 in pepper leaves, using Agrobacterium tumefaciens for gene delivery, results in hypersensitive cell death, specifically on plants carrying the resistance gene Bs3. In addition, for its intracellular recognition, AvrBs3 requires nuclear localization signals that are present in the C-terminal region of the protein. We propose that AvrBs3 is translocated into plant cells via the Xanthomonas Hrp type III secretion system and that nuclear factors are involved in AvrBs3 perception. The concept of Avirulence proteins injected by pathogenic bacteria into the plant cell, until then a hypothesis, was becoming a new paradigm. Rapidly, the “agroinfiltration protocol” became widely adopted, first to show that the concept of direct recognition of type III effectors within plant cells by plant resistance genes was valid for many other avirulence/resistance gene pairs, and later to study systemically-­‐spreading gene silencing by RNAi upon Agrobacterium-­‐mediated delivery of hairpin constructs in tobacco leaves (Voinnet and Baulcombe, 1997). After my diploma, I had the great opportunity to perform my “National Service” as a scientist in the laboratory of Jeff Dangl at the University of Chapel Hill in North Carolina. I applied the same approach to show that the Pseudomonas avirulence proteins AvrB and AvrRpm1 were also recognized by their cognate Arabidopsis resistance gene RMP1 after their transfer into the plant cell. In this case, my supervisor Suzanne Kjemtrup and myself were puzzled to discover a common amino acid motif at the N-­‐terminal end of many avirulence genes from Pseudomonas syringae, a motif we called the “MGCVSS”. We were very excited when a blast search performed using this motif as a query yielded a large number of animal proteins that all had N-­‐terminal myristoylation and palmitoylation in common. Then, the agroinfiltration protocol and other experiments (including labeling proteins with tritium-­‐labeled myristic acid) served to demonstrate that plant cells also performed this fatty acylation on avirulence proteins injected by the bacteria, and that this modification was required for membrane localization and full activity of these proteins inside the plant cell. I shared co-­‐first authorship with Zachary Nimchuk, the Ph.D. student who continued this project after I left, when we published these results in Cell in 2000 (Article 1, and abstract below): Cell. 2000 May 12;101(4):353-63. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Nimchuk Z, Marois E, Kjemtrup S, Leister RT, Katagiri F, Dangl JL. Department of Biology, University of North Carolina at Chapel Hill, 27599, USA. Abstract 12 Past Work Bacterial pathogens of plants and animals utilize conserved type III delivery systems to traffic effector proteins into host cells. Plant innate immune systems evolved disease resistance (R) genes to recognize some type III effectors, termed avirulence (Avr) proteins. On disease-susceptible (r) plants, Avr proteins can contribute to pathogen virulence. We demonstrate that several type III effectors from Pseudomonas syringae are targeted to the host plasma membrane and that efficient membrane association enhances function. Efficient localization of three Avr proteins requires consensus myristoylation sites, and Avr proteins can be myristoylated inside the host cell. These prokaryotic type III effectors thus utilize a eukaryote-specific posttranslational modification to access the subcellular compartment where they function. While I was performing this work in North Carolina, my Paris-­‐based DEA lab of Ulla Bonas was moving to the University of Halle (Germany), where I decided to do my Ph.D. (after considering staying in Jeff Dangl’s laboratory, but the prospects of working again with my favorite gene avrBs3 while discovering a new country/culture and learning German determined my choice). There, I used my cloning and leaf transfection skills to help my fellow Ph.D. student Boris Szurek to further dissect the protein motifs needed for AvrBs3 activity, including its domains interacting with the host nuclear import proteins and its transcription activation domain: Plant J. 2001 Jun;26(5):523-34. Eukaryotic features of the Xanthomonas type III effector AvrBs3: protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. Szurek B, Marois E, Bonas U, Van den Ackerveken G. Institut für Genetik, Martin-Luther Universität Halle-Wittenberg, 06099 Halle, Germany. Abstract The AvrBs3 protein of the phytopathogenic bacterium Xanthomonas campestris pv. vesicatoria is targeted to host-plant cells by the bacterial Hrp type III secretion system. In pepper plants containing the Bs3 resistance gene, AvrBs3 induces the hypersensitive response (HR). AvrBs3 recognition is thought to occur in the plant cell nucleus as HR induction is dependent on nuclear localization signals (NLSs) and an acidic transcription activation domain (AAD). In a search for AvrBs3-interacting pepper proteins using the yeast two-hybrid system, we have isolated eight different classes of cDNA inserts including two genes for importin alpha proteins. Importin alpha is part of the nuclear import machinery and interacts with AvrBs3 through an NLS in the carboxy-terminus of the protein, both in yeast and in vitro. The mechanism of AvrBs3 recognition was further studied by analysis of the C-terminal AAD. This putative transcriptionactivation domain was shown to be required for AvrBs3 HR-inducing activity, and could be functionally replaced with the VP16 AAD from the Herpes simplex virus. Our data support the model in which the AvrBs3 effector localizes to the nucleus, where the Bs3-mediated surveillance system of resistant plants detects AvrBs3 through its interference with host gene transcription. It was now clear that AvrBs3 resembled a transcription factor, probably made by bacteria to manipulate gene expression to their benefit in susceptible plants lacking the resistance gene Bs3. My most important task was thus to identify susceptible pepper genes induced specifically by AvrBs3. I used pepper leaf material infected with Xanthomonas bacteria expressing either AvrBs3 or an AvrBs3 mutant lacking its essential activation domain, extracted RNA, and used a complex differential screening procedure known as cDNA Amplified Fragment Length Polymorphism (cDNA-­‐AFLP). I cloned and sequenced differential AFLP products, obtained the sequences of the entire corresponding pepper gene cDNAs by “gene walking” on a phage cDNA library that I had 13 Past Work prepared, and contemplated a list of genes. These AvrBs3-­‐induced genes (that I called UPA, upregulated by AvrBs3) provided clues to better understand the virulence function of AvrBs3 in susceptible plants. Indeed, many of the AvrBs3-­‐induced genes are usually induced by auxin, a plant hormone controlling cell growth. In susceptible leaves, AvrBs3 induces cellular hypertrophy, believed to benefit Xanthomonas by promoting bacterial dissemination. It appeared that AvrBs3 was manipulating the plant genome to force the induction of an auxin-­‐dependent pathway in the absence of auxin. This work was published in Molecular Plant Microbe Interactions in 2002 (Article 2): Mol Plant Microbe Interact. 2002 Jul;15(7):637-46. The Xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Marois E, Van den Ackerveken G, Bonas U. Institut für Genetik, Martin-Luther-Universität Halle-Wittenberg, Halle, Germany. Abstract Xanthomonas campestris pv. vesicatoria bacteria expressing the type III effector protein AvrBs3 induce a hypersensitive response in pepper plants carrying the resistance gene Bs3. Here, we report that infection of susceptible pepper and tomato plants leads to an AvrBs3-dependent hypertrophy of the mesophyll tissue. Agrobacterium-mediated transient expression of the avrBs3 gene in tobacco and potato plants resulted in a similar phenotype. Induction of hypertrophy was shown to depend on the repeat region, nuclear localization signals, and acidic transcription activation domain (AAD) of AvrBs3, suggesting that the effector modulates the host's transcriptome. To search for host genes regulated by AvrBs3 in an AADdependent manner, we performed a cDNA-amplified fragment length polymorphism analysis of pepper mRNA populations. Thirteen AvrBs3-induced transcripts were identified and confirmed by reverse transcriptase-polymerase chain reaction. Sequence analysis revealed homologies to auxin-induced and expansin-like genes, which play a role in cell enlargement. These results suggest that some of the AvrBs3-induced genes may be involved in hypertrophy development and that xanthomonads possess type III effectors that steer host gene expression. With our discovery of a set of target genes, the idea that the type III effector AvrBs3 was a transcription factor invented by bacteria to remote-­‐control their host’s genome was gaining momentum. This work was expanded by Sabine Kay, a new Ph.D. student in Ulla’s laboratory, after my departure. Sabine identified UPA20, an additional AvrBs3-­‐induced transcription factor, the transient expression of which in pepper leaves was sufficient to trigger cellular hypertrophy. AvrBs3 directly induced the promoter of this (presumably auxin-­‐regulated) transcription factor, a major regulator of cell growth, which in turn caused the swelling of mesophyll cells. This work was the subject of our Science paper in 2007: Science. 2007 Oct 26;318(5850):648-51. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Kay S, Hahn S, Marois E, Hause G, Bonas U. Institute of Biology, Department of Genetics, Martin-Luther-University Halle-Wittenberg, Weinbergweg 10, D-06120 Halle (Saale), Germany. Abstract Pathogenicity of many Gram-negative bacteria relies on the injection of effector proteins by type III secretion into eukaryotic cells, where they modulate host signaling pathways to the pathogen's benefit. 14 Past Work One such effector protein injected by Xanthomonas into plants is AvrBs3, which localizes to the plant cell nucleus and causes hypertrophy of plant mesophyll cells. We show that AvrBs3 induces the expression of a master regulator of cell size, upa20, which encodes a transcription factor containing a basic helix-loophelix domain. AvrBs3 binds to a conserved element in the upa20 promoter via its central repeat region and induces gene expression through its activation domain. Thus, AvrBs3 and likely other members of this family provoke developmental reprogramming of host cells by mimicking eukaryotic transcription factors. Besides understanding the virulence function of AvrBs3, I was most excited by the prospect of finally understanding the molecular mechanism of AvrBs3-­‐mediated induction of plant gene promoters. Interestingly, a few of “my” UPA genes could still be induced by AvrBs3 in the presence of cycloheximide, a drug blocking protein synthesis. Therefore, these mRNAs must be directly upregulated by AvrBs3 rather than indirectly through another transcription factor. I began a quest towards the “UPA box”, a nucleotide sequence present in the promoter of induced genes that I imagined was directly targeted by AvrBs3. Based on comparisons between the 4 available “directly induced” promoters, no conserved box was immediately apparent. I selected a reliable AvrBs3-­‐induced promoter, cloned it in front of a luciferase reporter gene, and started truncating this promoter to test the minimal region responding to AvrBs3 activation (using Agrobacterium co-­‐transfections of the luciferase reporter construct and of AvrBs3 in tobacco leaves). When I left the laboratory in May 2002, I had restricted the promoter to a 130-­‐basepair fragment containing the UPA box, and it was frustrating for me to leave before getting to the very last essential nucleotides! I felt that identifying the UPA box and the way AvrBs3 interacted with it would open fascinating prospects. This was based on the fact that the 18 repeats of 34 aminoacids, composing the central domain of AvrBs3, were essential for gene activation, and that deletion of some repeats (e.g., 4 in our favorite AvrBs3 mutant) changed the specificity of gene induction: some targets were better induced by the mutant than by the native protein, others became completely uninduced. This argued for a tight correspondence between the repeat number and/or their identity and the targeted nucleotides in the promoter, although we were still unsure at the time if there was a direct physical interaction between AvrBs3 and the DNA. The identification by Sabine of new induced promoters, and the cloning of the resistance gene Bs3 which turned out to be a flavine monoxygenase “suicide gene” that AvrBs3 also induces transcriptionally (Römer et al., 2007), provided a larger dataset of promoters in which to look for a nucleotide consensus bound by AvrBs3. Suddendly, a putative UPA box of 19 nucleotides, corresponding to the number of the AvrBs3 repeats +1, became apparent. It was now possible to mutate the UPA box and examine the consequence on AvrBs3 or AvrBs3 mutants-­‐mediated induction. Just before the final illumination leading to “the code” (described below), we reported these promoter analyses in the Plant Journal (abstract below). Although still one step before the seminal paper that would open a new field in biotechnology, this was the paper I had dreamed for when leaving the Bonas lab, and I was extremely pleased by its publication! Plant J. 2009 Sep;59(6):859-71. Detailed analysis of the DNA recognition motifs of the Xanthomonas type III effectors AvrBs3 and 15 Past Work AvrBs3∆rep16. Kay S, Hahn S, Marois E, Wieduwild R, Bonas U. Institute of Biology, Department of Genetics, Martin Luther University Halle-Wittenberg, Weinbergweg 10, D-06120 Halle (Saale), Germany. Abstract The Gram-negative phytopathogenic bacterium Xanthomonas campestris pv. vesicatoria (Xcv) employs a type III secretion system to translocate effector proteins into plant cells where they modulate host signaling pathways to the pathogen's benefit. The effector protein AvrBs3 acts as a eukaryotic transcription factor and induces the expression of plant genes termed UPA (up-regulated by AvrBs3). Here, we describe 11 new UPA genes from bell pepper that are induced by AvrBs3 early after infection with Xcv. Sequence comparisons revealed the presence of a conserved AvrBs3-responsive element, the UPA box, in all UPA gene promoters analyzed. Analyses of UPA box mutant derivatives confirmed its importance for gene induction by AvrBs3. We show that DNA binding and gene activation were strictly correlated. DNase I footprint studies demonstrated that the UPA box corresponds to the center of the AvrBs3-protected DNA region. Type III delivery of AvrBs3 and mutant derivatives showed that some UPA genes are induced by the AvrBs3 deletion derivative AvrBs3∆rep16, which lacks four repeats. We show that AvrBs3∆rep16 recognizes a mutated UPA box with two nucleotide exchanges in positions that are not essential for binding and activation by AvrBs3. The next step in this story was conceptually so simple that it’s a conundrum why it wasn’t accomplished a bit sooner. Each of the 18 AvrBs3 repeats contains 34 amino acids that differ from the other repeats only by two consecutive variable amino acids. These residues are usually HD, NN, NI, NG or NS. Different genes in the AvrBs3 family show a different organization and number of the same repeats, and some of the target promoters of these homologues were beginning to be identified. Jens Boch, Sebastian Schornack and some of my other ex-­‐colleagues suddenly realized the significance of the observed parallelism between the number/identity of AvrBs3 repeats and the number/identity of bound nucleotides, and drew a correspondence table between repeat identity and promoter nucleotides. It appeared that the “HD” repeat almost always corresponded to a C nucleotide, while the “NG” repeat almost always to a T. “NI” repeats corresponded to A, “NN” to G or A, “NS” to any of the four nucleotides. The binding of the protein to the promoter appeared to follow a one repeat-­‐one base pattern; the protein-­‐to-­‐nucleotide code was cracked! In a seminal, experimentally-­‐rich Science paper (abstract below), of which I am not a co-­‐author as my contributions had been published before, the code was released. Back-­‐to-­‐back with this paper, a competing group published the same code identified using purely bioinformatics tools. These two papers are at the basis of the biotechnological “boom” of the Transcription Activator-­‐Like Nucleases (TALEN) field. AvrBs3 is the archetypical TAL (Transcription activator like) effector, and the 12 to 24 repeats in the TAL DNA-­‐binding domain can be re-­‐arranged at will (using the powerful technique of GoldenGate Cloning) to bind any desired target nucleotide sequence. If the DNA-­‐binding domain is fused to the FokI EndoNuclease domain, one obtains a TALEN, similarly to the previously existing (but harder to engineer) Zinc Finger Nucleases. A pair of TALENs that interact through dimerization of the two FokI domains can be used to cleave chromosomes at the desired locus and cause mutations. This led to a tremendous advance in gene engineering (Gaj et al., 2013), and I am quite proud to have played a role in this development. 16 Past Work Science. 2009 Dec 11;326(5959):1509-12. doi: 10.1126/science.1178811. Breaking the code of DNA binding specificity of TAL-type III effectors. Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Department of Genetics, Institute of Biology, Martin-Luther-University Halle-Wittenberg, Weinbergweg 10, D-06099 Halle (Saale) Germany. Abstract The pathogenicity of many bacteria depends on the injection of effector proteins via type III secretion into eukaryotic cells in order to manipulate cellular processes. TAL (transcription activator-like) effectors from plant pathogenic Xanthomonas are important virulence factors that act as transcriptional activators in the plant cell nucleus, where they directly bind to DNA via a central domain of tandem repeats. Here, we show how target DNA specificity of TAL effectors is encoded. Two hypervariable amino acid residues in each repeat recognize one base pair in the target DNA. Recognition sequences of TAL effectors were predicted and experimentally confirmed. The modular protein architecture enabled the construction of artificial effectors with new specificities. Our study describes the functionality of a distinct type of DNA binding domain and allows the design of DNA binding domains for biotechnology. III.2. Post-­Doctoral studies: A good dive in Drosophila biology Besides Plant Sciences, I have always been attracted to Developmental Biology. My post-­‐ doctoral studies offered me an opportunity to fulfill an old dream: go back to insects, use an attractive Genetics model organism (Drosophila), and contribute to dissect developmental processes. Our Biology classes about the Antennapedia, Ultrabithorax… homeotic mutants had fascinated me, as well as the concept of morphogen gradients. So I began working on understanding how the Wingless morphogen gradient is produced and shaped in the wing imaginal disc, in Suzanne Eaton’s laboratory at the Max-­‐Planck Institute for Cell Biology and Genetics in Dresden. I discovered the universe of antibody staining of proteins in the wing discs, of live confocal imaging of GFP, CFP, YFP…-­‐tagged proteins in that same tissue, all this in the wonderful scientific environment of the MPI. The endocytic pathway was suspected to play an important role in Wingless gradient formation, and Suzanne offered me the ambitious goal of generating a transgenic fly library of dominant negative of all Drosophila Rab genes. Rab proteins control the identity of vesicles in intracellular trafficking and their number is 35 in Drosophila. A mutation in a conserved domain present in each Rab converts these proteins to a dominant negative form. Thus, I intensely re-­‐used my site-­‐ directed mutagenesis abilities (acquired in North Carolina with avrRpm1 and avrB mutagenesis), built a nice pUAST-­‐derived expression vector for tissue-­‐specific and inducible expression (which used heat-­‐shock induced excision of an inhibitory FRT cassette) and developed skills for Drosophila transgenesis, as the in-­‐house service was too slow for my personal needs. For each of the 35 dominant negative Rab, I selected two or three independent P-­‐element insertion fly lines and began characterizing phenotypes. The inducible dominant negative Rab11 phenotype was very strong and allowed to better understand the process of protein recycling during morphogenetic junctional remodeling in pupal wings, a study that we published in Developemental Cell (abstract below). 17 Past Work Dev Cell. 2005 Dec;9(6):805-17. Hexagonal packing of Drosophila wing epithelial cells by the planar cell polarity pathway. Classen AK, Anderson KI, Marois E, Eaton S. Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstrasse 108, 01309 Dresden, Germany. Abstract The mechanisms that order cellular packing geometry are critical for the functioning of many tissues, but they are poorly understood. Here, we investigate this problem in the developing wing of Drosophila. The surface of the wing is decorated by hexagonally packed hairs that are uniformly oriented by the planar cell polarity pathway. They are constructed by a hexagonal array of wing epithelial cells. Wing epithelial cells are irregularly arranged throughout most of development, but they become hexagonally packed shortly before hair formation. During the process, individual cell boundaries grow and shrink, resulting in local neighbor exchanges, and Cadherin is actively endocytosed and recycled through Rab11 endosomes. Hexagonal packing depends on the activity of the planar cell polarity proteins. We propose that these proteins polarize trafficking of Cadherin-containing exocyst vesicles during junction remodeling. This may be a common mechanism for the action of planar cell polarity proteins in diverse systems. As expected from prior observations exploiting a dynamin temperature-­‐sensitive mutant (Strigini and Cohen, 2000), dominant negative expression of the endosome-­‐specific Rab5 also showed an extreme phenotype. Confocal imaging of imaginal discs at early time points after induction of dominant negative Rab5 allowed me to observe a direct link between Rab5 function and the Wingless morphogen gradient: as soon as Rab5 function was abolished, abnormally high levels of Wingless began to accumulate in the extracellular space. This favored a model in which Rab5-­‐mediated endocytosis usually restricts the movement of diffusing, secreted Wingless protein. In addition, I over-­‐expressed the heparan sulfate proteoglycan Dally-­‐like (already suspected to modulate Wingless signaling), which localizes to the lateral surface of disc epithelial cells, in subdomains of the wing disc, and observed that overexpressed Dally-­‐like sequestered elevated amounts of extracellular Wingless. We concluded in our Development paper (Article 4) that the Wingless gradient is shaped by endocytosis at the apical and basal epithelial surfaces, counter-­‐acted by Wingless sequestration with Dally-­‐like at the lateral cell surface: Development. 2006 Jan;133(2):307-17. Epub 2005 Dec 14. The endocytic pathway and formation of the Wingless morphogen gradient. Marois E, Mahmoud A, Eaton S. Max-Planck Institute for Cell Biology and Genetics, Pfotenhauerstr. 108, 01307 Dresden, Germany. Abstract Controlling the spread of morphogens is crucial for pattern formation during development. In the Drosophila wing disc, Wingless secreted at the dorsal-ventral compartment boundary forms a concentration gradient in receiving tissue, where it activates short- and long-range target genes. The glypican Dally-like promotes Wingless spreading by unknown mechanisms, while Dynamin-dependent endocytosis is thought to restrict Wingless spread. We have utilized short-term expression of dominant negative Rab proteins to examine the polarity of endocytic trafficking of Wingless and its receptors and to determine the relative contributions of endocytosis, degradation and recycling to the establishment of the Wingless gradient. Our results show that Wingless is internalized via two spatially distinct routes: one on 18 Past Work the apical, and one on the basal, side of the disc. Both restrict the spread of Wingless, with little contribution from subsequent degradation or recycling. As previously shown for Frizzled receptors, depleting Arrow does not prevent Wingless from entering endosomes. We find that both Frizzled and Arrow are internalized mainly from the apical membrane. Thus, the basal Wingless internalization route must be independent of these proteins. We find that Dally-like is not required for Wingless spread when endocytosis is blocked, and propose that Dally-like promotes the spread of Wingless by directing it to lateral membranes, where its endocytosis is less efficient. Thus, subcellular localization of Wingless along the apical-basal axis of receiving cells may be instrumental in shaping the Wingless gradient. Simulatenously, I developed vectors for transgenic RNAi by expressing long double-­‐ stranded RNAs with the Gal4-­‐UAS system in an inducible manner, a system that I described in a Methods in Molecular Biology book chapter: Methods Mol Biol. 2007;397:115-28. RNAi in the Hedgehog signaling pathway: pFRiPE, a vector for temporally and spatially controlled RNAi in Drosophila. Marois E, Eaton S. Abstract RNA interference (RNAi) has become an irreplaceable tool for reverse genetics in plants and animals. The universality and specificity of this phenomenon allows silencing of virtually any chosen gene to examine its involvement in biological processes. Many strategies exist to reduce the expression of a particular gene using RNAi. Some rely on delivering directly to cells the approximately 21-nucleotide long interfering double-stranded RNA (dsRNA) species that are central mediators of the silencing process. Others rely on the transgenic expression of longer dsRNA molecules, leaving it to the cellular machinery to process these hairpins into short active siRNA. In this chapter, we describe a transgenic method to deplete a chosen protein from a specific Drosophila tissue following induction of long dsRNA. It was used to uncover the role of lipidic particles in Hedgehog signaling by silencing lipophorin in the fat body (1), and we routinely use it to deplete specific proteins from wing imaginal disc subdomains (2). The method, certainly not restricted to the study of Hedgehog signaling, allows fast and efficient construction of a plasmid incorporating various Drosophila genetic tools to allow heat-shock-induced expression of dsRNA at the desired time and in the desired tissue. For protocols involving injection of in vitro synthesized dsRNA in embryos to study Hedgehog signaling, see for example (3). For genomic screens to identify Hedgehog pathway components in tissue culture cells by transfection of small interfering RNAs, see refs (4,5). Using these tools, I began to explore the effects of knocking down lipophorin in fly larvae. Lipophorin is a lipid transporter protein, scaffolding cholesterol and fatty acid particles analogous to vertebrate LDL. Suzanne hypothesized that lipophorin particles could serve as a transport platform (argosome) for morphogens, whose lipidic modifications (cholesterol and palmitate for Hedgehog, palmitate for Wingless) make them hydrophobic and should prevent their free diffusion from cell to cell. We observed that depleting lipophorin reduced the range of morphogen signaling, and Daniela Panáková, Ph.D. student, showed that a fraction of Hedgehog and Wingless proteins was indeed found in association with lipidic particles using larval extract fractionation on potassium bromide density gradients. Therefore, lipophorin particles did appear to be a vehicle for at least some morphogens, an important finding that advanced the understanding of morphorgen gradient formation that we could report in Nature (Article 5): 19 Past Work Nature. 2005 May 5;435(7038):58-65. Lipoprotein particles are required for Hedgehog and Wingless signalling. Panáková D, Sprong H, Marois E, Thiele C, Eaton S. Source Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstrasse-108, 01307 Dresden, Germany. Abstract Wnt and Hedgehog family proteins are secreted signalling molecules (morphogens) that act at both long and short range to control growth and patterning during development. Both proteins are covalently modified by lipid, and the mechanism by which such hydrophobic molecules might spread over long distances is unknown. Here we show that Wingless, Hedgehog and glycophosphatidylinositol-linked proteins copurify with lipoprotein particles, and co-localize with them in the developing wing epithelium of Drosophila. In larvae with reduced lipoprotein levels, Hedgehog accumulates near its site of production, and fails to signal over its normal range. Similarly, the range of Wingless signalling is narrowed. We propose a novel function for lipoprotein particles, in which they act as vehicles for the movement of lipidlinked morphogens and glycophosphatidylinositol-linked proteins. III.3. From flies to mosquitoes For my next switch in field of studies in 2006 I stuck to Diptera, from Drosophila to Anopheles mosquitoes. At the same time, I returned to Immunity, a field that I had in a way touched before in plants. For some years, I had kept an eye on the work performed in the Strasbourg CNRS laboratory created by Jules Hoffmann on insect immunity. In particular, my entomologist’s sensitive cord had been touched by the idea of exploiting insect antimicrobial peptides to find novel antimicrobial drugs, a concept that had led to the creation of the company Entomed. It felt natural to me to apply to join one of the research teams in Strasbourg as a post-­‐doc, and I became aware of the laboratory of Elena Levashina, working in Strasbourg since 2002 on the immune response of Anopheles mosquitoes to malaria parasites. After one year of post-­‐doctoral studies in Lena’s laboratory, I successfully applied for CNRS and INSERM staff researcher positions. After some hesitation, I chose INSERM and became a “CR1” researcher on October 1, 2007. Helping Stéphanie Blandin and Lena to write a review for Cell Host Microbe was a good opportunity to get more familiar with the protein that Lena’s lab had been characterizing in depth since 2000, the antiparasitic, vertebrate complement factor C3-­‐like, Thioesther Containing Protein I (TEP1), as well as with the known components of the mosquito anti-­‐ Plasmodium response: Cell Host Microbe. 2008 Jun 12;3(6):364-74. doi: 10.1016/j.chom.2008.05.007. Antimalarial responses in Anopheles gambiae: from a complement-like protein to a complementlike pathway. 20 Past Work Blandin SA, Marois E, Levashina EA. UPR9022, CNRS, 15 rue Descartes, F-67084 Strasbourg, France. Abstract Malaria transmission between humans depends on the ability of Anopheles mosquitoes to support Plasmodium development. New perspectives in vector control are emerging from understanding the mosquito immune system, which plays critical roles in parasite recognition and killing. A number of factors controlling this process have been recently identified, and key among them is TEP1, a homolog of human complement factor C3 whose binding to the parasite surface targets it for subsequent killing. Here, we review our current knowledge of mosquito factors that respond to Plasmodium infection and elaborate on the activity and mode of action of the TEP1 complement-like pathway. Just at the time when I applied for a post-­‐doc in Lena’s laboratory, a competing group in the UK had found a modulating role for Anopheles lipophorin in the Plasmodium-­‐Anopheles interaction (Vlachou et al. 2005). Lena and I made a connection to my previous work in Suzanne Eaton’s laboratory, where I contributed to show that lipophorin particles are a vehicle for proteins involved in development. Why not in addition a vehicle for immune factors? This idea would prove to be probably wrong, as all the potassium bromide density gradients that Martin Rono (Ph.D student), Miranda Whitten (post-­‐doc) and myself performed in Strasbourg never revealed any association of lipophorin with the most interesting anti-­‐Plasmodium factors (with the exception of prophenoloxidase) but this beautiful hypothesis allowed me to “get my hands wet” in mosquito biology. Instead of finding a physical association between immune factors and lipophorin, we observed that lipophorin expression influenced the levels of a second nutrient transporter, vitellogenin, the knockdown of which showed exactly the same phenotype as the knockdown of lipophorin. When both nutrient transporters (which fill mosquito eggs with supplies required for their development) are depleted, not only do mosquito females fail to reproduce, but they also show increased resistance to Plasmodium. Further, knocking down the inhibitor Cactus of NF-­‐kB factor Rel1 caused vitellogenin levels to plummet, while elevating the levels of the antiparasitic factor TEP1. Thus, we uncovered a network of genetic interactions between immunity and reproduction that provided a molecular explanation contributing to understand the long-­‐observed trade-­‐off between these two processes. We reported these results in PLoS Biology, and Lena offered me senior authorship on this article — a first time for me (Article 5): PLoS Biol. 2010 Jul 20;8(7):e1000434. doi: 10.1371/journal.pbio.1000434. The major yolk protein vitellogenin interferes with the anti-Plasmodium response in the malaria mosquito Anopheles gambiae. Rono MK, Whitten MM, Oulad-Abdelghani M, Levashina EA, Marois E. INSERM, U963, Strasbourg, France. Abstract When taking a blood meal on a person infected with malaria, female Anopheles gambiae mosquitoes, the major vector of human malaria, acquire nutrients that will activate egg development (oogenesis) in their ovaries. Simultaneously, they infect themselves with the malaria parasite. On traversing the mosquito midgut epithelium, invading Plasmodium ookinetes are met with a potent innate immune response predominantly controlled by mosquito blood cells. Whether the concomitant processes of mosquito reproduction and immunity affect each other remains controversial. Here, we show that proteins that 21 Past Work deliver nutrients to maturing mosquito oocytes interfere with the antiparasitic response. Lipophorin (Lp) and vitellogenin (Vg), two nutrient transport proteins, reduce the parasite-killing efficiency of the antiparasitic factor TEP1. In the absence of either nutrient transport protein, TEP1 binding to the ookinete surface becomes more efficient. We also show that Lp is required for the normal expression of Vg, and for later Plasmodium development at the oocyst stage. Furthermore, our results uncover an inhibitory role of the Cactus/REL1/REL2 signaling cassette in the expression of Vg, but not of Lp. We reveal molecular links that connect reproduction and immunity at several levels and provide a molecular basis for a longsuspected trade-off between these two processes. Following this publication, I was invited by Isabelle Tardieux, editor at Current Opinion in Microbiology, to write a review on the mosquito anti-­‐Plasmodium response, the first article that I wrote alone (Article 6): Curr Opin Microbiol. 2011 Aug;14(4):429-35. doi: 10.1016/j.mib.2011.07.016. Epub 2011 Jul 27. The multifaceted mosquito anti-Plasmodium response. Marois E. INSERM U963, CNRS UPR9022, 15 rue René Descartes, 67084 Strasbourg, Cedex, France. E.Marois@unistra.fr Abstract Plasmodium development within its mosquito vector is an essential step in malaria transmission, as illustrated in world regions where malaria was successfully eradicated via vector control. The innate immune system of most mosquitoes is able to completely clear a Plasmodium infection, preventing parasite transmission to humans. Understanding the biological basis of this phenomenon is expected to inspire new strategies to curb malaria incidence in countries where vector control via insecticides is unpractical, or inefficient because insecticide resistance genes have spread across mosquito populations. Several aspects of mosquito biology that condition the success of the parasite in colonizing its vector begin to be understood at the molecular level, and a wealth of recently published data highlights the multifaceted nature of the mosquito response against parasite invasion. In this brief review, we attempt to provide an integrated view of the challenges faced by the parasite to successfully invade its mosquito host, and discuss the possible intervention strategies that could exploit this knowledge for the fight against human malaria. While working on these topics, I quickly realized that compared to Drosophila, the Anopheles gambiae field of studies was in a prehistoric state as far as the gene engineering toolbox was concerned. Knowing how much transgenesis and genetic manipulations contributed to the understanding of Drosophila biology, I felt invested by the divine mission of developing gene engineering tools to increase experimental possibilities in Anopheles. In more practical terms, Lena was seeking to develop transgenesis in her laboratory. Then began a long period of technical engineering, continuing today, that led to major technological advances. 22 Current Projects IV. CURRENT RESEARCH AND PROJECTS IV.1. GETTING TRANSGENESIS TO WORK IN A. GAMBIAE It took time, effort, and investment into state-­‐of-­‐the-­‐art equipment to establish A. gambiae transgenesis in Strasbourg. Probably less than 8 laboratories worldwide can successfully transform the major African malaria vector Anopheles gambiae. About 15 are able to transform the Indian malaria vector A. stephensi, which is less delicate and whose eggs better survive the microinjection process. The transgenesis protocol is essentially the same as the classical Drosophila protocol, whereby freshly laid insect eggs are aligned on a microscope slide and injected into their posterior pole with plasmid DNA. In contrast to Drosophila but also to A. stephensi or Aedes mosquito eggs, A. gambiae eggs are killed if dechorionated. As a consequence, DNA must be injected through the eggshell using quartz rather than glass capillaries. Lena devoted a large investment effort to furbish our laboratory with state-­‐of-­‐the-­‐art injection equipment (a quartz capillary needle puller, an inverted light microscope with excellent optics to visualize aligned eggs, an Eppendorf FemtoJet injector and a Complex Object Parametric Sorter and Analyzer (COPAS) machine to automatically sort large numbers of fluorescent transgenic larvae according to transgene copy number). We largely owe our current success in A. gambiae transgenesis to Lena’s vision and investment efforts. Julien Soichot, then our insectary technician, played a key role in establishing the transgenesis techniques in Strasbourg by traveling to the laboratory of Mark Benedict in Vienna to learn some of the tricks necessary for success. Flaminia Catteruccia, another major expert in the field, also advised us during a visit in our laboratory. Subsequently, thanks to Julien’s great patience and perseverance, he and I managed to transform A. gambiae here. After some months of unsuccessful attempts despite my own injection experience with Drosophila eggs, I felt somewhat discouraged and it was Julien’s persistence that made us move forwards. Meanwhile, I had investigated alternative transgenesis approaches that had failed, including electroporation of eggs, injection of in vivo tranfection mixes into the body of live adult mosquitoes, and even Agrobacterium-­‐ mediated egg transformation. Finally, only the classical protocol yielded positive results. Over the past 6 years, we developed a series of improvements to the existing mosquito transgenesis procedures. Classically, transgenes are cloned within the terminal repeats of a transposon (usually the P element in Drosophila, usually the PiggyBac element for mosquitoes and many other insects). A helper plasmid encoding transposase is co-­‐injected. To ensure that PiggyBac transposase was efficiently co-­‐delivered with modified transposons in our initially unsuccessful experiments, I subcloned the transposase-­‐coding helper gene into the backbone of the transposon-­‐containing transgenesis vector itself. It seems that this was key for success, as we then started to obtain transgenic mosquitoes (April 2007), and when transposase-­‐containing and non-­‐containing plasmids were co-­‐ injected, transgenics were only obtained from the former. Therefore, the transposase helper gene acts more efficiently in cis than in trans. This may be due to a very small number of plasmid copies, perhaps often only one, integrating a forming germ cell, as suggested by the 23 Current Projects observation of transient expression in surviving larvae injected as embryos with a mix of plasmids encoding RFP and GFP: many transiently expressing cells in the larva show only one of the two fluorescence colors, in a red/green mosaic pattern. Intriguingly, the transposase gene itself sometimes co-­‐integrated in the genome of transformed mosquitoes, suggesting that the orientation of the PiggyBac terminal repeats is unimportant and that the transposase-­‐encoding vector backbone flanked by these repeats can also be recognized as a transposon! Inspired by novel technological improvements in Drosophila, I then started using PiggyBac transposon-­‐mediated transformation to insert attP docking sites, derived from the Streptomyces phage ΦC31 (Bischof et al., 2007), into the A. gambiae genome. An integrated attP site can then serve as a docking site for new injected plasmids containing an attB site from Streptomyces. Docking-­‐site transgenesis offers several advantages over transposons. First, transgenes land in a well-­‐defined locus chosen to be homozygous viable. Second, different lines carrying various transgenes (e.g., encoding variants of a given protein) can be compared if all are inserted at the same locus. Third, should a precious transgenic line be lost, it can be produced identically using the same docking line. To establish my A. gambiae docking lines at a time when transgenesis success rates were low, I made sure to include an attP docking site in all transgenesis plasmids that I was generating for various projects. The other genetic elements relevant for these various projects, as well as the transgenesis fluorescent reporter, were all cloned on a large cassette flanked by loxP sites. On obtaining homozygous transgenic lines, I was able to excise that lox cassette by injecting a plasmid transiently expressing Cre recombinase into the transgenic embryos. This allowed me to generate “empty” docking lines, containing just an attP site, in which any new fluorescence transgenesis selection marker could then be inserted. Our main current acceptor line, called X1, was generated in this manner and thus carries an unmarked attP site on chromosome II. We also obtained docking sites on the X and third chromosome. Before we could publish any of these, the laboratory of Paul Eggleston at the University of Keele reported A. gambiae docking lines for the first time, generated by PiggyBac insertion with a CFP marker transgene (Meredith et al., 2011). Besides the attP locus position in the genome, the Strasbourg lines differ from the Keele lines by the absence of CFP expression in the eyes, and by the presence of a unique loxP site left over from the cassette excision event. I further constructed an efficient docking-­‐site transgenesis helper plasmid by cloning phage ΦC31 integrase under the control of a germline-­‐specific A. gambiae promoter. In docking site transgenesis, the predictability of the transgene insertion site now allows us to inject multiple constructs simultaneously, typically 4, respectively labeled with a CFP, GFP, YFP, and RFP selection marker. To facilitate the assembly of complex transgenic constructs in the attB site-­‐containing transgenesis plasmids allowing this fluorescence-­‐based selection, I prepared a collection of destination vectors compatible with GoldenGate cloning, a powerful technique allowing the ligation of any number of inserts in the desired order (Engler et al., 2008; Geissler et al., 2011). These vectors carry an SV40 terminator to stop transcription of cloned transgenes, as well as CFP, GFP, YFP, DsRed or puromycin acetyl transferase (see below) transgenesis selection markers. We also started a collection of 24 Current Projects promoters to drive transgene expression in specific tissues such as midgut, fat body, germ cells or blood-­‐fed midgut. Mosquito larvae arising from the transgenesis procedure are visually screened under the fluorescence microscope. The fluorescence color of the identified transgenic individuals keeps track of the transgene’s identity. Generally, a successful microinjection experiment yields transgenic individuals for all 4 of the simultaneously injected constructs. This approach cuts the time and work required for obtaining transgenic mosquitoes by a factor of 4. Furthermore, we established an additional, novel selection marker for transgenic mosquitoes: a puromycin resistance cassette, which allows screening for transgenic larvae by adding the antibiotic directly to the water of the larval culture (Figure 1). This can greatly facilitate the selection process and is intended for use in sophisticated genetic manipulations such as gene knockout. It is also useful as a mere transgenesis selection marker that does not fluoresce, in order to save all fluorescent protein options for tissue-­‐ specific reporter genes. Figure 1. Puromycin resistance as a transgenic selection marker. Transgenic A. gambiae larvae expressing a puromycin resistance gene can be easily selected in a Petri dish containing water and antibiotic, much like E. coli growing on selective medium. Non-­‐ transgenic larvae die within 3 days and eventually serve as food for the transgenic ones. From September 2010 to September 2013, these tools have allowed us to generate 41 different transgenic lines, including many fluorescent reporter lines to determine the expression pattern of cloned promoters, constructs designed to manipulate TEP1 expression, mutagenic proteins for gene targeting, and constructs that will potentially affect Plasmodium development in the mosquito. A further key innovation has allowed us to speed up the process of obtaining stable transgenic lines: the use of the automated COPAS sorter, a flow cytometry machine that allows accurate selection of larvae based on their fluorescence. Thus, homozygous (the most fluorescent) larvae can immediately be obtained from the F2 progeny of a transgenic mosquito, while less-­‐fluorescent heterozygotes and non-­‐fluorescent wild types can be discarded (Figure 2B). No efficient balancer chromosome-­‐containing lines are available for A. gambiae, but we efficiently circumvent this deficit using the COPAS machine. Since transgenic lines obtained in docking sites are always single insertions (so far we never observed non-­‐specific integration at secondary genomic sites), we can simply measure the level of fluorescence of larvae from an F2 progeny and determine which larvae are negative, heterozygous or homozygous. 25 Current Projects A Figure 2: COPAS-­assisted selection of transgenic larvae. Diagram A plots the larvae (and contaminating dust) population according to time of flight (size) and extinction (opacity). Diagram B resolves the cloud of larvae gated in A according to red and green fluorescence. Here, larvae carry 0 (wild-­‐type), 1 (heterozygotes) or 2 (homozygotes) copies of a red-­‐marked transgene. Diagram C shows separation of F2 larvae from a cross with two transgenes (marked with YFP and RFP), the gated larvae are the double-­‐homozygotes. B C From the COPAS sorter, we thus obtain stable homozygous populations within minutes, rendering the process actually simpler than in Drosophila. We can also combine different fluorescence marker colors to establish doubly-­‐homozygous lines (Figure 2C), or to select trans-­‐heterozygous lines. This technological advance is particularly interesting for the prospect of mass production of single-­‐sex mosquito populations, a prospect that our collaborator Flaminia Catteruccia (Harvard Medical School of Public Health) has long been interested in. Field interventions against insect pests, such as the Sterile Insect Technique (SIT), or the Release of Insects carrying a Dominant Lethal (RIDL), or the future potential release of males carrying genes conferring resistance to Plasmodium, need mass production of male populations rendered possible by this tool. We reported the proof-­‐of-­‐principle of such sortings in the Malaria Journal in 2012 (Article 7): Malar J. 2012 Aug 28;11:302. doi: 10.1186/1475-2875-11-302. High-throughput sorting of mosquito larvae for laboratory studies and for future vector control interventions. Marois E, Scali C, Soichot J, Kappler C, Levashina EA, Catteruccia F. Institut de Biologie Moléculaire et Cellulaire, INSERM U963, CNRS UPR9022, 15 rue René Descartes, 67084 Strasbourg, France. Abstract BACKGROUND: Mosquito transgenesis offers new promises for the genetic control of vector-borne infectious diseases such as malaria and dengue fever. Genetic control strategies require the release of large number of male mosquitoes into field populations, whether they are based on the use of sterile males (sterile insect technique, SIT) or on introducing genetic traits conferring refractoriness to disease transmission 26 Current Projects (population replacement). However, the current absence of high-throughput techniques for sorting different mosquito populations impairs the application of these control measures. METHODS: A method was developed to generate large mosquito populations of the desired sex and genotype. This method combines flow cytometry and the use of Anopheles gambiae transgenic lines that differentially express fluorescent markers in males and females. RESULTS: Fluorescence-assisted sorting allowed single-step isolation of homozygous transgenic mosquitoes from a mixed population. This method was also used to select wild-type males only with high efficiency and accuracy, a highly desirable tool for genetic control strategies where the release of transgenic individuals may be problematic. Importantly, sorted males showed normal mating ability compared to their unsorted brothers. CONCLUSIONS: The developed method will greatly facilitate both laboratory studies of mosquito vectorial capacity requiring high-throughput approaches and future field interventions in the fight against infectious disease vectors. A powerful additional application of the COPAS is that we can now maintain up to four distinct transgenic constructs (labeled with each of the four fluorescent proteins) within the same mosquito population. A given transgenic construct required for experiments can easily be extracted from the main population when needed in one COPAS sorting step. Considering the large amount of space, handling, and resources required for maintaining mosquito lines, this advance can cut these budgets by 4. IV.2. MOSQUITO GENOME ENGINEERING – AND WHAT FOR? Genetically modified mosquitoes for research and public health The long-­‐term projects I would like to develop revolve around two topics that I am particularly interested in: (i) the biology of host-­‐pathogen interactions, using malaria mosquitoes and Plasmodium as a model, and (ii) gene engineering and genetically modified (GM) organisms. Beyond their great utility in basic biological studies and their more controversial commercial applications in crop plants and farm animals, can GM mosquitoes become a significant part of the arsenal in the public health fight against malaria? Vector control through the use of insecticides has promoted the spread of insecticide resistance in insect populations, causes damage to ecosystems and has unquantified consequences on human health. Could the release of genetically modified mosquito males propagating malaria resistance genes, or suppressing wild mosquito populations, alleviate the need for insecticide treatments or at least complement them for better vector control? Our successes 27 Current Projects in Anopheles transgenesis, and my continued interest in designing sophisticated genetic engineering tools for mosquitoes, place us in a perfect position to explore these questions. We are currently successfully establishing tools to produce and characterize GM mosquitoes. In a second phase, I would like to participate in producing mosquito lines truly refractory to P. falciparum and mobilize the necessary infrastructure and people network to test their potential in the genetic control of vector populations. I am already involved in this aspect through my participation in the European FP7-­‐funded project INFRAVEC, coordinated by Andrea Crisanti at Imperial College London, which Elena Levashina contributed to define before transferring the responsibility of CNRS participation to me. The aim of this European project (http://www.infravec.eu/) is to develop the infrastructure and technologies to study and implement GM mosquitoes for vector control, and to establish a network of scientists representing European countries (where most of the GM research is performed) and endemic countries (especially in Africa). The management of this project has met a number of obstacles, which made me highly aware of the difficulties inherent to organizing large-­‐scale international projects. Still, I feel committed to this great initiative and find particularly important that scientists from endemic countries are involved. In my future career, I plan to pay special attention to this networking with endemic countries, including through the hosting and training of Ph.D. and post-­‐doctoral fellows. One goal of such training would be to reach a common language and understanding from molecular biology to the situation in the field. In this context, thanks to INFRAVEC funding, I am currently training Ferdinand Nanfack, a highly motivated young scientist from Cameroon, who would like to join the lab as a Ph.D. student and is currently helping me develop gene targeting in the mosquito genome. Working in the frame of the INFRAVEC and ANR projects was also an opportunity for me to develop a fruitful collaboration with Tony Nolan and Nikolai Windbichler (Imperial College London) on homologous recombination-­‐based gene knockout, from which several publications are in preparation. To date, we have not yet managed to establish gene knockout following the method established in Drosophila (Rong and Golic, 2000), due to the apparent inactivity of FLP recombinase in the A. gambiae germ line (even after codon optimization of the FLP transgenes). INFRAVEC funding was initially due to terminate in August 2013. In the frame of its recently announced extension to February 2014 (conferring us less coordination and more laboratory work), and of the ANR project described below, we are now attempting to achieve gene knockout using Cre recombinase instead of FLP. The long term goal of using GM mosquitoes in the fight against malaria, and perhaps my own future participation in field release experiments, have to be based on a strong technical and theoretical basis. The combination of our well-­‐working docking site-­‐mediated transgenesis protocol with the possibility to COPAS-­‐sort large populations of larvae carrying desired transgene combinations is a basis for us to establish more sophisticated gene manipulation tools and perform feasibility experiments. To establish these tools, I wrote a young researcher grant proposal to the ANR, which was successful (see Annex I). The two main goals of this ANR are (i) to establish gene knockout and gene exchange in A. gambiae (inspired from the existing Drosophila protocols, with mosquito-­‐specific improvements), in order to be able to characterize genes of interest via their loss-­‐of-­‐ 28 Current Projects function phenotype, and (ii) to create synthetic transcription factors to manipulate the expression of mosquito genes involved in the interaction with Plasmodium. Such a tool would be useful for functional characterization of genes of interest, and for use in potential malaria control strategies. I proposed to base these synthetic transcription factors on Xanthomonas TALEs, which I knew intimately from my Ph.D. work described in the first chapter, and of which the binding specificity code had just been published by my former colleagues. As the project started, two papers (Miller et al., 2011; Cermak et al., 2011) introduced TALENs, custom-­‐designed nucleases in which the TAL DNA-­‐binding domain is fused to the endonuclease domain of FokI. Since our initial attempts to use homologous recombination to target TEP1 were being unsuccessful, I decided to test TALENs to obtain TEP1 mutations and establish targeted mutagenesis in the mosquito genome. To carry out the ANR projects, I was pleased to work again with TAL proteins derived from Xanthomonas. Concerning manpower, we were lucky to be contacted by my former mentor Jeff Dangl at the University of North Carolina, recommending Andrea Smidler, a very dynamic and motivated young scientist applying for a laboratory internship in a European country. Thanks to European funding raised earlier by Elena Levashina, we could have her work on the TALEN project. Andie was the perfect person for this work, coming from a laboratory studying type III effectors from plant pathogenic bacteria. She loved the idea to make use of type III effector-­‐derived proteins to manipulate the Anopheles-­‐Plasmodium interaction. After 8 months in our lab, which also gave her the opportunity to obtain her Master 2 from the University of Strasbourg, Andie and I had managed to obtain TALEN-­‐ generated TEP1 mutants and begin their characterization. We recently published this work in PLoS One (Article 8): PLoS One. 2013 Aug 15;8(8):e74511. doi: 10.1371/journal.pone.0074511. Targeted Mutagenesis in the Malaria Mosquito Using TALE Nucleases. Smidler AL, Terenzi O, Soichot J, Levashina EA, Marois E. Institut National de la Santé et de la Recherche Médicale U963, Strasbourg, France ; Centre National de la Recherche Scientifique UPR9022, Strasbourg, France. Abstract Anopheles gambiae, the main mosquito vector of human malaria, is a challenging organism to manipulate genetically. As a consequence, reverse genetics studies in this disease vector have been largely limited to RNA interference experiments. Here, we report the targeted disruption of the immunity gene TEP1 using transgenic expression of Transcription-Activator Like Effector Nucleases (TALENs), and the isolation of several TEP1 mutant A. gambiae lines. These mutations inhibited protein production and rendered TEP1 mutants hypersusceptible to Plasmodium berghei. The TALEN technology opens up new avenues for genetic analysis in this disease vector and may offer novel biotechnology-based approaches for malaria control. At the beginning of our TALEN work, comforted by my previous knowledge of TAL proteins, I felt we were among the pioneers in the field and hoped we would be the first to publish TALEN-­‐generated mutants in a complex organism. However, mosquito transgenesis and mutant selection are time-­‐consuming. In the meantime, many other laboratories working with simpler model systems, often having previous experience with Zinc Finger Nucleases, 29 Current Projects had adopted the TALEN technology and started reporting mutagenesis experiments similar to ours. The field underwent a supernova-­‐like literature explosion. Consequently, there was some slight disappointment on my part when publishing our results, since the system was not so novel any longer. Ironically, as TALENs were already all the rage, a new, similar, custom-­‐designed nuclease technology emerged and underwent a similar literature explosion as the TALENs. The CRISPR-­‐Cas system is a customizable nuclease guided by a small RNA that is easy to engineer, requiring less bench work than the TALENs. CRISPR-­‐Cas will probably allow mutagenesis experiments with the possibility to multiplex (though an off-­‐target rate that seems higher than that of TALENs may complicate this matter). Of course, we are now investigating the potential of the CRISPR-­‐Cas system to more quickly achieve targeted mutagenesis in the A. gambiae genome, in coordination with the laboratory of Jean-­‐Marc Reichhart exploring the same in Drosophila in our institute. Under my supervision, Ferdinand Nanfack from Cameroon is working on this project, an opportunity for him to be trained in high-­‐level molecular biology. Outline of the CRIPSR-­Cas project To obtain heritable mutations in target genes, the mutagenesis event has to occur in germ line cells that will differentiate into sperm or oocytes. In Anopheles gambiae, we would subsequently leg-­‐PCR-­‐screen individuals in the progeny of the mutagenized mosquitoes to identify mutants as described in the TALEN paper above. Several methods exist to deliver the synthetic mutagenic constructs into germ cells. In most animal systems, they are delivered by embryonic microinjection in the form of messenger RNA or plasmid DNA encoding them. This approach has been successfully applied with TALENs in Aedes mosquitoes (Degennaro et al., 2013; Aryan et al., 2013). In plants and in our own experiments with A. gambiae, mutagenic TALENs were expressed transgenically. This ensures that all germline cells are expressing the mutagenic enzymes, while injection in mosquito eggs is not well controlled and most injections probably miss the right cells and the ideal stage in time (as judged by the low efficiency of transgenesis). An additional advantage offered by transgenic expression of the mutagens is that one can decide to make them act over several successive generations of mosquitoes to enrich for mutants, which greatly facilitated the obtaining of TEP1 mutants in our TALEN experiments. We therefore reasoned that the RNA-­‐guided nuclease Cas9 would be more efficient if expressed in the germ cells of a stable transgenic mosquito line, rather than injected as plasmid or mRNA in every mutagenesis experiment. Thus, we generated mosquitoes in which Cas9 is under the control of the germ line vasa promoter (Papathanos et al., 2009). We are now testing three different options to deliver the mutagenic RNA component of the nuclease: (i) transient expression under the control of the U6 promoter by injection of expression plasmids; (ii) direct injection of mutagenic RNA synthesized in vitro from a T7 promoter-­‐containing PCR template; (iii) transgenic expression of the mutagenic RNA under the control of the U6 promoter. For the reasons outlined above, I expect that the latter approach will be the most efficient: expression will concern all germ cells of all mosquitoes, will be durable in time, and mutagenesis can be maintained for several successive 30 Current Projects generations by keeping the Cas9 and mutagenic RNA constructs together using COPAS selection. Our preliminary experiments with the first approach have failed so far; we are now attempting the two others. For these initial mutagenesis experiments using Cas9, we selected target genes suspected to play a role in anti-­‐Plasmodium immunity (Imd pathway components and TEP family members other than TEP1); in Plasmodium development and transmission (Saglin; xanthurenic acid biosynthetic pathway enzyme) and olfactory genes studied by Julien Pelletier and Rick Ignell, collaborators of ours in Malmö, Sweden. IV.3. Conclusion and perspectives These projects and our PLoS One paper above perfectly illustrate the current nature of my research, which mainly consists in engineering sophisticated genetic manipulation tools for mosquitoes. However, the middle-­‐term goal is to progress beyond the tools to make them truly serve basic research, in order to gain a better understanding of the vector / parasite interaction, in particular through the dissection of the mosquito innate immune system. In other words, the gene engineering tools will serve to characterize molecules suspected to play roles in the mosquito anti-­‐Plasmodium immune response in order to understand the “big picture” of that response. In the TALEN paper, we could show using heterozygous TEP1 mutants that levels of the antiparasitic factor TEP1 can be limiting in killing P. berghei. Importantly, we could also observe that albeit central in the antiparasitic response, the TEP1 pathway cannot be the only mechanism by which mosquitoes kill Plasmodium, since a few mosquito individuals homozygous for a TEP1 loss-­‐of-­‐function mutation still displayed good resistance to parasites. This resistance could, for instance, be mediated by specific bacterial strains present in the gut microbiota of some mosquitoes as suggested by the recent literature (Cirimotich et al., 2011a; Cirimotich et al., 2011b; Boissière et al., 2012); or by genetic factors other than TEP1. Our TEP1 mutants will be useful to identify antiparasitic pathways that are usually masked by TEP1 activity, illustrating how the mutagenesis tools can facilitate basic biological studies. In the future, I hope to apply “my” genetic engineering arsenal to basic research through tight collaborations with Stéphanie Blandin and Elena Levashina, who are currently using genetic screens and bioinformatics tools to identify novel candidate molecules acting in the antiparasitic response. Promising candidates will be mutagenized and over-­‐expressed thanks to our gene engineering toolbox, enabling the genetic characterization of these candidates as well as their biochemical characterization. In vivo protein-­‐protein interaction studies will be greatly facilitated by the availability of knock-­‐out mutant controls. To develop these future prospects, and particularly parasite-­‐refractory mosquito stains, we will have the great advantage to benefit from a new insectary facility whose construction is 31 Current Projects about to begin next to the IBMC in Strasbourg. Construction of this “I2MC insectarium” is funded by a “Plan Campus” benefitting the University of Strasbourg; the new laboratories will be furbished by an obtained EquipEx grant coordinated by Jean-­‐Luc Imler (to the redaction of which I brought a small contribution). I2MC will include P2+ safety level laboratories to work with the human parasite P. falciparum and P3 laboratories to develop projects involving mosquito-­‐borne viral pathogens. Therefore, I expect to be able to contribute to dissecting P. falciparum resistance pathways and perform preliminary tests of engineered mosquito resistance to P. falciparum here in Strasbourg. Promising engineered lines may then be tested in semi-­‐field facilities to test the behavior and dynamics of transgenes/mutations in more natural conditions. Within the INFRAVEC project, a facility has been built in Perugia (Italy) allowing mosquito population experiments in a relatively large confined space. A subsequent step would be to test genetically modified lines by collaborating with semi-­‐field facilities built in Kenya, that re-­‐create the local environmental conditions in a larger confined environment that includes animals, plants and human settlements. Being able to obtain and publish results from these future studies in the context of public research, rather than the private sector, is a strong motivation for me. GM insects may have a great potential in future public health interventions. Western public perception, for example of the release of GM Aedes mosquitoes by the company Oxitec in the Cayman Islands, Malaysia and soon Brazil, has been largely skeptical and suspicious, in part because a private company has performed the work. Public research is still perceived to be more impartial and transparent, and hopefully will guide the debate around implementing GM mosquitoes in the fight against malaria. 32 References REFERENCES Aryan, A., Anderson, M.A., Myles, K.M., and Adelman, Z.N. (2013). TALEN-­‐Based Gene Disruption in the Dengue Vector Aedes aegypti. PLoS One 8, e60082. Bischof, J., Maeda, R.K., Hediger, M., Karch, F., and Basler, K. (2007). An optimized transgenesis system for Drosophila using germ-­‐line-­‐specific phiC31 integrases. Proc Natl Acad Sci U S A 104, 3312-­‐3317. Boissiere, A., Tchioffo, M.T., Bachar, D., Abate, L., Marie, A., Nsango, S.E., Shahbazkia, H.R., Awono-­‐Ambene, P.H., Levashina, E.A., Christen, R., et al. (2012). Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog 8, e1002742. Cermak, T., Doyle, E.L., Christian, M., Wang, L., Zhang, Y., Schmidt, C., Baller, J.A., Somia, N.V., Bogdanove, A.J., and Voytas, D.F. (2011). Efficient design and assembly of custom TALEN and other TAL effector-­‐based constructs for DNA targeting. Nucleic Acids Res. Cirimotich, C.M., Dong, Y., Clayton, A.M., Sandiford, S.L., Souza-­‐Neto, J.A., Mulenga, M., and Dimopoulos, G. (2011a). Natural microbe-­‐mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 332, 855-­‐858. Cirimotich, C.M., Ramirez, J.L., and Dimopoulos, G. (2011b). Native microbiota shape insect vector competence for human pathogens. Cell Host Microbe 10, 307-­‐310. Degennaro, M., McBride, C.S., Seeholzer, L., Nakagawa, T., Dennis, E.J., Goldman, C., Jasinskiene, N., James, A.A., and Vosshall, L.B. (2013). orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature. Engler, C., Kandzia, R., and Marillonnet, S. (2008). A one pot, one step, precision cloning method with high throughput capability. PLoS One 3, e3647. Gaj, T., Gersbach, C.A., and Barbas, C.F., 3rd (2013). ZFN, TALEN, and CRISPR/Cas-­‐based methods for genome engineering. Trends Biotechnol 31, 397-­‐405. Geissler, R., Scholze, H., Hahn, S., Streubel, J., Bonas, U., Behrens, S.E., and Boch, J. (2011). Transcriptional activators of human genes with programmable DNA-­‐specificity. PLoS One 6, e19509. Harris, A.F., McKemey, A.R., Nimmo, D., Curtis, Z., Black, I., Morgan, S.A., Oviedo, M.N., Lacroix, R., Naish, N., Morrison, N.I., et al. (2012). Successful suppression of a field mosquito population by sustained release of engineered male mosquitoes. Nat Biotechnol 30, 828-­‐ 830. 33 References Lacroix, R., McKemey, A.R., Raduan, N., Kwee Wee, L., Hong Ming, W., Guat Ney, T., Rahidah, A.A.S., Salman, S., Subramaniam, S., Nordin, O., et al. (2012). Open field release of genetically engineered sterile male Aedes aegypti in Malaysia. PLoS One 7, e42771. Meredith, J.M., Basu, S., Nimmo, D.D., Larget-­‐Thiery, I., Warr, E.L., Underhill, A., McArthur, C.C., Carter, V., Hurd, H., Bourgouin, C., et al. (2011). Site-­‐specific integration and expression of an anti-­‐malarial gene in transgenic Anopheles gambiae significantly reduces Plasmodium infections. PLoS One 6, e14587. Miller, J.C., Tan, S., Qiao, G., Barlow, K.A., Wang, J., Xia, D.F., Meng, X., Paschon, D.E., Leung, E., Hinkley, S.J., et al. (2011). A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29, 143-­‐148. Murray, C.J., Rosenfeld, L.C., Lim, S.S., Andrews, K.G., Foreman, K.J., Haring, D., Fullman, N., Naghavi, M., Lozano, R., and Lopez, A.D. (2012). Global malaria mortality between 1980 and 2010: a systematic analysis. Lancet 379, 413-­‐431. Papathanos, P.A., Windbichler, N., Menichelli, M., Burt, A., and Crisanti, A. (2009). The vasa regulatory region mediates germline expression and maternal transmission of proteins in the malaria mosquito Anopheles gambiae: a versatile tool for genetic control strategies. BMC Mol Biol 10, 65. Romer, P., Hahn, S., Jordan, T., Strauss, T., Bonas, U., and Lahaye, T. (2007). Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318, 645-­‐648. Rong, Y.S., and Golic, K.G. (2000). Gene targeting by homologous recombination in Drosophila. Science 288, 2013-­‐2018. Vlachou, D., Schlegelmilch, T., Christophides, G.K., and Kafatos, F.C. (2005). Functional genomic analysis of midgut epithelial responses in Anopheles during Plasmodium invasion. Curr Biol 15, 1185-­‐1195. Voinnet, O., and Baulcombe, D.C. (1997). Systemic signalling in gene silencing. Nature 389, 553. 34 ANR proposal ANR young researcher project (November 2011-­‐October 2014): Outils génétiques pour la manipulation du genome d'Anopheles gambiae (GEMM) document scientifique PROGRAMME JCJC EDITION 2011 Projet GEMM DOCUMENT SCIENTIFIQUE Nom et prénom du coordinateur / coordinator’s name Marois Eric Acronyme / Acronym GEMM Titre de la proposition de projet Outils génétiques pour la manipulation du génome d'Anopheles gambiae Proposal title Tools for the genetic engineering of the malaria mosquito Anopheles gambiae Comité d’évaluation / Evaluation committee JCJC-SVSE 3 X Recherche Fondamentale / Basic Research Recherche Industrielle / Industrial Research Développement Expérimental / Experimental Development Durée de la proposition de 292 148 € 36 mois projet / Proposal duration Type de recherche / Type of research Aide totale demandée / Grant requested ANR-GUI-AAP-04 – Doc Scientifique 2011 1/26 PROGRAMME JCJC EDITION 2011 1. 2. Projet GEMM DOCUMENT SCIENTIFIQUE RESUME DE LA PROPOSITION DE PROJET / PROPOSAL ABSTRACT ...................... 3 CONTEXTE, POSITIONNEMENT ET OBJECTIFS DE LA PROPOSITION / CONTEXT, POSITIONNING AND OBJECTIVES OF THE PROPOSAL ..................................... 3 2.1. 2.2. Contexte de la proposition de projet / Context of the proposal ........................ 3 État de l'art et position de la proposition de projet / state of the art and positionning of the proposal...................................................................... 5 2.3. Objectifs et caractère ambitieux et/ou novateur de la proposition de projet / Objectives, originality and/or novelty of the proposal ................................. 7 3. PROGRAMME SCIENTIFIQUE ET TECHNIQUE, ORGANISATION DE LA PROPOSITION DE PROJET / SCIENTIFIC AND TECHNICAL PROGRAMME, PROPOSAL ORGANISATION .............................................................................. 8 3.1. Programme scientifique, structuration de la proposition de projet/ Scientific programme, proposal structure ................................................................. 8 3.1.1 3.1.2 3.1.3 3.1.4 3.1.5 Tâche 1 / Task 1 : coordination Tâche 2 / Task 2 : establishment and testing of the enzyme line Tâche 3 / Task 3 : establishment of the donor lines Tâche 4 / Task 4 : selection of mosquito larvae recombinant at the target locus Tâche 5 / Task 5 : characterization of mosquitoes resulting from gene exchange recombination 3.1.6 Tâche 6 et 7 / Task 6 and 7: obtention of synthetic transcription factors 3.1.7 Tâche 8 and 9 / Task 8 and 9 : characterization of transgenic mosquitoes carrying synthethic transcription factors 3.1.8 Tâche 10 / Task 10: evaluation of the specificity of synthetic TFs 3.1.9 Tâche 11 / Task 11: experiments in semi-field conditions 3.1.10 Tâche 12 / Task 12: valorization 3.2. 11 11 12 13 13 14 15 15 15 16 Calendrier des tâches, livrables et jalons / Tasks schedule, deliverables and milestones ............................................................................................17 4. STRATEGIE DE VALORISATION, DE PROTECTION ET D’EXPLOITATION DES RESULTATS / DISSEMINATION AND EXPLOITATION OF RESULTS, INTELLECTUAL PROPERTY .................................................................................. 19 5. DESCRIPTION DU PARTENARIAT / CONSORTIUM DESCRIPTION ....................... 19 5.1. Description, adéquation et complémentarité des partenaires / Partners description and relevance, complementarity ..............................................19 5.2. Qualification du coordinateur de la proposition de projet/ Qualification of the proposal coordinator .........................................................................19 5.3. Qualification, rôle et implication des participants / Qualification and contribution of each partner ....................................................................20 6. JUSTIFICATION SCIENTIFIQUE DES MOYENS DEMANDES / SCIENTIFIC JUSTIFICATION OF REQUESTED RESSOURCES ........................................... 20 7. ANNEXES / ANNEXES ..................................................................... 21 7.1. Références bibliographiques / References ...................................................21 7.2. Biographies / CV, resume .........................................................................23 7.3. Lettre de soutien du directeur du laboratoire ...............................................24 ANR-GUI-AAP-04 – Doc Scientifique 2011 2/26 PROGRAMME JCJC EDITION 2011 Projet GEMM DOCUMENT SCIENTIFIQUE 1. RESUME DE LA PROPOSITION DE PROJET / PROPOSAL ABSTRACT The ability to precisely engineer the genome of the malaria mosquito Anopheles gambiae would open unprecedented opportunities for research in the biology of this disease vector. In particular, it would help to dissect the mosquito immune system, which plays a key role in eliminating malaria parasites. In addition to this tremendous potential for fundamental studies, transgenic mosquitoes may in the long run be used in strategies to combat malaria. Field trials involving transgenic mosquitoes are already under way to fight the dengue virus. However, academic research on the feasibility of such approaches and on their possible associated risks is scarce. New tools must be developed to address these emerging questions. Recently, our laboratory has achieved several advances in mosquito transgenesis, including the easy production of transgenic A. gambiae using docking lines as well as innovative tools for the selection of rare transposition events. These results give us a head start to design more ambitious methods to finely manipulate the mosquito genome. Goal 1 of this proposal is to establish homologous recombination-based gene exchange, and its variant gene knockout, in A. gambiae. This work will combine our novel transgenesis tools with strategies adapted from the Drosophila research field. Goal 2 is to engineer synthetic transcription factors that specifically bind chosen DNA sequences in mosquito gene promoters, in order to stimulate or repress the expression of antiparasitic genes. This part of the work will exploit our knowledge of a novel type of bacterial transcription factor that is amenable to biotechnology. As a proof-of-principle of the technologies developed in goals 1 and 2, we will test these new tools on the TEP1 antiparasitic factor. Mosquitoes engineered to express mutant versions of TEP1, and elevated levels of genes in the TEP1 pathway, will be evaluated for their resistance to Plasmodium. Overall, this project will provide a suite of applications for functional gene analysis permitting transcriptional overexpression/supression of endogene expression and knock-in/knock-out substitutions. It will help establish knowledge and strategies to increase malaria resistance in mosquitoes, and will be transferable to other insect species amenable to transgenesis. 2. CONTEXTE, POSITIONNEMENT ET OBJECTIFS DE LA PROPOSITION / CONTEXT, POSITIONNING AND OBJECTIVES OF THE PROPOSAL 2.1. CONTEXTE DE LA PROPOSITION DE PROJET / CONTEXT OF THE PROPOSAL Malaria today is the second-most important human disease after AIDS, killing about 800 000 people annually, the majority of whom are infants in sub-Saharan Africa. 225 million people are infected every year in 108 countries, half the human population is at risk (1). Global warming is expected to promote the spread of malaria, and other mosquito-borne illnesses, from tropical regions to higher latitudes. 5 different species of Plasmodium are the causal agent of malaria, the most deadly being P. falciparum. About 20 species of mosquitoes in the genus Anopheles are known vectors of Plasmodium, transmitting the parasite during their blood meals on humans. In sub-Saharan Africa, Anopheles gambiae is the major malaria vector. Malaria mosquitoes of the genus Anopheles are routinely reared in many laboratories to study the biology of the interaction between Plasmodium parasites and their mosquito vectors. Anopheles mosquitoes are easy to rear and have a short generation time (about 2 weeks from ANR-GUI-AAP-04 – Doc Scientifique 2011 3/26 PROGRAMME JCJC EDITION 2011 Projet GEMM DOCUMENT SCIENTIFIQUE egg to egg), making them amenable to Genetics studies and manipulation. Another Dipteran insect, Drosophila melanogaster, is arguably the best-established animal model for Genetics and Developmental studies. Drosophilists possess an extensive toolbox of exquisite precision to dissect biological processes in their model organism (2). Routine and efficient transgenesis in Drosophila has played a central role in allowing the subsequent development of many other tools, and has lead to today’s high degree of understanding of the fruit fly’s biology. In contrast, few Genetics tools have been developed for mosquitoes. Transgenesis in many mosquito species, especially the major malaria vector A. gambiae, has proved to be technically much more challenging than in its distant Drosophila cousin. However, the ability to routinely insert synthetic transgenes or mutant versions of mosquito genes by transgenesis would greatly facilitate the further unraveling of mosquito biological processes, especially those involved in the resistance/susceptibility to Plasmodium parasites. Further, it would allow to import some of the most useful tools available in Drosophila, such as gene knockout / gene exchange or the Gal4/UAS system, in order to enable genetic analyses and investigate mosquito-specific biological questions. In addition to providing powerful basic research tools, another potential future application of mosquito transgenesis is vector control. Historically, the destruction of mosquito vector populations and protecting humans against vector bites with insecticide-impregnated bednets have been more effective means to combat malaria transmission than medical prophylaxis and treatment. DDT and other insecticides have played, and are still playing, an important role in reducing malaria, with unquantified adverse effects on human health and the environment. Over time, mosquito populations have also shown a great ability to develop resistances against these insecticides, rendering them ineffective. Mosquito transgenesis is considered as a serious alternative to achieve the same purpose with a more specific action, being directed only against the correct mosquito vector species. One possibility under consideration is to curb vector populations by punctually releasing lethal transgenes via laboratory-raised transgenic males (3). In this approach, transgenes are expected to rapidly disappear from the population due to natural selection, after having negatively impacted mosquito numbers. Another transgenic approach has been discussed ever since the recognition that mosquitoes mount a powerful immune response against malaria parasites, that renders most Plasmodium species unable to infect the vector (except in just a few unfortunate Plasmodium/Anopheles combinations). The strategy is to harness the power of the vector’s immune system to combat the parasite. Mosquitoes could be made refractory to Plasmodium in the laboratory and released in the field, under the provision that no adverse long-term effects of the transgenes would be identified in thorough preliminary experiments. In 2010, two field releases of transgenic male cohorts of the dengue fever mosquito Aedes aegypti have been conducted in Malaysia and the Cayman islands. The transgenic mosquitoes carried transgenes designed to impair female fitness, in the hope to decrease the local transmission of dengue fever. These field experiments were carried out by British private company Oxitec, without supervision from academic research. In the future, the public might demand that public research starts exploring both the potential benefits and the potential associated short and long-term risks of such approaches. We intend to develop the transgenic tools needed to address these issues in the laboratory with the main malaria vector A. gambiae. The suite of tools that we want to establish will provide the means to perform complete functional analysis of any gene of interest (tissue and time-specific overexpression, suppression of expression, gene elimination, allelic substitutions). In addition to its expected impact on basic research, it will attract more scientists to use these methods and develop assays for the analysis of genetically modified insects in semi-field and, eventually, field conditions. ANR-GUI-AAP-04 – Doc Scientifique 2011 4/26 PROGRAMME JCJC EDITION 2011 Projet GEMM DOCUMENT SCIENTIFIQUE 2.2. ÉTAT DE L'ART ET POSITION DE LA PROPOSITION DE PROJET / STATE OF THE ART AND POSITIONNING OF THE PROPOSAL Today, only a handful of laboratories are able to perform transgenesis in the malaria vector Anopheles gambiae, and not at a routine level. Transgenic lines are obtained at a low rate; probably only a few dozens transgenic A. gambiae lines have been obtained worldwide to date and fewer than 5 have been published, compared to tens of thousands in Drosophila. In the last 2 years, our laboratory has made tremendous progress in this field. Mosquito trangenesis has historically used the same general strategies as in Drosophila. While engineered P element transposons are the most widely used vector system for transgenesis in the fruit fly, they do not function in mosquitoes. Instead, most transgenic Anopheles lines obtained to date carry constructs prepared in the PiggyBac transposable element. The drawback of transposon-based transgenesis in Drosophila as well as in mosquitoes are the following: - the transposon insertion site is unpredictable. Each newly obtained transgenic insertion needs to be mapped at least roughly to a chromosome and stabilized by crosses to lines carrying balancer chromosome, which have not yet been established in the case of mosquitoes. - the obtention of stable homozygous transgenic lines is often complicated by multiple insertions and by lethality of many insertions in homozygous individuals. - Different versions of a given transgene cannot be accurately compared, as positional effects differentially influencing each insertion at different genomic loci may yield different spatio-temporal patterns and strength of transgene expression. Recently, two laboratories have generalized docking-site mediated transgenesis in Drosophila (4; 5; 6) relying on integration of new transgenes into specific chromosomal sites previously inserted using classical transgenesis. These docking sites are derived from the phage ΦC31 integration site (attP), which drives viral genome insertion into a well-defined target site (attB) in the chromosome of its Streptomyces bacterial host. For insect transgenesis, the phage attP site is integrated into the insect genome with the help of a transposon, and any synthetic DNA construct carrying the Streptomyces attB site can subsequently be inserted into the genomic attP locus. The advantages of this system are multiple: - docking lines are chosen to be homozygous viable and robust. - Multiple alleles of a given transgene can be inserted at the same locus in independent mosquito lines and accurately compared in their phenotypes. - The insertion site is known in advance and its location can be chosen on an appropriate chromosome for subsequent combinations with other loci of interest on other chromosomes. - If a precious transgenic line is lost, it can be generated again identically. Recently, we succeeded in adapting the docking-site transgenesis system to Anopheles gambiae. Each PiggyBac transgenic constructs that we used in several non-related projects carried an attP site, while the transgene under study and its fluorescence selection marker were flanked by loxP recombination sites. We could subsequently “recycle” the obtained transgenic lines and transform them into “empty” docking attP lines by micro-injecting embryos with a plasmid encoding Cre recombinase. This procedure led to excision of the loxP cassette at a high frequency, showing that the Cre-Lox system works in mosquitoes, and generated several A. gambiae docking lines that we are maintaining in our insectory. Initial attempts to obtain insertion of new, attB-containing transgenic constructs into our attP docking lines with the available ΦC31 integrase helper plasmid were successful only in 3 occasions. However, we recently placed the helper plasmid’s ΦC31 integrase gene under the control of a new, germline-specific mosquito promoter. Since then, we obtain new transgenic ANR-GUI-AAP-04 – Doc Scientifique 2011 5/26 PROGRAMME JCJC Projet GEMM EDITION 2011 DOCUMENT SCIENTIFIQUE lines easily following each injection attempt. This achievement is a tremendous advance that puts our laboratory at the lead for A. gambiae transgenesis. Thus far, the selection of transgenic mosquito larvae is based on visual screening for fluorescent larvae (expressing various GFP variants or RFP, see Fig. 1A) under the fluorescence microscope. A key development has allowed us to speed up the subsequent process of obtaining stable transgenic lines: the use of an automated COPAS sorter, a FACSlike machine that allows accurate selection of larvae based on their fluorescence. Thus, homozygous (the most fluorescent) larvae can immediately be obtained from the F2 progeny of a transgenic mosquito, while less-fluorescent heterozygotes and non-fluorescent wild types can be discarded. Recently, we have also developed a novel, non fluorescence-based tool for the selection of transgenic mosquito larvae: a genetic cassette encoding resistance to the antibiotic puromycin. Instead of screening thousands of larvae under the fluorescence microscope to find few transgenics, we can now add puromycin to the water and select larvae as simply as bacterial colonies on a Petri dish; only transgenic individuals carrying the antibiotic selection marker survive (Fig. 1B). This unique and exclusive tool in insect transgenesis paves the way for novel powerful genetic engineering procedures, such as gene exchange/ gene knockout in mosquitoes, which we now intend to develop. C Figure 1. Three modes of selection for transgenic mosquito larvae. A, transgenic larvae carry fluorescence markers (which can be combined in genetic crosses as seen in the rightmost larva) and are sorted under the fluorescence microscope. B, a single transgenic larva carried a puromycin resistance gene, which allowed it to survive in water containing the antibiotic. C, the F2 progeny of RFP fluorescent larvae were analyzed with the COPAS machine. In the the top diagram, objects are plotted according to their size (time of flight, X axis) and opacity (extinction, Y axis). Larvae were determined to be inside the selection. In the bottom diagram, the subset of objects gated in the top diagram are again plotted according to their fluorescence. (red fluorescence intensity on X axis, yellow autofluorescence on the Y). Negative larvae form a cloud on the left, heterozygous larvae (one copy of the RFP gene) form a cloud in the middle, homozygous larvae form the right cloud. Requesting these larvae (gated) from the machine immediately provides a stable homozygous line, circumventing the need for crosses with lines carrying balancer chromosomes. ANR-GUI-AAP-04 – Doc Scientifique 2011 6/26 PROGRAMME JCJC EDITION 2011 Projet GEMM DOCUMENT SCIENTIFIQUE 2.3. OBJECTIFS ET CARACTERE AMBITIEUX ET/OU NOVATEUR DE LA PROPOSITION DE PROJET / OBJECTIVES, ORIGINALITY AND/OR NOVELTY OF THE PROPOSAL We now want to build on our ability to easily produce transgenic A. gambiae and progress towards more ambitious Genetics tools. The recent advances described above (efficient docking line-based transgenesis, puromycin selection of transgenic mosquitoes, use of a COPAS machine to select vast numbers of fluorescent mosquitoes of the desired genotype) are placing our laboratory in a unique position in the community of mosquito research, and offer us the opportunity to further develop novel tools for the genetic engineering of the mosquito genome. In this proposal, we detail two major objectives: (i) the establishment of homologous recombination-based gene knockout in A. gambiae; (ii) the creation of synthetic transcription factors to up-regulate anti-plasmodial genes in transgenic A. gambiae. 2.3.1 Homologous recombination-based Gene Exchange / Gene Knockout in Anopheles gambiae Homologous recombination-based Gene knockout is well established in mice, zebrafish, and works in Drosophila. The establishment of gene exchange and knockout in A. gambiae would bring tremendous progress in the field of vector biology. It would lead to immediate applications in the investigation of the mosquito immune system (the long-standing focus of our laboratory) and other parasite-relevant biological processes. In the longer term, this procedure might help create malaria-refractory mosquitoes for potential field release. In Drosophila, the screening process for desired flies resulting from gene knockout or gene exchange is based on following the white selection marker moving from a donor synthetic construct (previously inserted into the genome by transgenesis) to the target endogenous locus on a different chromosome, and is notoriously tedious. Instead of a visible marker such as the Drosophila white gene, we propose to use our puromycin resistance cassette to render gene knockout in mosquitoes easier than in Drosophila, while retaining other features of the protocol established in the fruit fly by Rong and Golic (7; 8) (Figure 2). The selection process will thus be tremendously simplified and, therefore, will be possible at much larger scales than in Drosophila. This point might be crucial to identify the desired rare genetic events. As a proof-of-principle, we will use the gene exchange version of the procedure to first attempt substituting the endogenous, susceptible form of the antiparasitic TEP1 gene (in a malaria-susceptible mosquito strain) with a resistant allele fused to red fluorescent protein. In parallel, we will substitute TEP1 for a mutant in the thioester site of the protein, which our lab is interested in testing. This part of the work will be conducted in collaboration with the group of Elena Levashina, currently heading our laboratory, who will be developing research with P. falciparum at the Max-Planck Institute in Berlin from the Summer of 2011. E.L. has a long-standing interest in the thioester-containing TEP1 protein, a key antiplasmodial and immune factor homologous to vertebrate complement factor C3 (9; 10). 2.3.2. Synthetic transcription factors to manipulate gene expression in A. gambiae The coordinator’s Ph.D. work in the laboratory of Ulla Bonas in Halle, Germany, contributed to characterize a bacterial type III virulence factor, AvrBs3, which we showed to be a transcription factor (TF) made by Xanthomonas bacteria and injected into host plant cells to manipulate host gene expression (11; 12; 13). Since then, the Bonas laboratory has further characterized this family of transcription factors, which directly bind and activate host promoters (14). They also showed that it was functional in other eukaryotes cells than plants (J. Boch, pers. comm). The recently characterized, novel DNA-binding domain of the proteins consists of tandemly-arranged repeats of 34 amino acid (aa) with only two variable residues, ANR-GUI-AAP-04 – Doc Scientifique 2011 7/26 PROGRAMME JCJC EDITION 2011 Projet GEMM DOCUMENT SCIENTIFIQUE the identity of which determines to which DNA base a given 34-aa repeat will bind (15). This specificity offers the possibility to engineer novel proteins binding DNA at specific, chosen sites 10-20 nucleotides in length. This strategy is similar to the engineering of synthetic zinc finger TFs (16), but we expect that it will bind with higher specificity to the target DNA sequence and that it can be engineered at a much lower cost, given that the Halle laboratory has established a powerful method to assemble any sequence of repeats designed to bind the desired DNA sequence. In collaboration with the Bonas lab, we want to engineer an artificial TF and test its ability to upregulate chosen mosquito genes. Specifically, we chose the promoters of the antiparasitic factors TEP1, LRIM1 and APL1-C as targets for our artificial transcription factor. These three proteins act in concert to promote parasite killing (17; 18) and we have antibodies to follow their expression. Using our established mosquito attP docking lines, we will generate transgenic mosquitoes carrying different versions of the artificial TF varying in their transactivation domain and test which version works best in mosquitoes. We will monitor the efficiency of these TF by qRT-PCR and western blot on the target genes/proteins. To assess the specificity of the transcription factors, we will use microarray analysis of gene expression to identify potential variations in the expression level of mosquito genes other than the intended target genes. In addition, the effect of the synthetic TFs on mosquito resistance against Plasmodium will be evaluated by infection assays using our current laboratory model of malaria, the mouse pathogen Plasmodium berghei. If promising results are obtained, we will extend these experiments to P. falciparum. 3. PROGRAMME SCIENTIFIQUE ET TECHNIQUE, ORGANISATION DE LA PROPOSITION DE PROJET / SCIENTIFIC AND TECHNICAL PROGRAMME, PROPOSAL ORGANISATION 3.1. PROGRAMME SCIENTIFIQUE, STRUCTURATION DE LA PROPOSITION DE PROJET/ SCIENTIFIC PROGRAMME, PROPOSAL STRUCTURE Scientific Program for Goal 1 (gene knockout/gene exchange) : We will first construct a transgenic mosquito line expressing the two yeast enzymes, FLP recombinase and I-SceI nuclease, whose action upon the synthetic donor sequence will promote homologous recombination (Fig. 2). ANR-GUI-AAP-04 – Doc Scientifique 2011 8/26 PROGRAMME JCJC EDITION 2011 Projet GEMM DOCUMENT SCIENTIFIQUE Donor cassette Figure 2: outline of the gene exchang procedure. FLP recombinase excises the donor cassette containing the modified gene as a circular molecule, which is immediately linearized by I-SceI nuclease. The resulting linear DNA fragment promotes homologous recombination between the regions flanking the endogenous target locus and the same regions cloned inside the cassette. In the progeny of a cross between wild-type mosquitoes and mosquitoes carrying the donor cassette and the enzyme genes, selecting against GFP and for puromycin resistance yields larvae of the desired genotype. Molecular biology work will be required to assemble the transgenesis plasmid that can be injected into mosquito embryos to produce this line. We have all the fragments (promoters, codon-optimized genes) and the technology (Multisite Gateway cloning) needed for this step. Then, we will inject the resulting, attB-containing plasmid into eggs of one of our attP docking mosquito lines. Once obtained, the line will be made homozygous and amplified. This first step (which constitutes Task 2, as Task 1 is defined as the piloting and organizing the project) could take between 6 and 9 months. Task 3, which can be carried out in parallel to task 2, will be to obtain donor mosquito lines containing the synthetic construct upon which FLP and I-SceI will act. The required transgenesis plasmids have already been prepared, we thus only need to produce, stabilize and amplify the transgenic mosquitoes (46 months). Mosquitoes obtained in Tasks 2 and 3 will be crossed. Task 4 consists in the analysis of the progeny of this cross. Complex genetic events are likely to be observed and it will be interesting to analyze how the yeast enzymes behave in the mosquito. The selection of the desired recombination events themselves is also part of this task. Taken together, Task ANR-GUI-AAP-04 – Doc Scientifique 2011 9/26 PROGRAMME JCJC EDITION 2011 Projet GEMM DOCUMENT SCIENTIFIQUE 4 will last 6 to 8 months. In the next task (Task 5) we will perform infection assays on the newly obtained genetically engineered mosquitoes, first using Plasmodium berghei (4-6 months allowing for several repeats), then P. falciparum (4-6 months). Scientific program for goal 2 (synthetic transcription factors to manipulate mosquito gene expression) Using our directives, our collaborators at the University of Halle will assemble several synthetic transcription factor genes varying in their transactivation domain (TAD) (2 months). We will subsequently assemble the TF genes and other required sequences (promoter, selection marker) into a transgenesis vector, and generate transgenic mosquito lines using one of our attP docking lines. Together, this molecular biology/transgenesis task (Task 8) may take 6-8 months. This task overlaps with the testing of TFs in transfected cells (Task 7 —3 months). Subsequently in Task 9, transgenic mosquitoes expressing the chosen TFs will be characterized biochemically (western blots, qRT-PCR) and using infection assays with P. berghei, and if encouraging results are obtained, with P. falciparum. This Task 9 may therefore last 6-10 months, allowing for multiple repeats of the experiments. An additional task will be to search for potential off-target effects of the TFs by microarray analysis of the transgenic mosquitoes (Task 10 —5 months). Once resistance to Plasmodium of the newly established lines is confirmed and characterized, we will test the fitness and competitiveness of these lines relative to wild-type mosquitoes. This comparative analysis in semi-field conditions will give a first indication of how engineered transgenes might behave in the field after a potential release to fight malaria. In particular, given the interactions we observed between reproduction and immunity in A. gambiae (19), we wonder how well mosquitoes engineered to express higher immunity will fare in a natural environment. Mosquitoes carrying the substitution of the susceptible TEP1 allele with a refractory allele made for goal 1 of this project, and mosquitoes carrying a synthetic factor leading to upregulation of 3 immune genes in the TEP1 pathway established in goal 2, will be used along with wild type mosquitoes. Recapture experiments after many generations in semi-field conditions will tell how the frequencies of the transgenes change over time. The best set-up to carry out this part of the project is the semi-field environment of a large artificial habitat, such as the large population cages developed at the University of Peruggia, Italy, with which our group is collaborating in the frame of the CE FP7 INFRAVEC consortium. This work package is Task 11 of the project (6 months). We define Task 12 as the manuscript preparation work for the publication of results that will arise from the entire project. ANR-GUI-AAP-04 – Doc Scientifique 2011 10/26 PROGRAMME JCJC Projet GEMM EDITION 2011 DOCUMENT SCIENTIFIQUE Organigramme technique Task 1 Task 2 Task 4 Task 5 Task 3 Task 11 Task 8 Task 9 Task 12 Task 10 Task 6 Task 7 3.1.1 TACHE 1 / TASK 1 : COORDINATION The project coordinator is located at the IBMC in Strasbourg, where most of the work will take place (particularly mosquito rearing and transgenesis). He will, therefore, directly supervise the work and participate directly in the making of DNA constructs and transgenic mosquitoes. He will also remotely check the advancement of DNA constructs made by collaborators in Halle and of the tests performed by our collaborators T. Nolan and D. Naujoks in the laboratory of A. Crisanti in London, perform the downstream molecular biology work required to assemble final transgenesis plasmids, and perform mosquito embryo micro-injections. For mosquito micro-injection, the coordinator will team up with a technician, Julien Soichot, who is already trained in mosquito transgenesis, as it is more efficient to micro-inject when one person is aligning mosquito eggs onto a microscope slide, while the other injects them with the micro-manipulator. For mosquito rearing, given the anticipated extension in the number of cultured lines, we would like to get additional technical help from a second technician hired from this grant. We also request a postdoctoral position to participate to this project. 3.1.2 TACHE 2 / TASK 2 : ESTABLISHMENT AND TESTING OF THE ENZYME LINE To perform homologous recombination between the donor transgenic construct and the target endogenous locus in the mosquito genome, the synthetic construct is excised within mosquito germline cells by the combined activity of FLP recombinase and I-SceI nuclease. These two yeast enzymes need to be expressed from another transgenic construct in the same mosquito cells. Using Multisite Gateway technology to combine all necessary DNA sequences in a single transgenesis plasmid, we already obtained such a construct and tested it in a transgenic mosquito line. In this construct, the enzymes were subcloned from available Drosophila plasmids where they were under the control of the Drosophila heat-shock protein 70 promoter. Unfortunately, the enzymes were not expressed in A. gambiae under the control of this promoter. Our collaborators in London next placed them under the control of a germline-specific promoter that they had characterized, the A. gambiae vasa2 promoter (20). ANR-GUI-AAP-04 – Doc Scientifique 2011 11/26 PROGRAMME JCJC EDITION 2011 Projet GEMM DOCUMENT SCIENTIFIQUE The resulting transgenic mosquitoes now showed efficient I-SceI activity in our collaborator’s reporter system. However, it appeared that FLP still fails to be expressed in this second line. We analyzed the FLP coding sequence and realized that it is filled with codons that are rare in A. gambiae, to such an extent that the FLP gene appears to be the worst heterologous gene we ever tried to express in mosquitoes regarding codon usage. Therefore, we designed a codon-adapted synthetic FLP gene for expression in the mosquito. We are now cloning this optimized FLP gene under the control of the vasa2 promoter into attB-containing plasmids for mosquito transgenesis. The selection marker associated to the enzyme transgenic construct will be the classically-used, red fluorescent protein-encoding cassette, allowing to select transgenic larvae having red fluorescence in their eyes. Once a mosquito line carrying this optimized enzyme construct is obtained, we will (i) send this line to our London collaborators to test I-SceI activity in their reporter system. Specifically, they possess a transgenic line carrying a non-functional GFP reporter gene, which can become active upon action of I-SceI nuclease. The number of green fluorescent larvae in the progeny of a cross between our enzyme line and the reporter line will reflect ISceI activity. (ii) cross the enzyme line to donor lines (see Task 3) and check the progeny by PCR for evidence of FLP activity. Risk and contingency plan Should FLP recombinase for some reason still be inactive in mosquitoes, we will turn to Cre recombinase to achieve the same goal. We have already shown that Cre is working in mosquitoes. Cre was not our primary choice for the gene knockout/exchange procedure, because of the opportunity to directly import existing constructs from the Drosophila field. However, it is still possible to substitute the FLP recognition target sequences (FRT sites) for loxP sites in our donor constructs. Deliverables from Task 2: - new transgenesis plasmid for I-SceI and FLP expression in the germline (Month 2) - new enzyme-expressing mosquito line (Month 6) - evaluation of the activity of each enzyme within the germline. (Month 9) 3.1.3 TÂCHE 3 / TASK 3 : ESTABLISHMENT OF THE DONOR LINES As a proof-of-principle, we will test the gene exchange procedure with donor constructs designed to replace the endogenous susceptible allele of the antiparasitic TEP1 gene with either a thioester mutant version of TEP1 (Donor construct A), or a functional version of TEP1 fused in frame to a red fluorescent protein (Donor consruct B). The plasmids carrying Donor constructs A and B have already been constructed and will be injected into A. gambiae embryos. Resulting transgenic larvae will express GFP in the eyes and will be selected on this basis. Since the donor construct also carries our puromycin resistance cassette, we will verify that the donor lines grow in water containing the antibiotic. Risk and contingency plan Should homologous recombination prove to be impossible to achieve in mosquitoes (an unlikely event, since it works in mouse, zebrafish and flies), these donor lines will be used in experiments exploiting the fact that they express the TEP1 variants also before gene targeting. It will, anyway, be informative to study the effect of the TEP1 transgenes in the presence of the wild-type endogenous gene copies, and this alone might already lead to a publication in a high-impact journal. Deliverables from Task 3: ANR-GUI-AAP-04 – Doc Scientifique 2011 12/26 References Lacroix, R., McKemey, A.R., Raduan, N., Kwee Wee, L., Hong Ming, W., Guat Ney, T., Rahidah, A.A.S., Salman, S., Subramaniam, S., Nordin, O., et al. (2012). Open field release of genetically engineered sterile male Aedes aegypti in Malaysia. PLoS One 7, e42771. Meredith, J.M., Basu, S., Nimmo, D.D., Larget-­‐Thiery, I., Warr, E.L., Underhill, A., McArthur, C.C., Carter, V., Hurd, H., Bourgouin, C., et al. (2011). Site-­‐specific integration and expression of an anti-­‐malarial gene in transgenic Anopheles gambiae significantly reduces Plasmodium infections. PLoS One 6, e14587. Miller, J.C., Tan, S., Qiao, G., Barlow, K.A., Wang, J., Xia, D.F., Meng, X., Paschon, D.E., Leung, E., Hinkley, S.J., et al. (2011). A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29, 143-­‐148. Murray, C.J., Rosenfeld, L.C., Lim, S.S., Andrews, K.G., Foreman, K.J., Haring, D., Fullman, N., Naghavi, M., Lozano, R., and Lopez, A.D. (2012). Global malaria mortality between 1980 and 2010: a systematic analysis. Lancet 379, 413-­‐431. Papathanos, P.A., Windbichler, N., Menichelli, M., Burt, A., and Crisanti, A. (2009). The vasa regulatory region mediates germline expression and maternal transmission of proteins in the malaria mosquito Anopheles gambiae: a versatile tool for genetic control strategies. BMC Mol Biol 10, 65. Romer, P., Hahn, S., Jordan, T., Strauss, T., Bonas, U., and Lahaye, T. (2007). Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318, 645-­‐648. Rong, Y.S., and Golic, K.G. (2000). Gene targeting by homologous recombination in Drosophila. Science 288, 2013-­‐2018. Vlachou, D., Schlegelmilch, T., Christophides, G.K., and Kafatos, F.C. (2005). Functional genomic analysis of midgut epithelial responses in Anopheles during Plasmodium invasion. Curr Biol 15, 1185-­‐1195. Voinnet, O., and Baulcombe, D.C. (1997). Systemic signalling in gene silencing. Nature 389, 553. 34 PROGRAMME JCJC EDITION 2011 Projet GEMM DOCUMENT SCIENTIFIQUE - Two donor mosquito transgenic lines that can also serve to study TEP1 mutants in vivo (Month 6) - Information about the activity of the TEP1 thioester mutant and imaging of TEP1-RFP (Month 8) 3.1.4 TÂCHE 4 / TASK 4 : SELECTION OF MOSQUITO LARVAE RECOMBINANT AT THE TARGET LOCUS Transgenic lines established in tasks 2 and 3 will be crossed. In the germline of their progeny, FLP and I-SceI activity should excise the donor construct as a linear DNA molecule. This is interpreted by the DNA repair machinery of the cell as a broken chromosome and promotes homologous recombination. In some events, the 3’ and 5’ flanking regions of the construct, which were cloned from regions flanking endogenous TEP1, will recombine with their endogenous cognate sequences. The result of this double-cross over should be integration of the modified TEP1 gene and associated puromycin-resistance cassette in place of the endogenous TEP1 gene. We will cross this F1 progeny to wild-type mosquitoes. The progeny of this new cross will be sorted with the COPAS machine to eliminate all larvae that have inherited GFP fluorescence, linked to the donor locus from which the linear donor DNA molecule was excised. This step is intended to eliminate any donor locus (along with its puromycin resistance) that escaped enzymatic action and wasn’t excised. The non-GFP fluorescent larvae will be kept and grown in the presence of puromycin. Survivors must have integrated the antibiotic selection cassette at a new genomic location, distinct from the discarded GFP locus. By PCR using one primer binding in the synthetic construct and one binding in the target locus, we will screen for mosquitoes in which the engineered sequence has integrated into the intended target locus. Their progeny will be saved to establish new homozygous lines, using high puromycin concentration for selection of homozygotes in F2 generations. Deliverables from Task 4: - assessment of the frequency of the desired targeting event (Month 14) - a better knowledge of FLP and I-SceI activity in the mosquito (Month 14) - engineered mosquito lines with their endogenous TEP1 gene replaced by modified versions of the gene (Month 14) 3.1.5 TACHE 5 / TASK 5 : CHARACTERIZATION OF MOSQUITOES RESULTING FROM GENE EXCHANGE RECOMBINATION Successful exchange of the TEP1 gene will provide us with sister mosquito lines for further functional analyses: (i) wild-type line with the susceptible TEP1 allele, puromycin-sensitive; (ii) line differing from the wild-type only at the TEP1 locus and puromycin-resistant. We will test for the susceptibility / refractoriness phenotypes of the sister lines by counting parasites in individual mosquitoes that were offered a blood meal on a mouse infected with P. berghei. If the genetically modified mosquitoes are more resistant, it will be a proof that polymorphism in the TEP1 gene is sufficient to explain differences in susceptibilities among mosquito strains. If the modified mosquitoes only show a minor or no difference in resistance to the sister line, it will mean that polymorphism at TEP1 is associated with polymorphisms in other genes and that the susceptibility/refractoriness character is determined by combinations of allelic variants in a number of genes. Engineered mosquitoes obtained from donor construct B (TEP1 fused to RFP) should, in addition, allow live imaging studies that will provide new spatial and timing information on the TEP1-mediated parasite killing process. Finally, we will extend this analysis to P. falciparum in collaboration with E. L. ANR-GUI-AAP-04 – Doc Scientifique 2011 13/26 PROGRAMME JCJC EDITION 2011 Projet GEMM DOCUMENT SCIENTIFIQUE at the MPI in Berlin. These experiments will open up new perspectives for analysis of quantitative trait genes identified by forward genetic approaches. Resukts of these experiments should yield at least one paper in an important journal. Deliverables from Task 5: - Data on the sensititivity to Plasmodium of mosquito lines differing in the allelic forms of TEP1 (Month 26). 3.1.6 TACHE 6 ET 7 / TASK 6 AND 7: OBTENTION OF SYNTHETIC TRANSCRIPTION FACTORS We will build a synthetic transcription factor (TF) able to bind specifically the promoter of the antiparasitic genes TEP1, LRIM1 and APL1-C in collaboration with the coordinator’s former lab at the university of Halle, Germany. A region of homology common to all three promoters has been identified, and will determine the succession of nucleotide-specific tetratricopeptide repeats in the future TF. Repeats are available for specifically recognizing A, T, G, or C nucleotides. In addition, repeats binding either of any two nucleotides, and repeats able to bind all 4 nucleotides, are also available. This flexibility will help decide on a succession of repeats able to bind specifically the homologous region of the three promoters at once, in spite of some nucleotide differences between the three. For repeat assembly into a TF “backbone”, the Halle group will use their unpublished, rapid and efficient repeat-assembly protocol. The transcription activation domain (TAD) of the TF will be picked among a library of TADs, that includes Gal4, VP16 and the type III effector’s own TAD. In addition, we want to try the well-characterized TAD from the Drosophila HeatShock Factor (HSF). HSF also exists in A. gambiae, but the sequence of its TAD appears to be missing from the current genome release. Since TADs use extremely conserved interactions with the transcription machinery, we expect the Drosophila TAD to work well in other Dipteran insects including mosquitoes. A. gambiae TADs may be tested as well, as soon as some become characterized in detail. Thus, our custom DNA-binding domain will be combined with various TADs and transgenic mosquito lines will be established for each. To drive TF expression, we will clone the constitutive promoter of a prophenoloxidase gene, to ensure proper expression of the target antiparasitic factors in immune-competent hemocytes. To assemble a construct containing promoter, TF and a selection marker for transgenesis, we will use the Multisite Gateway system. The promoter must be cloned in entry plasmid pENTR L1-L4, the TF, once received from Halle, in a modified version of pENTR R4-R3 made compatible with the repeat-assembly protocol (needs to be built), and the selection marker in pENTR L3-L2 (already available). The Multisite Gateway LR reaction will assemble all three inserts from the entry plasmids into the appropriate attB-containing transgenesis vector (already available). The preparation of all necessary plasmids constitutes task 6. In Task 7, before generating transgenic mosquito lines, a cultured mosquito cell line resembling hemocytes (the Sua5* cells) that we maintain in our laboratory will be transfected with a mix of TF-encoding plasmid and a TEP1 promoter-RFP reporter plasmid. TFs with an active TAD should increase the level of red fluorescence upon transfection. This experiment, may allow choosing the best TAD in vitro even before making transgenic mosquitoes. Deliverables from Task 6-7: - Plasmids encoding synthetic, mosquito promoter-directed transcription factors (Month 2) - Data on the activity of synthetic TFs and of several TADs in mosquito cells (Month 7) ANR-GUI-AAP-04 – Doc Scientifique 2011 14/26 PROGRAMME JCJC EDITION 2011 Projet GEMM DOCUMENT SCIENTIFIQUE 3.1.7 TACHE 8 AND 9 / TASK 8 AND 9 : CHARACTERIZATION OF TRANSGENIC MOSQUITOES CARRYING SYNTHETHIC TRANSCRIPTION FACTORS Obtaining transgenic mosquitoes (using our attP docking lines) will be task 8. In task 9, we will characterize them by testing for expression of the synthetic TF and of its intended endogenous targets TEP1, LRIM1 and APL1-C simultaneously by qRT-PCR, comparing with wild-type mosquito controls. If an elevation in the expression of the antiparasitic genes is confirmed, resistance to P. berghei will be immediately evaluated in an infection assay, which we routinely perform in our insectary at IBMC. We expect the transgenic mosquitoes to show increased resistance, or even complete refractoriness, to P. berghei. If this is confirmed, the mosquito line will be examined by the laboratory of E.L. in Berlin, for evaluation of the mosquito resistance to P. falciparum. Risk and contingency plan: Should our synthetic TFs have no effect on the transcription of target genes, we will troubleshoot them using transfected mosquito cells until finding a working combination of DNA binding domain and TAD. In case some TFs prove to be unable to activate transcription of the target genes, we will examine the possibility that they even block normal gene expression because of their DNAbinding domain occupying and blocking the promoter region. We might then dedicate some time studying the possibility to block the expression of chosen genes (such as pro-parasitic genes) with AvrBs3-like DNA binding domains lacking a TAD. Deliverables from Task 8 and 9: - transgenic mosquitoes expressing immunity-promoting synthetic TFs (Month 14) - data on the resistance phenotype of mosquitoes with synthetically induced genes (Month 26) - proof-of-concept that the novel family of Xanthomonas TFs can be harnessed for genetic engineering applications (Month 26) 3.1.8 TACHE 10 / TASK 10: EVALUATION OF THE SPECIFICITY OF SYNTHETIC TFS Once we obtained mosquitoes expressing a synthetic TF that drives expression of its target genes, we will look for potential off-target affected genes. For this purpose, we will extract mosquito RNA samples at various stages of development and hybridize them to microarrays prepared using the latest release of the mosquito transcriptome. This work will be done in collaboration with the microarray platform at IGBMC, Illkirch. Deliverables from Task 10: - Evaluation of the number of endogenous genes whose expression is inadvertently affected by a given synthetic TF, providing data on how specific for a target gene a synthetic TF might be Month 26) 3.1.9 TACHE 11 / TASK 11: EXPERIMENTS IN SEMI-FIELD CONDITIONS Towards the end of the project, we will be ready to test the fitness of several of the transgenic mosquito lines obtained in the course of the project in a semi-natural environment. As part of ANR-GUI-AAP-04 – Doc Scientifique 2011 15/26 PROGRAMME JCJC EDITION 2011 Projet GEMM DOCUMENT SCIENTIFIQUE the INFRAVEC consortium funded by the EU program FP7, our laboratory will have access to a confined habitat reproducing the natural mosquito environment at the University of Peruggia, Italy, in collaboration with the laboratory of Prof. A. Crisanti. In these large cages, we will release mixes of large numbers of wild-type A. gambiae, “empty” docking attP lines (to see whether the fitness of “empty” attP lines is already altered compared to wild-type), mosquito line B arising from Goal1 of this project (in which susceptible TEP1 was replaced with the refractory allele), and a mosquito line expressing an immune gene-activating TF arising from Goal 2. After 10 mosquito generations (about 6 months), and if possible at later time points as well, we will sample the surviving mosquitoes and measure the frequency of each of the loci of interest. This experiment will enable us to deduce whether the transgenes confer any fitness advantage or disadvantage when transgenic mosquitoes co-inhabit with the wild-type in nearly natural conditions. These data will be valuable for the design of future transgenic mosquito release programs. Deliverables form Task 11: - Data on the dynamics of transgenes in semi-field conditions (Month 36) 3.1.10 TACHE 12 / TASK 12: VALORIZATION At any time during the project, when accumulated data become sufficient to assemble a coherent paper publishable in a high impact journal, we will start the redaction of manuscripts. We hope to publish at least two large articles describing comprehensively the results of Goal 1 and Goal 2 of the proposal, unless we then find more appropriate to subdivide into more, smaller articles describing intermediate advances. We believe that the proposal has a reasonable potential for several breakthrough papers. In addition, we will consider patenting appropriate major technical advances. The results of the work will be disseminated through oral and poster presentations at national and international meetings and conferences, and during public debates to inform the public at large on the advantages and safety issues of the transgenic mosquito technology. Once established, the protocols for homologous recombination and synthetic TFs will be made public and a workshop will be organized with the aim to share the expertise with colleagues working in vector biology. Deliverables from Task 12: - Publications (at least 2 comprehensive papers, or more smaller papers) (Month 36) ANR-GUI-AAP-04 – Doc Scientifique 2011 16/26 PROGRAMME JCJC Projet GEMM EDITION 2011 DOCUMENT SCIENTIFIQUE 3.2. CALENDRIER DES TACHES, LIVRABLES ET JALONS / TASKS SCHEDULE, DELIVERABLES AND MILESTONES Task 2 : establish enzyme transgenic line Task 4 : selection of recombinant mosquito lines Task 3 : establish donor lines A, B Task 6 : cloning synthetic TFs Task 7 : test TF in transfected cells Task 8 : establish TF transgenic lines … Month 1-12 … Task 4 Task 11 : manuscript prepaparation … Task 8 Task 5 : characterize mosquitoes arising from the gene exchange procedure … Task 9 : characterize mosquitoes expressing recombinant transcription factors Task 10 : microarray analysis of TF specificty Month 13-24 … Task 5 … Task 9 … Task 10 Task 11 : Experiments in semi-field conditions Task 12 : manuscript preparation Month 25-36 ANR-GUI-AAP-04 – Doc Scientifique 2011 17/26 PROGRAMME JCJC Projet GEMM EDITION 2011 DOCUMENT SCIENTIFIQUE Table of Deliverables Deliverable Task number Expected date New transgenesis plasmid for I-SceI and FLP expression in the germline 2 Month 2 EM New enzyme-expressing mosquito line 2 Month 6 EM Evaluation of the activity of each enzyme within the germline of transgenic mosquitoes. 2 Month 9 EM Two donor mosquito transgenic lines that can also serve to study TEP1 mutants in vivo 3 Month 6 EM Assessment of the frequency of the desired targeting event 4 Month 14 EM Engineered mosquito lines with their endogenous TEP1 gene replaced by modified versions of the gene 4 Month 14 EM Better knowledge of FLP and I-SceI activity in the mosquito 4 Month 14 EM Data on the sensititivity to Plasmodium of mosquito lines with an engineered TEP1 gene 5 Month 26 EM Plasmids encoding synthetic, mosquito promoter-directed transcription factors 6 Month 2 EM Data on the activity of synthetic TFs and of several TADs in cells 7 Month 6 EM Transgenic mosquitoes synthetic TFs immunity-promoting 8 Month 14 EM Assessment of the resistance phenotype of mosquitoes with synthetically-upregulated immune genes 9 Month 26 EM proof-of-concept that the novel family of Xanthomonas TFs can be harnessed for genetic engineering applications Evaluation of how specific for a target gene a synthetic TF might be 9 Month 26 EM 10 Month 26 EM Data on the dynamics of transgenes in semi-field conditions 11 Month 36 EM Publications 12 Month 36 EM expressing Responsible Milestones Month 6 : I-SceI nuclease and FLP recombinase shown to be active in recombinant mosquitoes. If FLP recombinase is inactive, activation of corresponding contingency plan (redesign constructs to use Cre recombinase instead) Month 14 : Gene knockout/ exchange is established. Month 26 : Mosquito lines with an exchanged TEP1 gene fully characterized. Month 14 : Synthetic transcription factors shown to be active in mosquitoes. Month 26 : Phenotype of synthetic TF-carrying mosquitoes fully analyzed. ANR-GUI-AAP-04 – Doc Scientifique 2011 18/26 PROGRAMME JCJC EDITION 2011 Projet GEMM DOCUMENT SCIENTIFIQUE 4. STRATEGIE DE VALORISATION, DE PROTECTION ET D’EXPLOITATION DES RESULTATS / DISSEMINATION AND EXPLOITATION OF RESULTS, INTELLECTUAL PROPERTY Since our project is of basic research, the first expected outcome is the publication of our research in highly ranked journals. We hope to publish at least two large articles describing comprehensively the results of Goal 1 and Goal 2 of the proposal, unless we then find more appropriate to subdivide into more, smaller articles describing intermediate advances. We believe that the proposal has a reasonable potential for several breakthrough papers. In addition, we will consider patenting major technical advances where appropriate. The results of the work will be disseminated through oral and poster presentations at national and international meetings and conferences, and during public debates to inform the public at large on the advantages and safety issues of the transgenic mosquito technology. Once established, the protocols for homologous recombination and synthetic TFs will be made public and a workshop will be organized with the aim to share the expertise with colleagues working in vector biology, particularly in an endeavor to transfer our technology to vector mosquitoes transmitting viral diseases (dengue, chikungunya…). From then on, we expect this work to have a major scientific impact in the field of vector biology. We hope this work will contribute to medical and socio-economical progress in regions plagued with malaria or mosquito-borne viral diseases. One way this could be achieved is through the development of well-designed, well-tested vector control strategies involving genetic engineering. Such interventions would require vast campaigns of public information. Another way to approach this goal is by providing a better understanding of the biology of mosquito-parasite interactions thanks to laboratory analyses of genetically engineered mosquitoes. The knowledge gained could then be used to orient vector control measures in targeted and efficient interventions. 5. DESCRIPTION DU PARTENARIAT / CONSORTIUM DESCRIPTION 5.1. DESCRIPTION, ADEQUATION ET COMPLEMENTARITE DES PARTENAIRES / PARTNERS DESCRIPTION AND RELEVANCE, COMPLEMENTARITY This project does not involve a consortium. It only involves international collaborations with colleagues that do not request funding from this grant. 5.2. QUALIFICATION DU COORDINATEUR DE LA PROPOSITION DE PROJET/ QUALIFICATION OF THE PROPOSAL COORDINATOR I carried out my Ph.D. work at the University of Halle in Ulla Bonas’ laboratory, working on the project that is at the basis of Goal 2 of this proposal. I am deeply familiar with the Xanthomonas AvrBs3 protein and its transcription factor function, since I was the first to identify pepper plant genes induced by this TF of bacterial origin. I modified several domains of the protein (in particular, I substituted the endogenous TAD for that of VP16) to see the effect that modifications would have on target gene expression. When I left U.B.’s laboratory, I had cloned a 130-bp plant promoter fragment which I knew, from reporter gene assays, contained the DNA sequence recognized by AvrBs3. Now that this sequence has ANR-GUI-AAP-04 – Doc Scientifique 2011 19/26 PROGRAMME JCJC Projet GEMM EDITION 2011 DOCUMENT SCIENTIFIQUE been identified and the amino acid code conferring Avrs3 binding specificity to a given DNA sequence has been cracked, I am excited by the idea to resume engineering this protein and use its properties in an innovative and original way for mosquito biotechnology. Since obtaining my Ph.D. in 2002, I kept an excellent relationship with U. Bonas and with Jens Boch, the associate professor who will be directly involved in our collaboration. Before moving to the mosquito field of research, I spent 4 exciting years as a post-doc studying lipoproteins and morphogen gradient formation of the Wingless protein in Drosophila in the laboratory of Suzanne Eaton in Dresden. There, I developed innovative tools for transgenic RNAi in the fruit fly, which were instrumental in obtaining an important breakthrough (ref). This internship was a great opportunity to gain knowledge of Drosophila genetics and especially of the transgenesis technology, since I preferred (for the sake of time) to generate myself the transgenic flies I needed rather than using the available service. This expertise has helped me enormously to develop transgenesis in A. gambiae. Therefore, I am trained to successfully carry out Goal 1 of this project. Finally, it is worth mentioning that this proposal is coming at a turning point of my career, since I will be heading the Strasbourg mosquito research group while E. Levashina moves to the Max-Planck Institute of Infection Biology in Berlin to set up a P. falciparum facility at the end of 2011. She and I intend to take advantage of each other’s competences and of the facilities available in each institute to successfully carry out the collaborations described in Goal 1 and 2. 5.3. QUALIFICATION, ROLE ET IMPLICATION DES PARTICIPANTS / QUALIFICATION AND CONTRIBUTION OF EACH PARTNER Partenaire / partner Nom / Name Coordinateur/responsable Marois Autres membres Prénom / First name Eric Emploi actuel / Position Discipline / Personne. Rôle/Responsabilité dans la Field of mois* / proposition de projet/ Contribution to research Person.mo the proposal nth 4 lignes max CR1 Mosquito Immunity 30 General piloting ; Molecular cloning, mosquito transgenesis, training of students and post-doc, Plasmodium berghei infection assays, etc. Levashina Elena DR2 Mosquito Immunity 3.6 Expertise in TEP1 allelic forms and P. falciparum infections Soichot Technicien Mosquito rearing 7.2 Mosquito rearing and transgenesis Julien * à renseigner par rapport à la durée totale du projet 6. JUSTIFICATION SCIENTIFIQUE DES MOYENS DEMANDES / SCIENTIFIC JUSTIFICATION OF REQUESTED RESSOURCES • Équipement / Equipment Our laboratory is well furbished and we request essentially salary money, but we would like to budget 15,000 € in case we need to participate in replacing a piece of equipment (centrifuges, incubators, PCR machine…) or decide to buy a new one. ANR-GUI-AAP-04 – Doc Scientifique 2011 20/26 PROGRAMME JCJC EDITION 2011 Projet GEMM DOCUMENT SCIENTIFIQUE • Personnel / Staff Hiring a full-time post-doctoral fellow (36 person.months) at a cost of 53,841 €/year results in 161,523 € for the total duration of the project. Six months after the project begins, the expected increase in the number of mosquito lines to be maintained demands hiring a second, full-time technician, hence 30 person.months at an average cost of 28,520 €/year=70625 €. Total salaries=232,148 € • Missions / Travel We plan 3000 € /year for travels of the coordinator and of the post-doctoral fellow to our collaborator’s laboratories for P. falciparum work, to meetings, workshops and conferences. • Dépenses justifiées sur une procédure de facturation interne / Costs justified by internal invoices - Microarrays (600 EUR/chip) : we expect to need 10 chips, 6000 €. - 3-year licence for the microarray analysis software Genespring : 9258 € - Kits for Multiste Gateway cloning, for qRT-PCR and molecular biology reagents and consumables : 3000 €per year - Mosquito rearing cages, containers and consumable: 3000 € per year - COPAS machine maintenance contract : 4000 € per year Hence a total of 45,258 € for 3 years 7. ANNEXES / ANNEXES 7.1. REFERENCES BIBLIOGRAPHIQUES / REFERENCES 1. WHO, World Malaria Report 2010. http://whqlibdoc.who.int/publications/2010/9789241564106_eng.pdf 2. Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. (2010) Refinement of tools for targeted gene expression in Drosophila. Genetics 186(2):73555. 3. Nolan T., Papathanos P. Windbichler N., Magnusson K., Benton J., Catteruccia F., Crisanti A. (2010). Developing transgenic Anopheles mosquitoes for the sterile insect technique. Genetica Sep 7. [Epub ahead of print] 4. Groth, A.C., Fish, M., Nusse, R., Calos, M.P. (2004) Construction of Transgenic Drosophila by Using the Site-Specific Integrase From Phage C31. Genetics 166:17751782 5. Bischof J, Maeda RK, Hediger M, Karch F, Basler K. (2007) An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 104(9):3312-7. 6. Venken KJ, He Y, Hoskins RA, Bellen HJ. (2006) P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314(5806):1747-51. Epub 2006 Nov 30. 7. Rong YS, Golic KG. (2000) Gene targeting by homologous recombination in Drosophila. Science 288(5473):2013-8. ANR-GUI-AAP-04 – Doc Scientifique 2011 21/26 PROGRAMME JCJC EDITION 2011 Projet GEMM DOCUMENT SCIENTIFIQUE 8. Rong YS, Golic KG. (2001) A targeted gene knockout in Drosophila. Genetics 157(3):1307-12. 9. Blandin S, Shiao SH, Moita LF, Janse CJ, Waters AP, Kafatos FC, Levashina EA. (2004) Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 116(5):661-70. 10. Volohonsky G, Steinert S, Levashina EA. (2010) Focusing on complement in the antiparasitic defense of mosquitoes.Trends Parasitol. 2010 26(1):1-3. 11. Marois E., Van den Ackerveken G., Bonas U. (2002) The Xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Mol. Plant Microbe Interact. 15:637-46 12. Szurek B., Marois E., Bonas U., Van den Ackerveken G. (2001) Eukaryotic features of the Xanthomonas type III effector AvrBs3: protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. Plant J. 5:523-34. 13. Kay S, Hahn S, Marois E, Hause G, Bonas U. (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318(5850):648-51. 14. Kay S, Hahn S, Marois E, Wieduwild R, Bonas U. (2009) Detailed analysis of the DNA recognition motifs of the Xanthomonas type III effectors AvrBs3 and AvrBs3Deltarep16. Plant J. 59(6):859-71. 15. Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009 Dec 11;326(5959):1509-12 16. Sera T. (2009) Zinc-finger-based artificial transcription factors and their applications. Adv Drug Deliv Rev.61(7-8):513-26. 17. Fraiture M, Baxter RH, Steinert S, Chelliah Y, Frolet C, Quispe-Tintaya W, Hoffmann JA, Blandin SA, Levashina EA. (2009) Two mosquito LRR proteins function as complement control factors in the TEP1-mediated killing of Plasmodium. Cell Host Microbe 19;5(3):273-84. 18. Povelones M, Waterhouse RM, Kafatos FC, Christophides GK. (2009) Leucine-rich repeat protein complex activates mosquito complement in defense against Plasmodium parasites. Science 324(5924):258-61. 19. Rono, M., Whitten, M., Oulad-Abdelghani, M., Levashina, E., and Marois, E. (2010) The Major Yolk Protein Vitellogenin Interferes with the Anti-Plasmodium Response in the Malaria Mosquito Anopheles gambiae. PLoS Biology 8(7):e1000434. 20. Papathanos PA, Windbichler N, Menichelli M, Burt A, Crisanti A. (2009) The vasa regulatory region mediates germline expression and maternal transmission of proteins in the malaria mosquito Anopheles gambiae: a versatile tool for genetic control strategies. BMC Mol Biol. 2009 Jul 2;10:65 ANR-GUI-AAP-04 – Doc Scientifique 2011 22/26 Article 1: 2000 Cell Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Nimchuk Z.*, Marois E.*, Kjemtrup S., Leister R.T., Katagiri F., Dangl J.L. *co-­‐first authors Cell, Vol. 101, 353–363, May 12, 2000, Copyright ©2000 by Cell Press Eukaryotic Fatty Acylation Drives Plasma Membrane Targeting and Enhances Function of Several Type III Effector Proteins from Pseudomonas syringae Zachary Nimchuk,1,6 Eric Marois,1,6,7 Susanne Kjemtrup,1,6 R. Todd Leister,3 Fumiaki Katagiri,3,4 and Jeffery L. Dangl1,2,5 1 Department of Biology 2 Curriculum in Genetics and Molecular Biology University of North Carolina at Chapel Hill Chapel Hill, North Carolina 27599 3 Department of Biological Sciences University of Maryland Baltimore, Maryland 21250 4 Novartis Agricultural Discovery Institute, Inc. 3115 Merryfield Row San Diego, California 92121 Summary Bacterial pathogens of plants and animals utilize conserved type III delivery systems to traffic effector proteins into host cells. Plant innate immune systems evolved disease resistance (R) genes to recognize some type III effectors, termed avirulence (Avr) proteins. On disease-susceptible (r) plants, Avr proteins can contribute to pathogen virulence. We demonstrate that several type III effectors from Pseudomonas syringae are targeted to the host plasma membrane and that efficient membrane association enhances function. Efficient localization of three Avr proteins requires consensus myristoylation sites, and Avr proteins can be myristoylated inside the host cell. These prokaryotic type III effectors thus utilize a eukaryotespecific posttranslational modification to access the subcellular compartment where they function. Introduction Plants can specifically resist infection by bacterial pathogens through the interaction of host resistance (R) genes and pathogen avirulence (avr) genes. The simplest mechanistic interpretation of these genetic systems is that Avr and R proteins interact directly, although this has been difficult to generalize experimentally (see Scofield et al., 1996; Tang et al., 1996 for the exception). Interaction of Avr and R proteins, potentially in a multiprotein complex, results in disease resistance responses characterized by a suite of biochemical events often culminating in both host cell death (hypersensitive response, HR) at the site of infection and cessation of pathogen growth (Yang et al., 1997; Scheel, 1998). If alternate alleles of either the R or avr genes are expressed during this interaction, then there is no recognition and successful infection of the plant by the bacteria ensues. 5 To whom correspondence should be addressed (e-mail: dangl@ email.unc.edu). 6 These authors contributed equally to this work. 7 Present address: Institute of Genetics, Martin Luther University, Halle D-06120, Germany. It is puzzling that phytopathogens express Avr proteins which can condition the pathogen’s demise. Some avr genes contribute to successful infections on susceptible hosts, ensuring a continued advantage for bacteria containing them (Kearney and Staskawicz, 1990; Lorang et al., 1994). For example, avrRpm1 from Pseudomonas syringae pv. maculicola strain M2 (PsmM2) is a virulence factor. PsmM2 requires avrRpm1 to grow optimally on Arabidopsis plants lacking the corresponding RPM1 R gene (Ritter and Dangl, 1995). It is generally true that a given avr gene is not widely dispersed among isolates of Pseudomonads. This suggests that a battery of genes, dispersed among pathogen isolates, contributes quantitatively to the virulence of any given strain. For example, avrRpm1 is present in only 5 of 20 P. syringae pv. maculicola strains analyzed (Dangl et al., 1992), yet strains that lack it are still pathogenic. Bacterial avr genes are part of the hrp (hypersensitive response and pathogenicity) regulon (Huynh et al., 1989; reviewed by Alfano and Collmer, 1997). In P. syringae, this regulon encodes linked transcriptional regulators and the structural proteins for an evolutionarily conserved type III secretion apparatus. Bacterial virulence factors that modulate or usurp host mammalian cell functions are trafficked to the interior of host cells via the type III pilus. These type III effectors have targets inside eukaryotic host cells (see Cornelis and Wolf-Watz, 1997; Galan and Collmer, 1999 for reviews). Both Avr proteins and known type III effectors from animal pathogens can be secreted from phytopathogenic bacterial cells in a type III–dependent manner (Anderson et al., 1999; Rossier et al., 1999). Thus, Avr proteins are type III effector proteins. Avr-R recognition can occur inside the plant cell. Following expression of a bacterial avr gene using plant transcriptional control signals, Avr proteins can elicit an HR-like cell death in plant cells expressing the appropriate R gene (reviewed in Mudgett and Staskawicz, 1998). Curiously, expression of Avr proteins in diseasesusceptible plants can lead to delayed, weak cytotoxic effects, suggesting that Avr proteins may have additional targets inside the plant cell (Gopalan et al., 1996; McNellis et al., 1998). Based on analogy to mammalian pathosystems, we and others infer that type III effectors from phytopathogens are translocated into the host cell, although direct demonstrations of this are lacking. For example, a cleaved form of the P. syringae AvrRpt2 protein is detected inside plant cells and not in bacterial lysates following type III–dependent delivery (Mudgett and Staskawicz, 1999). This result argues strongly for delivery of AvrRpt2 into the plant cell before, or concomitant with, its proteolytic cleavage. Despite these recent advances, little is known about the subcellular localization, and hence site of action, of the phytopathogen type III effector proteins inside the plant cell. The related Xanthomonas proteins AvrBs3 and PthA have nuclear localization sequences that are required for both avirulence function when delivered in a type III–dependent manner, and for nuclear localization following expression inside the plant cell (Yang and Gabriel, 1995; Van den Cell 354 Table 1. P. syringae Avirulence Genes Contain N-Terminal Consensus Eukaryotic Fatty Acylation Sequences 123456789 avrRpm1 avrB avrPphBa MGCVSSTSR MGCVSSKST GCASSGVS SOS3 Ca sensor MGCSVSKKK avrC avrPto MGNVCFRPS MGNICVGGS CPK1 MGNTCVGPS References for each sequence are (top to bottom): Dangl et al., 1992; Tamaki et al., 1988; Jenner et al., 1991; J.-K. Zhu, personal communication; Tamaki et al., 1988; Salmeron and Staskawicz, 1993; Ellard-Ivey et al., 1999. a Represents N terminus of processed AvrPphB protein as determined by Puri et al., 1997. Ackerveken et al., 1996). Presumably, these Avr proteins interact with host nuclear factors, potentially influencing host defense gene transcription (Zhu et al., 1999). We noted a subset of P. syringae type III effectors (Table 1) with predicted N-terminal eukaryotic consensus sequences for fatty acylation, modifications that promote plasma membrane association. This subset includes both AvrRpm1 and AvrB from P. syringae pv. maculicola and P. syringae pv. glycinea, respectively, which are recognized in Arabidopsis by RPM1 (Bisgrove et al., 1994; Grant et al., 1995). AvrRpm1 and AvrB share no homology other than this N-terminal sequence, including the G2 residue known to be the target for covalent myristoylation (a C14:0 acyl group; Johnson et al., 1994). An apparent exception to the G2 rule is the AvrPphB protein from P. syringae pv. phaseolicola. AvrPphB expresses a glycine not at its translational N terminus, but rather at the N terminus of an intramolecular cleavage product (Puri et al., 1997). Myristoylation often occurs in conjunction with palmitoylation, a C16:0 lipid attachment at C3 or C5 (Resh, 1994), and the proteins in Table 1 feature this residue. Both fatty acylation events are specific to eukaryotes, notably G␣ subunits and Src family tyrosine kinases (Johnson et al., 1994; Resh, 1994). Arabidopsis proteins in Table 1 use these N-terminal sequences as acylation sites (Ellard-Ivey et al., 1999; J. K. Zhu, personal communication). Thus, plant consensus acylation sites are typical of those from other eukaryotes. We addressed whether or not these amino acid residues are important for Avr protein function, and whether these pathogen effectors are targeted to the host plasma membrane. RPM1 is enriched in plasma membrane vesicles from plant cells (Boyes et al., 1998), making it likely that the Avr proteins it recognizes would also localize there. Results Consensus Eukaryotic Fatty Acylation Sites Mediate Type III–Dependent Delivery of AvrRpm1 and AvrB Function We introduced site-specific alanine exchanges G2A, C3A, S5A, and S6A into both AvrRpm1 and AvrB and expressed these from the native avrRpm1 promoter, Figure 1. Expression of Native and HA Epitope–Tagged AvrRpm1 and AvrB in P. syringae DC3000 Extracts were prepared from Pst DC3000 expressing empty vector (V) or the wild-type (wt) and mutant Avr proteins indicated at top. Duplicate blots were probed with either native antisera to AvrRpm1 or AvrB or the monoclonal anti-HA antibody, as listed on the right. Lanes were equally loaded. Top two blots: AvrRpm1, bottom two blots: AvrB. with or without a C-terminal HA epitope tag (Experimental Procedures). We assessed whether these exchanges affected protein production in P. syringae pv. tomato (Pst) DC3000. Figure 1 demonstrates that either antisera to each Avr protein, or the anti-HA epitope monoclonal antibody, recognized proteins of the correct apparent molecular weight (AvrRpm1 at 29 kDa, AvrB at 36 kDa) that are not present in bacterial extracts made from cells carrying an empty vector. The G2A mutant of AvrRpm1 accumulated to variably lower levels than wild type, but we did not observe similar decreases for the AvrB G2A mutant protein. The C3A exchanges in either gene were as stable as wild type. Surprisingly, S6A exchange in either AvrRpm1 or AvrB significantly reduced protein accumulation, and interpretation of subsequent functional data must bear this in mind. Structural signals comprising at least 15 codons mediate type III–dependent secretion or delivery of effector proteins into host cells (Anderson et al., 1999). We expressed wild-type and mutant proteins in an E. coli strain expressing either a wild-type or mutant Erwinia chrysanthemi type III secretion system previously used to monitor AvrB secretion (Ham et al., 1998). We monitored hrp-dependent AvrB secretion and found that wild-type and mutant proteins were secreted equally in an hrp-dependent manner (data not shown). However, we were unable to observe secretion of wild-type AvrRpm1, consistent with the observation that different type III effectors are secreted with different efficiencies (e.g., Ham et al., 1998). We tested delivery of AvrRpm1 or AvrB avirulence function to an Arabidopsis accession (inbred line), Col-0, which expresses RPM1. We monitored in planta growth of strains expressing the various avirulence protein derivatives (Figure 2A, top). Virulent Pst DC3000 carrying an empty vector grow ⵑ1000-fold over three days. Expression of wild-type avrRpm1 or avrB in Pst DC3000 decreases pathogen growth by ⵑ100 fold. This reflects recognition of AvrRpm1 or AvrB via RPM1. In contrast, the G2A derivative of either AvrRpm1 or AvrB is not recognized efficiently by the host, and pathogen growth is unhindered. The S6A mutation eliminated avirulence, but interpretation of this data is compromised by the diminished levels of S6A accumulation noted above. We also monitored the onset of HR following Host Modification of Prokaryotic Virulence Factors 355 conclude from Figure 2A that the consensus myristoylation sites of both AvrRpm1 and AvrB are required for full avirulence function following type III–mediated delivery to Arabidopsis. Several P. syringae type III effector proteins, including AvrRpm1, serve as virulence factors during infection of plant genotypes lacking the appropriate R gene product. Loss of function Col-0 rpm1 mutant alleles exist, and several Arabidopsis accessions have a naturally occurring deletion allele (rpm1 null; Grant et al., 1995). PsmM2 is pathogenic on rpm1 null Arabidopsis accessions like Mt-0, Fe-1, and Cvi-0. A Tn3spice insertion into avrRpm1 in Psm M2 (giving rise to strain CR299) decreased virulence in a dose-dependent manner (Ritter and Dangl, 1995). The essence of this finding is displayed in Figure 2B, left, where expression of wild-type avrRpm1 rescues CR299 to full virulence on rpm1 nulls Mt-0 and Fe-1. Figure 2B demonstrates that both G2A and C3A exchanges significantly reduce the virulence function of wild-type avrRpm1 as measured by CR299 growth. We conclude from this experiment that both myristoylation and palmitoylation consensus sites are important for maximal AvrRpm1 virulence function when delivered via the type III system to Arabidopsis. Similar experiments with AvrB were not performed, as no obvious virulence activity has been ascribed to AvrB on Arabidopsis. Figure 2. Maximal Type III–Dependent AvrRpm1 and AvrB Effector Functions Are Mediated by Consensus Acylation Sites (A) The G2A and C3A exchanges reduce the avirulence functions of AvrRpm1 and AvrB. Col-0 (RPM1) leaves were inoculated with Pst DC3000 carrying either empty vector (V), the wild-type (wt) or mutant avr derivatives as listed on the x axis (initial inoculum of ⵑ1 ⫻ 105 cfu/ml). Bacterial titers three days post inoculation (dpi) are graphed on the y axis. Mean and standard deviation from 3 independent experiments. Day 0 titers ranged from log10 ⫽ 2.5–3.2. The number of HR⫹ leaves/total inoculated leaves scored at 5 hr post inoculation (hpi) using high initial inoculum (OD600 ⫽ 0.05 for AvrB and OD600 ⫽ 0.1 for AvrRpm1) is displayed below the corresponding growth data. Data are summed from three different experiments. (B) The G2A and C3A exchanges reduce the virulence function of AvrRpm1. In planta growth assay on rpm1 null accessions Mt-0 and Fe-1 plants as in (A) but with an initial inoculum Psm CR299 of 1.0 ⫻ 103 cfu/ml. Mean and standard deviation from four independent experiments at 3 dpi Student’s t test for significance of differences to vector control was p ⬍ 0.1 (*) and p ⬍ 0.15 (**). inoculation of a high dose of P. syringae expressing the various avirulence proteins. Expression of either wildtype avrRpm1 or avrB in Pst DC3000 triggers HR on Col-0 at 5 hr post inoculation, while empty vector did not (Figure 2A, bottom). G2A exchange in either AvrRpm1 or AvrB significantly lowers the percentage of leaves responding. While the effects of G2A exchanges are not complete in this assay, they are at the lower titers used for in planta growth. Additionally, C3A exchange in AvrB and AvrRpm1 reproducibly resulted in both fewer responding leaves, and a slightly delayed response. We Consensus AvrRpm1 and AvrB Acylation Sites Are Required for Maximal RPM1 Function by Following avr Expression Inside Host Cells Our genetic experiments suggested that acylation of the AvrRpm1 and AvrB type III effector proteins might mediate their function. Unfortunately, it has proven impossible to directly detect type III–dependent delivery of effector proteins from plant pathogenic bacteria to plant host cells. However, expression of putative type III effector proteins like AvrRpm1 an AvrB inside RPM1 accessions like Col-0 results in an HR-like response (Gopalan et al., 1996; Leister et al., 1996). We used Agrobacterium to deliver the avr genes and a dexamethasone (DEX)-inducible vector system to conditionally express them from the transferred T-DNA (Aoyama and Chua, 1997; Experimental Procedures). If mutant derivatives of AvrRpm1 and AvrB unable to trigger RPM1-dependent resistance when delivered from P. syringae also proved unable to initiate an RPM1-dependent response when delivered from Agrobacterium, then two conclusions could be drawn: first, that the mutant phenotypes were unlikely to be a consequence of altered delivery via the P. syringae type III system and second, that localization of Avr proteins delivered via Agrobacterium should reflect the natural localization during P. syringae infection. Figure 3A demonstrates that DEX-induced transient expression of either AvrRpm1 or AvrB initiates an RPM1dependent response. We used two sets of isogenic plants for this experiment: first, wild-type Col-0 (RPM1) and an isogenic loss-of-function rpm1-fs allele, and second, the rpm1 null accession Fe-1 and a transgenic Fe-1 expressing RPM1. Figure 3B illustrates that G2A exchange in either Avr protein significantly reduced the ability to trigger an RPM1-dependent response. Avr protein levels in these leaves, however, were undetect- Cell 356 able (not shown). Our inability to detect Avr protein in this assay suggests that low Avr protein levels are sufficient to trigger RPM1-dependent cell death. DEX treatment does not induce Avr protein accumulation in cultured Agrobacterium (see Experimental Procedures). The Consensus Myristoylation Site Mediates Maximal rpm1-Independent Cytotoxicity Triggered by AvrB Expression Inside Host Cells We assayed a series of rpm1 loss-of-function or null alleles using the same expression system. We reasoned that Avr proteins might be sufficient to trigger a cellular response indicative of their virulence function, based on the slow cytotoxicity indicative of type III effector action in susceptible animal cells (see Introduction). Figure 4A demonstrates that AvrB is sufficient to trigger chlorosis mediated by a slow cytotoxic response in the absence of RPM1 (measured by Trypan blue uptake, not shown). Surprisingly, these responses are polymorphic with respect to host genotype. Thus, AvrB expression initiates a chlorotic response on all accessions tested (including Nd-0; see Gopalan et al., 1996) except the rpm1 null Cvi-0. AvrRpm1 can sometimes trigger a response in the rpm1 null accessions Mt-0 and Aa-0, but this phenotype has proven unreliable and will not be discussed further. The fact that AvrRpm1 and AvrB protein both accumulated in Cvi-0 (see below) argues against general cytotoxicity as the cause of the rpm1independent response. Response to AvrB in a cross between the Col-0 rpm1-fs allele and Cvi-0 indicates either one or two host genes segregating which control this trait (F1: 8/8 responding; F2: 154 responding, 36 not responding, 2 for 3:1 ⫽ 3.42, p ⫽ 0.05; 2 for 13:3 ⫽ 0.0, p ⫽ 0). We mapped a locus near the chromosome 5 marker SPL2 which controls this response (linkage ⫽ 14.7 map units; Z. N. and J. L. D., unpublished). Note that RPM1 maps to chromosome 3, proving that this response is not due to residual RPM1 activity in the rpm1-fs allele. While we cannot exclude a weak R gene effect as an explanation of this phenotype, preliminary transcriptional profiling suggests that the rpm1-independent response is not related to the RPM1-dependent response (Z. N. et al., unpublished). Figure 4B demonstrates that both AvrRpm1 and AvrB accumulate in a DEX-dependent manner in Mt-0 and Fe-1 leaves. Figure 4C demonstrates that the rpm1independent response to AvrB is abolished by G2A exchange. AvrB protein levels in plants expressing rpm1-independent phenotypes decline rapidly, preceding onset of the visible phenotype. However, Figure 4D demonstrates that the phenotypic difference between wild-type and G2A exchange AvrB proteins is not due to differential intrinsic stability, at least over a 48 hr induction time course in the nonresponding Cvi-0 accession. We conclude that AvrB can consistently initiate a slow, rpm1-independent response in some, but not all, Arabidopsis genetic backgrounds. G2A exchange greatly diminishes this response, like the RPM1-dependent responses described above. Because this rpm1-independent response is also greatly enhanced by consensus acylation sites, we conclude that it reflects both a normal function and cellular localization of this type III effector protein. Figure 3. Myristoylation Sites Are Required for Maximal Avirulence Function inside Plant Cells (A) AvrRpm1 and AvrB initiate RPM1 action inside plant cells. Agrobacterium carrying the genes listed at right were inoculated at OD600 ⫽ 0.5 and leaves sprayed with 20 M DEX 48 hpi. RPM1dependent responses were photographed 24 hr post DEX treatment. Plant accessions listed across top are: Fe-1 (rpm1 null), Fe-1::RPM1 (transgenic Fe-1 expressing RPM1), rpm1-fs (a Col-0 mutant RPM1 allele), Col-0 (wild type, RPM1). (B) Consensus myristoylation sites greatly enhance AvrB and AvrRpm1 effector function inside the plant cell. Col-0 (RPM1) leaves were inoculated with Agrobacterium (OD600 ⫽ 0.4) carrying wild type (wt) or G2A mutant (top) of either avrRpm1 or avrB (listed at right). Experiment done as in (A). Numbers above or below each leaf refer to RPM1-dependent response positive leaves/total number inoculated; data pooled from four experiments. In all cases, native avr gene constructs gave identical phenotypes compared to HA epitope–tagged constructs. Host Modification of Prokaryotic Virulence Factors 357 Figure 4. Expression of AvrRpm1 and AvrB in rpm1 Plants (A) Definition of an RPM1-independent, polymorphic response to AvrB expression in a series of rpm1 null plants. Agrobacterium carrying the genes listed at left were inoculated into rpm1 null accessions Mt-0, Fe-1, or Cvi-0, or the rpm1-fs mutant listed across the top. DEX induction as in Figure 3A, but photographed at 72 hr post DEX treatment. GUS expression in planta was utilized as a control for transformation. (B) DEX-dependent expression of AvrRpm1 and AvrB results in detectable protein accumulation in rpm1 null plants. Protein extracts (10 g) taken from two leaf discs from either rpm1 null accessions Mt-0 or Fe-1, at 8 hr post DEX treatment, were subjected to SDS-PAGE and immunodetected using anti-HA monoclonal antibody. Molecular weight standards are marked at left. (C) Robust AvrB-induced host response requires G2. The rpm1 null accession Mt-0 was inoculated with Agrobacterium carrying vector, wild type (wt) or mutant avrB derivatives as listed on the top. Numbers below the leaves represent response ⫹/total inoculated pooled from three independent experiments. (D) Wild-type and G2A mutant AvrRpm1 and AvrB Are Localized to the Host Cell Plasma Membrane, and Efficient Localization Requires Consensus Acylation Sites Our results suggested that Avr protein localization dictates both RPM1-dependent responses to both AvrRpm1 and AvrB, and the rpm1-independent response to AvrB. We localized both HA-tagged Avr proteins in Cvi-0 (Figure 5A) and Mt-0 (not shown) at 12 hr post DEX induction. We chose this time point because wild-type AvrRpm1 was undetectable at later time points in all tested accessions (not shown), and wild-type AvrB was undetectable in Mt-0 beyond this time point. We collected total extracts and prepared soluble and 100,000 ⫻ g microsomal fractions after DEX induction (Experimental Procedures). The anti-HA epitope monoclonal antibody detected bands of the correct apparent molecular weight (AvrRpm1 at 29 kDa, AvrB at 36 kDa) almost exclusively in the microsomal membrane fraction (antisera to AvrRpm1 and AvrB confirmed these results, not shown). These bands are not present in extracts from empty vector controls. G2A or C3A exchange had significant effects on membrane localization of both AvrRpm1 and AvrB. First, G2A exchange essentially eliminated membrane localization. Second, the C3A exchange significantly reduced membrane association. These results are precisely those expected given the requirement for myristoylation to occur before palmitoylation, but not vice versa, in various dually acylated proteins (see Discussion). These data also demonstrate that the localization differences observed are not due to differential stability of the mutant proteins, at least at 12 hr post DEX induction. Antisera against the tonoplast membrane protein ␥-TIP served as a control for fractionation (Daniels et al., 1994). We performed two-phase membrane vesicle separation to determine if the plasma membrane contains wildtype AvrRpm1 and AvrB (Experimental Procedures). Western blots (Figure 5B) demonstrate that both Avr proteins were enriched in these vesicles to the same extent as a known plasma membrane marker. This is consistent with previous enrichment of RPM1 protein in plasma membrane vesicles (Boyes et al., 1998). Marker proteins for various subcellular membranes confirmed the two-phase separation efficiency (Experimental Procedures). We used an independent method to confirm these results. Wild type and G2A exchange derivatives of either AvrRpm1 or AvrB were fused with green fluorescence protein (GFP) at their carboxyl termini and expressed from the strong cauliflower mosaic virus 35S promoter in Arabidopsis rpm1 mutant protoplasts (see Experimental Procedures). Stacked laser confocal micrographs (Figure 5C) clearly demonstrate that the wildtype AvrRpm1 and AvrB proteins were enriched in the plasma membrane while their respective G2A derivatives localized like a nontargeted GFP control, mostly to the cytoplasm. AvrB proteins are equally stable. The rpm1 null accession Cvi-0 was inoculated with Agrobacterium carrying vector, wild-type (WT) or the G2A mutant avrB derivative as listed on the top. Total protein extracts (10 g) from two leaf discs were harvested at 12 hr, 24 hr, and 48 hr post DEX treatment, subjected to SDS-PAGE, and immunodetected using anti-HA monoclonal antibody. Similar results were seen in separate experiments. Cell 358 Myristoylation of AvrRpm1 and AvrB In Vivo Requires Consensus G2 Acylation Sites Our functional and plasma membrane localization data strongly support the contention that AvrRpm1 and AvrB are acylated, and thus tethered into the eukaryotic host plasma membrane. We tested myristoylation directly by radiolabeling leaves with [3H]myristic acid subsequent to Agrobacterium inoculation and DEX induction. We prepared total protein extracts for fractionation and immunoprecipitation (Figure 6). The wild-type and G2A exchange mutants for either Avr protein were equally represented in the extracts, yet 3H was only incorporated into the wild-type AvrRpm1 and AvrB proteins. Thus, these proteins can be myristoylated in vivo. Coupled with the functional role for a G2 residue for complete expression of all tested functions and for efficient plasma membrane localization of both AvrRpm1 and AvrB, it is likely that myristoylation is essential for both optimal function and localization of AvrRpm1 and AvrB. Figure 5. AvrRpm1 and AvrB Localize to a Plant Cell Membrane Fraction (A) Localization is facilitated by consensus myristoylation and palmitoylation sites. The rpm1 null accession Cvi-0 was inoculated with Agrobacterium carrying vector (V), wild type (WT) or the mutant avr derivatives listed across the top. DEX induction as in Figure 3A. Leaves were harvested 12 hr post DEX treatment. Total (T) extracts were separated into soluble (S) and 100,000 ⫻ g microsomal pellet (M) fractions, subjected to SDS-PAGE, blotted, and probed with anti-HA epitope monoclonal antibody to detect either AvrRpm1 (experiment in top set of two blots) or AvrB (bottom set of two blots), or with antisera against the tonoplast membrane marker ␥-TIP (both sets of blots). All apparent molecular weights are correct (AvrRpm1 at 29 kDa, AvrB at 36 kDa, ␥-TIP at 27 kDa). (B) AvrRpm1 and AvrB are highly enriched in plasma membrane vesicles. The rpm1 null accession Cvi-0 was inoculated with Agrobacterium carrying vector (V) or wild-type avr genes listed across the top. Total (T) vesicles were separated by two-phase enrichment into intracellular (I) and plasma membrane enriched (P) pools. Equal yields were electrophoretically separated, blotted, and probed with the anti-HA monoclonal Proteolytic Processing of a P. syringae Type III Effector in Host Cells Exposes a Eukaryotic N-Myristoylation Consensus Site The 35 kDa AvrPphB protein from P. syringae pv. phaseolicola is rapidly cleaved between K62 and G63, in both E. coli and P. syringae (Puri et al., 1997). The longer, 28 kDa product of this cleavage exposes a potentially myristoylated free glycine at its N terminus (Table 1). To generalize our findings with AvrRpm1 and AvrB, we constructed a G63A avrPphB mutation for expression of either native or HA-tagged derivatives in the DEXinducible Agrobacterium system. The 28 kDa cleavage product consistently accumulates following DEX-induced transient expression of AvrPphB in leaves. We have occasionally observed very low levels of the 35 kDa translation product in soluble fractions (not shown), suggesting that it is also rapidly processed in plant cells. The 28 kDa cleavage product localizes to a membrane fraction in a G63-dependent manner (Figure 7). This residue also greatly enhances recognition of AvrPphB by RPS5 (Warren et al., 1998) following Agrobacterium delivery (responding leaves: 59/70 for wild type and 13/74 for G63A), consistent with a functional role for myristoylation in membrane localization of AvrPphB in the plant cell. We conclude that AvrPphB can be cleaved in the plant cytoplasm to an active 28 kDa form, which utilizes a myristoylation site for both localization to a membrane compartment and recognition by RPS5. Discussion Bacterial pathogens of both plants and animals use type III secretion systems to deploy effector proteins into to detect either AvrRpm1 or AvrB, or with antisera against markers known to reside in the cellular compartments listed at right (Cyt is cytosol, ER is endoplasmic reticulum, PM is plasma membrane). All apparent molecular weights are correct (Bip at 70 kDa; ␥-TIP at 27 kDa; RD28 at 27 kDa). (C) AvrRpm1 and AvrB green fluorescence fusion proteins localize to the protoplast plasma membrane. Protoplasts from rpm1 plants were transformed with plasmids that express either wild type (WT) or G2A derivatives of AvrRpm1 or AvrB. Control transformations were with GFP alone. Stacked laser confocal micrographs are presented. The experiment was repeated many times with the same result (see Experimental Procedures). Host Modification of Prokaryotic Virulence Factors 359 Figure 7. An Embedded Consensus Myristoylation Site at G63 Mediates AvrPphB Membrane Localization Figure 6. AvrRpm1 and AvrB Are Myristoylated In Vivo in a G2Dependent Manner The rpm1 null accession Cvi-0 was inoculated with Agrobacterium carrying vector (V), wild type (WT), or G2A derivatives of avrRpm1 or avrB listed at the top. At 48 hpi, an ⵑ5 M [3H]myristic acid/20 M dexamethasone solution was hand inoculated into the Agrobacterium-infiltrated leaves. Total extracts were prepared 12 hr later. Total extract from two leaf discs was immunoprecipitated with antiHA monoclonal. Equal yield aliquots were either immunoblotted as in Figure 5, or analyzed by fluorographically enhanced autoradiography for 3 weeks. eukaryotic host cells. These effector proteins are positioned to interact with and regulate specific components of host signaling networks, and are targets for modification by host cellular components. Avr proteins are type III–dependent effectors of disease and triggers of plant disease resistance. We identified eukaryotic N-terminal myristoylation and palmitoylation consensus sequences on AvrRpm1, AvrB, AvrC, and AvrPto, and we noted that posttranslational cleavage exposes a eukaryotic consensus acylation site on AvrPphB. We demonstrated that consensus myristoylation sites are required for maximal function of AvrRpm1 and AvrB when delivered from P. syringae to Arabidopsis. We also demonstrated that both the consensus myristoylation and palmitoylation sites of AvrRpm1 are required for maximal virulence of the P. syringae strain Psm CR299. Additionally, the myristoylation sites of AvrRpm1, AvrB, and AvrPphB enhance R gene–specific responses when each Avr protein is expressed inside host cells. Thus, the genetically defined role for these consensus acylation sites is independent of the type III secretion machinery. Function correlates with membrane localization and myristoylation of the Avr proteins, and the major membrane system targeted for AvrRpm1 and AvrB is the plasma membrane. We documented a host genotype– specific, rpm1-independent response to AvrB expression. Importantly, this response is also greatly enhanced by membrane association mediated by the consensus myristoylation site. We propose that this slow cytotoxic response could reflect the AvrB function in promoting disease on rpm1 plants. We were unable to detect either AvrB or AvrRpm1 protein in host cells at the time when rpm1-independent responses were observed. However, there is no intrinsic difference in the stability of wildtype and mutant AvrRpm1 or AvrB proteins at either a time point preceding the onset of this response, or for AvrB in a nonresponding plant at time points beyond the onset of the rpm1-independent response. Thus, the differential phenotypes of wild-type and mutant Avr proteins are probably not due to differential intrinsic stability of each protein in host cells. Accession La-er (rps5) leaves were inoculated with Agrobacterium carrying empty vector (V), wild type (WT) or a G63A exchange derivative of avrPphB. DEX induction as in Figure 3A. Samples were harvested 6 hr post induction and processed as in Figure 5A. M5 refers to 5⫻ more than the equal yield amounts loaded in the other lanes. We provide compelling in vivo evidence that AvrRpm1 and AvrB are myristoylated in a G2-dependent manner. Protein palmitoylation is widely believed to be posttranslational and palmitoyltransferase activity is enriched in plasma membranes (Dunphy et al., 1996). Palmitoylation often requires previous myristoylation (Berthiaume and Resh, 1995). Myristoylation is viewed as a cotranslational process: cyclohexamide treatment abolishes myristoylation (Olson and Spizz, 1986); nascent polypeptide chains associated with tRNAs have myristate covalently attached (Wilcox et al., 1987); and a significant fraction of N-myristoyltransferase is associated with ribosomes (Glover et al., 1997). Pathogen effector proteins delivered into host cells via the type III apparatus would thus appear to be unsuitable substrates for myristoylation. Yet, the necessity of cotranslational myristoylation was initially challenged by incorporation of radiolabelled myristate when protein synthesis is inhibited (da Silva and Klein, 1990) and by the finding of N-myristoyltransferases in both the endoplasmic reticulum and the cytosol (Boutin, 1997). The proteolytic cleavage product of AvrPphB associates with a membrane fraction in a G63-dependent manner following processing from the full-length wild-type protein. Pulse–chase studies in P. syringae revealed that full-length AvrPphB is rapidly processed (Puri et al., 1997) and our data suggest that the same is true in plant cells. Myristoylation of the exposed N-terminal glycine of the larger AvrPphB cleavage product would thus follow synthesis of full-length protein, and subsequent cleavage exposing G63. These constraints make cotranslational acylation of AvrPphB unlikely and support the notion that some proteins are capable of posttranslational myristoylation. We cannot, however, exclude cotranslational myristoylation of AvrB and AvrRpm1. Exposure of G2 by removal of the initiator methionine for AvrRpm1 and AvrB could be achieved by methionine aminopeptidases inside either the host cell or the bacteria prior to delivery, as reported for the type III effector proteins TIR and SopE (Wood et al., 1996; Kenny et al., 1997). AvrPto (Table 1) can localize to a plasma membrane fraction (T. L. and F. K., unpublished; X. Tang personal communication). Pto, which recognizes AvrPto, contains a G2 residue. However, site-directed G2A exchange did not alter Pto function when overexpressed (Loh et al., 1998). Wild-type Pto expressed from its own promoter has not been localized. Yet, the Pto-related Fen kinase has a demonstrated G2 requirement for function, and a chimeric Fen possessing a Pto N terminus Cell 360 still functions. Thus, the N terminus of Pto is capable of providing a myristoylation site required for Fen function (Rommens et al., 1995). Cytoplasmic Pto may sequester incoming AvrPto, before the latter is localized. Alternatively, overexpressed, nonmyristoylated Pto may be recruited to the membrane by AvrPto as observed for G␣z/ B␥ subunit interactions (Morales et al., 1998). This would be consistent with our unpublished observations that constitutive overexpression of the G2A Avr derivatives eliminated phenotypic differences with wild type. Subcellular localization of P. syringae type III effector proteins in a host cell probably facilitates virulence. The decreased functions of G2A and C3A mutants of AvrRpm1, for Psm M2 virulence, and of AvrB, for the rpm1-independent response, support this view. Plasma membrane localization may mirror that of the host targets of AvrRpm1 and AvrB disease effector function. Localization of RPM1 to the Arabidopsis plasma membrane is consistent with either direct or indirect recognition of membrane-associated AvrRpm1 or AvrB. Other P. syringae Avr proteins lack acylation sequences, suggesting variable subcellular sites of action for incoming type III effectors. This variation could favor evolution of host recognition complexes, anchored by the relevant R product, with different subcellular localizations. If this model is correct, it predicts that a major function of R products is to “guard” subcellular targets of type III effector proteins. This model, first elaborated by Van der Biezen and Jones (1998) further predicts that the cellular targets of effector action during disease onset would also be found in an R protein complex in resistant host genotypes. By contrast, if each structural class of R protein assembled into a defense-dedicated protein complex, then common elements should be found in the signalasome of various R proteins. Thus far, yeast two-hybrid analyses with various NBS-LRR class R proteins have failed to identify such a uniform set of proteins. Interestingly, while the putative palmitoylation site at C3 is not absolutely required for AvrRpm1 triggering of RPM1 function, it is required for the virulence function of AvrRpm1 on rpm1 null hosts. Mutation at C3 partially reduces, but does not abolish, AvrRpm1 membrane localization, while the G2A exchange has a severe localization defect. This is consistent with studies demonstrating that myristoylation defects prevent subsequent palmitoylation, but that the reverse is not true (McCabe and Berthiaume, 1999). RPM1 function is probably sensitive to very small quantities of Avr protein at the membrane, congruent with recent evidence that increasing the level of Avr or R protein can increase the sensitivity of the overall disease resistance response (Bendahmane et al., 1999). In contrast, the virulence function of AvrRpm1 might operate via a more stable association with the membrane, achieved through additional palmitoylation, or quantitatively by delivery of more protein to the membrane. Alternatively, the putative palmitoylation may serve a regulatory role as demonstrated for the modulation of GAP activity by palmitoylated G␣ subunits (Tu et al., 1997). In this scenario, palmitoylation of AvrRpm1 C3 could differentially affect RPM1-dependent and rpm1-independent functions. The host targets of type III effectors of phytopathogen virulence, and the means by which they modulate or usurp host cell signal pathways to promote disease, are largely unknown. That acylation and membrane localization are not general features of type III effectors is illustrated by our finding that, of 46 putative or known type III effectors from animal and plant pathogens (Hueck, 1998), none carry consensus acylation sequences at their N termini. Of course, some of these, like AvrPphB, could be proteolytically processed to reveal an acylation site. We demonstrated that expression of AvrB induces cytotoxicity in some, but not all, rpm1 null plants. These cytotoxic effects may be indicative of the virulence function of AvrB in host cells. Note that the inability to ascribe a virulence function to AvrB in the context of Pst DC3000–induced disease does not alter this conclusion, as it could be the case that AvrB is redundant to an unknown type III effector in DC3000. The polymorphic nature of the host response, and our ability to map a locus responsible for it, indicates that this is a specific effect. Several other Avr type III effectors can induce cytotoxic effects on disease-susceptible host cells using similar expression-based systems. In animal systems, expression of various type III effector molecules in host cells is known to phenocopy aspects of the host disease response and in many cases this effect can be traced to interactions with relevant host cell targets (Hueck, 1998). Our ability to isolate Arabidopsis mutants in the rpm1 null Mt-0 background that fail to respond to AvrB expression (Z. N. and J. L. D., unpublished) should enrich our understanding of the role these proteins play in inducing disease and altering host cell physiology. Experimental Procedures Construction of avr Clones and Mutants Site-directed mutants (Chameleon System) were sequenced for verification. DEX-inducible HA-tagged wild-type and mutant avr genes were cloned into pTA7002 (Aoyama and Chua, 1997) for expression in planta. For expression in P. syringae, avrB and avrRpm1 constructs are cloned behind the native avrRpm1 promoter (Ritter and Dangl, 1995) in pVSP61 (Bisgrove et al., 1994). avrB and avrRpm1 were cloned into pDSK519 for expression in E. coli. Details available upon request. P. syringae HR and In Planta Growth Assays For in planta inoculations (Debener et al., 1991), P. syringae were resuspended to OD600 ⫽ 0.1 (corresponding to ⵑ5 ⫻ 107 cfu/ml) for HR assays or diluted to 1 ⫻ 105 cfu/ml for growth curves. HR was scored from 5 hr for avrRpm1 and avrB. P. syringae Protein Extraction and E. coli Secretion Assays 2.5 ml overnight Pst DC3000 cultures in KB with the appropriate antibiotics were pelleted, washed with hrp gene–inducing media (Ritter and Dangl, 1995), resuspended in 2.5 ml of the same, and induced for 5 hr. Cultures were spun down and resuspended in 500 l 65⬚C, 3⫻ Laemmli buffer. Samples were boiled 2 min and 5 l loaded on an SDS-PAGE. Agrobacterium Transient Expression Assays 2 ml overnight Agrobacterium cultures were grown at 30⬚C in YEB (5 g bacto beef extract, 1 g bacto yeast extract, 5 g bacto peptone, 5 g sucrose, 2 mM MgSO4, pH 7.2, per liter) containing 100 g/ml each of rifampicin, kanamycin, and gentamycin for strain GV3101. The following day, 150 l of saturated culture was inoculated into 3 ml of YEB plus antibiotics, and grown for 13 hr. Two milliliters was collected and resuspended in 3 ml Agrobacterium induction medium (10.5 g K2HPO4, 4.5 g KH2PO4, 1 g (NH4)2SO4, 0.5 g (NaCitrate), 1 mM MgSO4, 1 g glucose, 1 g fructose, 4 ml glycerol, 10 mM MES, pH Host Modification of Prokaryotic Virulence Factors 361 5.6, per liter, 50 g/ml acetosyringone), grown at 23⬚C for 5–7 hr, collected and resuspended in infiltration medium (1/2 MS-MES) to an OD600 of 0.4. The underside of 3-week-old leaves were inoculated using a needleless syringe. Plants were grown in ⬎120 E of light and sprayed with 20 M DEX (Sigma, St. Louis, MO) 48 hr after inoculation, except for in planta myristoylation assays (see below). RPM1-dependent or RPS5-dependent responses were scored 24 hr later, and the rpm1-independent responses scored 2–3 days later. All phenotypes noted were confirmed to be dependent on T-DNA transfer by testing constructs in a GV3101 strain cured of the vir plasmid (data not shown). Plant Protein Extractions Two 6 mm diameter leaf discs were ground in a 1.5 ml Eppendorf tube in 50 l protein extraction buffer (20 mM Tris-HCL, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 1⫻ plant protease inhibitor cocktail (PIC; Sigma, St. Louis, MO), and 10 l 6⫻ Laemmli buffer (final concentration 1⫻) was added. Samples were vortexed, boiled for 3 min, then spun briefly. Ten microliters was loaded on an SDS-PAGE gel. Total Plant Membrane Fractionation Fifteen 6 mm leaf discs per sample were ground in 200 l membrane extraction buffer (MEB) (10 mM Tris-HCl pH 7, 0.33 M sucrose, 1 mM EDTA, 1⫻ PIC) in a 1.5 ml Eppendorf tube. Three hundred microliters of MEB was added, and samples vortexed and cleared at 8,000 ⫻ g for 3–4 min. Four hundred fifty microliters of supernatant was transferred to an Eppendorf tube containing 10 l 1 M CaCl2. A 50 l aliquot was removed as the “total extract” fraction and the remainder was spun 50,000 ⫻ g for 1.5 hr. The resulting supernatant (soluble fraction) was prepared for SDS-PAGE. The pellet (membrane fraction) was resuspended in 460 l TE with PIC. For each fraction, 10 l of 6⫻ Laemmli buffer was added to a 50 l aliquot and 10 l of this sample was loaded to obtain equal yield. Proteins were separated using SDS-PAGE on a 15% polyacrylamide gel. Plasma Membrane Purification Plasma membrane and intracellular vesicles were prepared using aqueous two-phase partitioning as described in Boyes et al. (1998). Equal protein levels from each fraction were loaded on the gel. Antibodies used for membrane localization were raised against RD28 (plasma membrane), ␥-TIP (tonoplast), (gifts of Maarten Chrispeels), BiP (intercellular membranes and cytosol; gift of Rebecca Boston), and an antibody to the HA epitope (Roche Biochemicals, Indianapolis, IN). All antibodies were used at a dilution of 1:1000. In Planta Myristoylation Assay Agrobacterium cultures were grown and induced as described above. Avr induction and labeling of total proteins with [3H]myristate was performed by coinoculating a solution of 20 M dexamethasone and 500 Ci/ml [9,10–3H(N)] myristic acid (American Radio Chemicals, St. Louis, MO) directly into the underside of leaves using a needleless syringe 48 hr after Agrobacterium inoculation. After 12 hr, two leaf discs per sample were ground in IP buffer (20 mM TrisHCl, pH 7.5, 1 mM EDTA, 150 mM NaCl2, 1% Triton X-100 [v/v]), and 1% plant PIC. Crude lysate was spun down (2 min, 4⬚C, 10,000 ⫻ g) and the supernatant was immunoprecipitated using 30 l of High Affinity HA-Antibody coupled matrix (Roche Diagnostics, Indianapolis, IN) and end-over-end tumbling on a rotary motor for 2 hr at 4⬚C. Matrix was pelleted by spinning 2 min, 4⬚C, 10,000 ⫻ g. Matrix was then washed and pelleted twice with 500 l ice-cold IP solution and finally resuspended in 50 l SDS-protein extraction buffer (see above) and 10 l of 6⫻ gel loading buffer. Ten-microliter aliquots were loaded onto gels and separated as described above. Proteins were identified on Western blots with mouse, anti-HA antibody and subsequent detection with mouse-secondary conjugated to peroxidase as described above. For radiolabel detection, gels were subjected to fluorographic impregnation using Amplify (Amersham International), dried down, and exposed to X-ray film. Plasmid Construction for GFP Microscopy Transient expression constructs are in pKEx4tr (Leister et al., 1996). C-terminal green fluorescent protein (GFP) fusions were obtained by fusing the synthetic (GFP) coding sequence to the coding sequences in pExavrRpm1 and pExavrB. pExGFP was constructed by cloning the GFP coding sequence into pKEx4tr. Site-directed mutagenesis of the G2A residues in avrRpm1 and avrB used the respective wild-type avr-GFP fusions and specific primers as template. Details of the cloning procedures are available upon request. Protoplast Preparation and Transformation Arabidopsis plants from a cross between ecotype Niederzenz (Nd-0) (rpm1/rpm1) and rps2–101C (rps2–101/rps2–101C; Col-0 background), were used for all protoplast studies as in Leister et al. (1996). Arabidopsis leaf mesophyll protoplasts were prepared from 5-week-old plants and transformed using polyethylene glycol. Transformation efficiency (i.e., percentage of GFP-positive protoplasts) was 30%–40% with pExGFP. Protoplasts were transformed with either 3.0 g pExGFP, 3.0 g pavrBG2A-GFP, 6.0 g pavrBGFP, 6.0 g pavrRpm1-GFP, or 6.0 g pavrRpm1G2A-GGFP. pKEx4tr-dGFP (a defective GFP; Leister et al., 1996) was added where needed to keep the total amount of DNA equal for each treatment. Following transformation, the protoplasts were incubated overnight in the light at room temperature. Microscopy Protoplasts were examined on a Leica model DMIRBE confocal microscopy system appropriate for the detection of S65T GFP using an argon laser for excitation (488 nm) and the BP-FITC detector setting for collection. Images were generated using the extended focus option under 3D-image processing. Each image represents the scaled summation of ten optical sections beginning 5 m from the bottom of the cell and continuing toward the center of the cell. Each optical section is approximately 0.5 m in thickness. Acknowledgments This research is supported by DOE Grant DE-FG05–95ER20187 to J. L. D., NIH-NRSA Fellowship F32GM17612 to S. K., and NSF Grant MCB-9604830 to F. K. We thank Drs. Ulla Bonas, Alan Collmer, and John Mansfield for gifts of strains and reagents. We thank Jen Sheen for GFP constructs, and Chere Petty for assistance with confocal microscopy. Experimental Fu provided by Ben F. Holt III and Dr. Petra Epple. Received January 6, 2000; revised April 17, 2000. References Alfano, J.R., and Collmer, A. (1997). The type III secretion pathway of plant pathogenic bacteria: trafficking harpins, avr proteins and death. J. Bacteriol. 179, 5655–5662. Anderson, D., Fouts, D., Collmer, A., and Schneewind, O. (1999). Reciprocal secretion of proteins by the bacterial type III machines of plant and animal pathogens suggests universal recognition of mRNA targeting signals. Proc. Natl. Acad. Sci. USA 96, 12839– 12843. Aoyama, T., and Chua, N.-H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. Bendahmane, A., Kanyuka, K., and Baulcombe, D.C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11, 781–791. Berthiaume, L., and Resh, M.D. (1995). Biochemical characterization of a palmitoyl acyltransferase activity that palmitoylates myristoylated proteins. J. Biol. Chem. 38, 22399–22405. Bisgrove, S.R., Simonich, M.T., Smith, N.M., Sattler, N.M., and Innes, R.W. (1994). A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell 6, 927–933. Boutin, J.A. (1997). Myristoylation. Cell. Signal. 9, 15–35. Boyes, D.C., Nam, J., and Dangl, J.L. (1998). The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl. Acad. Sci. USA 95, 15849–15854. Cell 362 Cornelis, G.R., and Wolf-Watz, H. (1997). The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol. Microbiol. 23, 861–887. Loh, Y.-T., Zhou, J., and Martin, G.B. (1998). The myristylation motif of Pto is not required for disease resistance. Mol. Plant Microbe Interact. 11, 572–576. Dangl, J.L., Ritter, C., Gibbon, M.J., Wood, J.R., Mur, L.A.J., Goss, S., Mansfield, J.W., Taylor, J.D., and Vivian, A. (1992). Functional homologs of the Arabidopsis RPM1 disease resistance gene in bean and pea. Plant Cell 4, 1359–1369. Lorang, J.M., Shen, H., Kobayashi, D., Cooksey, D., and Keen, N.T. (1994). avrA and avrE in Pseudomonas syringae pv. tomato PT23 play a role in virulence on tomato plants. Mol. Plant Microbe Interact. 7, 208–215. Daniels, M.J., Mirkov, T.E., and Chrispeels, M.J. (1994). The plasma membrane of Arabidopsis thaliana contains a mercury-insensitive aquaporin that is a homolog of the tonoplast water channel protein TIP. Plant Physiol. 106, 1325–1333. McCabe, J.B., and Berthiaume, L.G. (1999). Functional roles for fatty acylated amino-terminal domains in subcellular localization. Mol. Biol. Cell 10, 3771–3776. da Silva, A.M., and Klein, C. (1990). A rapid posttranslational myristylation of a 68-kD protein in D. discoideum. J. Cell Biol. 111, 401–407. Debener, T., Lehnackers, H., Arnold, M., and Dangl, J.L. (1991). Identification and molecular mapping of a single Arabidopsis thaliana locus determining resistance to a phytopathogenic Pseudomonas syringae isolate. Plant J. 1, 289–302. Dunphy, J.T., Greentree, W.K., Manahan, C.L., and Linder, M.E. (1996). G-protein palmitoyltransferase activity is enriched in plasma membranes. J. Biol. Chem. 271, 7154–7159. Ellard-Ivey, M., Hopkins, R.B., White, T.J., and Lomax, T.L. (1999). Cloning, expression and N-terminal myristoylation of CpCPK1, a calcium-dependent protein kinase from zucchini (Curcurbita pepo L.). Plant Mol. Biol. 39, 199–208. Galan, J., and Collmer, A. (1999). Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284, 1322– 1328. Glover, C.J., Hartman, K.D., and Felsted, R.L. (1997). Human N-myrsitoyltransferase amino-terminal domain involved in targeting the enzyme to the ribosomal subcellular fraction. J. Biol. Chem. 272, 28680–28689. Gopalan, S., Bauer, D.W., Alfano, J.R., Lonlello, A.O., He, S.Y., and Collmer, A. (1996). Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell 8, 1095–1105. Grant, M.R., Godiard, L., Straube, E., Ashfield, T., Lewald, J., Sattler, A., Innes, R.W., and Dangl, J.L. (1995). Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269, 843–846. Ham, J.H., Bauer, D.W., Fouts, D.E., and Collmer, A. (1998). A cloned Erwinia chrysanthemi Hrp (type III protein secretion) system functions in Escherichia coli to deliver Pseudomonas syringae Avr signal to plant cells and to secrete Avr proteins in culture. Proc. Natl. Acad. Sci. USA 95, 10206–10211. McNellis, T.W., Mudgett, M.B., Li, K., Aoyama, T., Horvath, D., Chua, N.-H., and Staskawicz, B.J. (1998). Glucocorticoid-inducible expression of a bacterial avirulence gene in transgenic Arabidopsis thaliana induces hypersensitive cell death. Plant J. 14, 247–258. Morales, J., Fishburn, C.S., Wilson, P.T., and Bourne, H.R. (1998). Plasma membrane localization of G␣z requires two signals. Mol. Biol. Cell 9, 1–14. Mudgett, M., and Staskawicz, B. (1998). Protein signaling via type III secretion pathways in phytopathogenic bacteria. Curr. Opin. Microbiol. 1, 109–114. Mudgett, M., and Staskawicz, B. (1999). Characterization of the Pseudomonas syringae pv. tomato AvrRpt2 protein: demonstration of secretion and processing during bacterial pathogenesis. Mol. Microbiol. 32, 927–941. Olson, E.N., and Spizz, G. (1986). Fatty acylation of cellular proteins. Temporal and subcellular difference between palmitate and myrsitate acylation. J. Biol. Chem. 261, 2458–2466. Puri, N., Jenner, C., Bennett, M., Stewart, R., Mansfield, J., Lyons, N., and Taylor, J. (1997). Expression of avrPphB, an avirulence gene from Pseudomonas syringae pv. phaseolicola, and the delivery of signals causing the hypersensitive reaction in bean. Mol. Plant Microbe Interact. 10, 247–256. Resh, M.D. (1994). Myristoylation and palmitoylation of Src family members: the fats of the matter. Cell 76, 411–413. Ritter, C., and Dangl, J.L. (1995). The Pseudomonas syringae pv. maculicola avrRpm1 gene is required for virulence on Arabidopsis. Mol. Plant Microbe Interact. 8, 444–453. Rommens, C.M.T., Salmeron, J.M., Baulcombe, D.C., and Staskawicz, B.J. (1995). Use of a gene expression system based on Potato Virus X to rapidly identify and characterize a tomato Pto homolog that controls fenthion sensitivity. Plant Cell 7, 249–257. Rossier, O., Wengelnik, K., Hahn, K., and Bonas, U. (1999). The Xanthomonas Hrp type III system secretes proteins from plant and mammalian bacterial pathogens. Proc. Natl. Acad. Sci. USA 96, 9368–9373. Hueck, C.J. (1998). Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62, 379–433. Salmeron, J.M., and Staskawicz, B.J. (1993). Molecular characterization and hrp dependence of the avirulence gene avrPto from Pseudomonas syringae pv. tomato. Mol. Gen. Genet. 239, 6–16. Huynh, T.V., Dahlbeck, D., and Staskawicz, B.J. (1989). Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245, 1374–1377. Scheel, D. (1998). Resistance response physiology and signal transduction. Curr. Opin. Plant Biol. 1, 305–310. Jenner, C., Hitchin, E., Mansfield, J.W., Walters, K., Betteridge, P., Teverson, D., and Taylor, J.D. (1991). Gene-for-gene interactions between Pseudomonas syringae pv. phaseolicola and Phaseolus. Mol. Plant Microbe Interact. 4, 553–562. Johnson, D.R., Bhatnagar, R.S., Knoll, L.J., and Gordon, J.I. (1994). Genetic and biochemical studies of protein N-myristoylation. Annu. Rev. Biochem. 63, 869–914. Kearney, B., and Staskawicz, B.J. (1990). Widespread distribution and fitness contribution of Xanthomonas campestris avirulence gene avrBs2. Nature 346, 385–386. Kenny, B., DeVinney, R., Stein, M., Reinscheid, D.J., Frey, E.A., and Finlay, B.B. (1997). Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91, 511–520. Leister, R.T., Ausubel, F.M., and Katagiri, F. (1996). Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1. Proc. Natl. Acad. Sci. USA 93, 15497–15502. Scofield, S.R., Tobias, C.M., Rathjen, J.P., Chang, J.H., Lavelle, D.T., Michelmore, R.W., and Staskawicz, B.J. (1996). Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274, 2063–2065. Tamaki, S., Dahlbeck, D., Staskawicz, B.J., and Keen, N.T. (1988). Characterization and expression of two avirulence genes cloned from Pseudomonas syringae pv. glycinea. J. Bacteriol. 170, 4846– 4854. Tang, X., Frederick, R.D., Zhou, J., Halterman, D.A., Jia, Y., and Martin, G.B. (1996). Physical interaction of avrPto and the Pto kinase defines a recognition event involved in plant disease resistance. Science 274, 2060–2063. Tu, Y., Wang, J., and Ross, E.M. (1997). Inhibition of brain Gz GAP and other RGS proteins by palmitoylation of G protein ␣ subunits. Science 278, 1132–1135. Van den Ackerveken, G., Marois, E., and Bonas, U. (1996). Recognition of the bacterial AvrBs3 protein occurs inside the plant cell. Cell 87, 1307–1316. Host Modification of Prokaryotic Virulence Factors 363 Van der Biezen, E.A., and Jones, J.D.G. (1998). Plant disease resistance genes and the “gene-for-gene” concept. Trends Biochem. Sci. 23, 454–456. Warren, R.F., Henk, A., Mowery, P., Holub, E., and Innes, R.W. (1998). A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell 10, 1439–1452. Wilcox, C., Hu, J.-S., and Olson, E.N. (1987). Acylation of proteins with myristic acid occurs cotranslationally. Science 238, 1275–1278. Wood, M.W., Rosqvist, R., Mullan, P.B., Edwards, M.H., and Galyov, E.E. (1996). SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via sip-dependent mechanism and promotes bacterial entry. Mol. Microbiol. 22, 327–338. Yang, Y., and Gabriel, D.W. (1995). Xanthomonas avirulence/pathogenicity gene family encodes functional plant nuclear targeting signals. Mol. Plant Microbe Interact. 8, 627–631. Yang, Y., Shah, J., and Klessig, D.F. (1997). Signal perception and transduction in plant defense responses. Genes Dev. 11, 1621–1639. Zhu, W., Yang, B., Kurata, N., Johnson, L.B., and White, F.F. (1999). The C terminus of AvrXa10 can be replaced by the transcriptional activation domain of VP16 from the herpes simplex virus. Plant Cell 11, 1665–1674. Article 2: 2002 Molecular Plant Microbe Interaction The Xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Marois E., Van den Ackerveken G., Bonas U. MPMI Vol. 15, No. 7, 2002, pp. 637–646. Publication no. M-2002-0507-01R. © 2002 The American Phytopathological Society The Xanthomonas Type III Effector Protein AvrBs3 Modulates Plant Gene Expression and Induces Cell Hypertrophy in the Susceptible Host Eric Marois, Guido Van den Ackerveken, and Ulla Bonas Institut für Genetik, Martin-Luther-Universität Halle-Wittenberg, 06099 Halle, Germany. Submitted 4 February 2002. Accepted 7 March 2002. Xanthomonas campestris pv. vesicatoria bacteria expressing the type III effector protein AvrBs3 induce a hypersensitive response in pepper plants carrying the resistance gene Bs3. Here, we report that infection of susceptible pepper and tomato plants leads to an AvrBs3-dependent hypertrophy of the mesophyll tissue. Agrobacterium-mediated transient expression of the avrBs3 gene in tobacco and potato plants resulted in a similar phenotype. Induction of hypertrophy was shown to depend on the repeat region, nuclear localization signals, and acidic transcription activation domain (AAD) of AvrBs3, suggesting that the effector modulates the host’s transcriptome. To search for host genes regulated by AvrBs3 in an AAD-dependent manner, we performed a cDNA-amplified fragment length polymorphism analysis of pepper mRNA populations. Thirteen AvrBs3-induced transcripts were identified and confirmed by reverse transcriptase-polymerase chain reaction. Sequence analysis revealed homologies to auxin-induced and expansinlike genes, which play a role in cell enlargement. These results suggest that some of the AvrBs3-induced genes may be involved in hypertrophy development and that xanthomonads possess type III effectors that steer host gene expression. Additional keywords: avirulence, bacterial spot disease, SAUR (small auxin up RNA), transcription factor, upa (upregulated by AvrBs3). Phytopathogenic bacteria are responsible for a great variety of diseases in plants, causing important agricultural losses. Many gram-negative plant and animal pathogenic bacteria share a common mechanism to attack and exploit their eukaryotic hosts: the type III secretion system, which is required to deliver bacterial proteins into host cells. The delivered proteins, termed type III effectors, are thought to be involved in virulence by targeting specific steps of the host cell metabolism for the benefit of the bacterial invader (Cornelis and van Gijsegem 2000). In plant pathogens, one class of type III effectors are avirulence proteins (Bonas and Van den Ackerveken 1999). The term “avirulence” (avr) defines bacterial genes that determine specific recognition of the bacteria by plants possessing a matching resistance (R) gene. Plant R gene-mediated Corresponding author: U. Bonas, Institut für Genetik, Martin-Luther Universität, 06099 Halle, Germany; Telephone: +49 (0) 345 552 6290; Fax: +49 (0) 345 552 7277; Email: bonas@genetik.uni-halle.de. Current address of G. Van den Ackerveken:: University of Utrecht, Dept. of Molecular and Cellular Biology, P.O. Box 80 056, 3508 TB Utrecht, The Netherlands. GenBank accession numbers for upa1 to upa11 are AF492625 to AF492635. recognition of an Avr protein leads to the induction of plant defense reactions that generally include the hypersensitive response (HR), a rapid localized cell death associated with the arrest of pathogen ingress (Morel and Dangl 1997). Thus, Avr proteins restrict the pathogen’s host range, an effect that is deleterious to the bacteria and is probably not their primary function. In fact, a growing number of Avr proteins appear to play a role in bacterial virulence (i.e., bacterial growth and symptom formation) in susceptible host plants (Bai et al. 2000; Chen et al. 2000; Kearney and Staskawicz 1990; Kjemtrup et al. 2000; Lorang et al. 1994; Ritter and Dangl 1995; Shan et al. 2000; Tsiamis et al. 2000; Yang et al. 1994, 1996, 2000). We study the gram-negative bacterial pathogen Xanthomonas campestris pv. vesicatoria, the causal agent of bacterial spot disease of pepper and tomato plants. The ability of X. campestris pv. vesicatoria to cause disease depends on the type III protein secretion system encoded by the hrp gene cluster (Bonas et al. 1991; Rossier et al. 1999). A number of avirulence genes have been isolated from X. campestris pv. vesicatoria (Jones et al. 1998), including avrBs3 (Bonas et al. 1989). Pepper plants carrying the Bs3 resistance gene are able to recognize X. campestris pv. vesicatoria strains expressing avrBs3 (Bonas et al. 1989; Minsavage et al. 1990). The AvrBs3 protein, which is secreted in a type III-dependent manner (Rossier et al. 1999), is recognized inside the plant cell (Van den Ackerveken et al. 1996), suggesting that it is translocated into the plant cell by the X. campestris pv. vesicatoria type III secretion system. AvrBs3 is a member of a large family of highly related proteins found in many Xanthomonas spp., the AvrBs3 family (Gabriel 1999; Vivian and Arnold 2000). In addition to an avirulence activity, some family members are involved in disease symptom formation (e.g., PthA from X. citri [citrus canker; Swarup et al. 1991], Avrb6 from X. campestris pv. malvacearum [increased watersoaking of cotton leaves; Yang et al. 1996] and AvrXa7 from X. oryzae [leaf lesion length in rice; Bai et al. 2000]). In the case of AvrBs3 from X. campestris pv. vesicatoria, an activity in susceptible plants has not been reported. The most striking feature of members of the AvrBs3 protein family is their central region composed of 12.5 to 25.5 nearly identical tandem repeats of a 34-amino-acid (aa) motif. Domain swapping experiments have shown that the repeat region determines both virulence and avirulence specificities (Yang et al. 2000). The 17.5 repeats of AvrBs3 were found to be essential for recognition by the Bs3 resistance gene, because AvrBs3 repeat deletion mutants no longer were recognized by Bs3 but unmasked new resistance genes in other pepper and tomato genotypes (Herbers et al. 1992). The N- and C-terminal protein regions are highly conserved among AvrBs3 family members and are functionally interchangeable (Ballvora et al. 2001; Zhu et al. 1998). Vol. 15, No. 7, 2002 / 637 The C-terminus of the proteins contains functional nuclear localization signals (NLSs) and an acidic transcription activation domain (AAD) (Van den Ackerveken et al. 1996; Yang and Gabriel 1995b; Zhu et al. 1998). Both types of motifs are required for activity (Szurek et al. 2001; Van den Ackerveken et al. 1996; Yang et al. 2000; Yang and Gabriel 1995b; Zhu et al. 1998), and the AvrBs3 NLSs interact in yeast and in vitro with pepper importin a (Szurek et al. 2001). Studies of AvrXa7, an AvrBs3 family member from X. oryzae, have suggested a direct interaction of the protein with AT-rich DNA sequences (Yang et al. 2000). Here, we describe AvrBs3-induced mesophyll cell hypertrophy in different solanaceous plants. To investigate the role of AvrBs3 as a modulator of host gene transcription, we studied pepper gene expression using the cDNA-amplified fragment length polymorphism (AFLP) technique. Many of the identified induced genes appear to be related to cell expansion. RESULTS AvrBs3 delivered by X. campestris pv. vesicatoria induces hypertrophy of mesophyll cells in susceptible plants. Inoculation of virulent X. campestris pv. vesicatoria strains into leaves of pepper (Capsicum annuum) and tomato (Lycopersicon esculentum) plants at high inoculum leads to the for- Fig. 1. AvrBs3 causes hypertrophy in plant leaves. a, A susceptible pepper leaf was inoculated with Xanthomonas campestris pv. vesicatoria strain I74A (empty pDSK602) ("–AvrBs3") and with X. pv. vesicatoria I74A expressing avrBs3 from pDSF300 ("+AvrBs3"). Inoculation density was 5 108 cfu per ml. Only the avrBs3-expressing strain causes hypertrophy. The photograph was taken 4 days postinoculation (dpi). b, Transient expression of avrBs3 in Nicotiana clevelandii. Agrobacterium strain GV3101 (at 7 dpi, OD600 = 0.8) delivering the avrBs3 gene from pVSF300. c, Leaf section through a susceptible pepper plant (ECW) leaf infected with X. campestris pv. vesicatoria strain I74A expressing avrBs3, observed with light microscopy. Spongy mesophyll cells appear to be vertically elongated in the infected area, pushing out the abaxial epidermis. Note the dilution of chloroplasts in the infected mesophyll due to cell hypertrophy. d and e, Sections at 4 dpi through pepper leaves infected with X. campestris pv. vesicatoria strain I74A expressing d, avrBs3DAAD (plasmid pDSF341) and e, functional avrBs3 (plasmid pDSF340). f and g, Section at 7 dpi through N. clevelandii leaf infected with Agrobacterium delivering f, an empty vector control (plasmid pVB60) and g, the avrBs3 gene (pVSF300). h and i, Section at 10 dpi through N. clevelandii leaf infected with Agrobacterium delivering h, the GUS gene used as negative control (plasmid p35SGUSINT) (Vancanneyt et al. 1990) and i, the avrBs3 gene (pVSF300). j and k, Section at 7 dpi through N. clevelandii leaf infected with Agrobacterium delivering j, the avrBs4 gene and k, the upa7a gene (putative expansin). All genes delivered by Agrobacterium are expressed under the control of the 35S* promoter (Mindrinos et al. 1994). Bar represents 200 µm. 638 / Molecular Plant-Microbe Interactions mation of watersoaked lesions that later become necrotic (day 4 to 5). Growth curve experiments revealed that multiplication in planta of X. campestris pv. vesicatoria strains differing only in the presence or absence of the avrBs3 gene is identical (Bonas et al. 1989). However, susceptible pepper plants infected with X. campestris pv. vesicatoria strains naturally containing avrBs3 (e.g., strain 82-8) often develop pustules on the abaxial leaf surface, in a type III secretion-dependent manner. Deletion of the avrBs3 gene abolished pustule induction (data not shown). Introduction of a plasmid-borne avrBs3 copy into strains naturally lacking avrBs3, such as 85-10, resulted in pustule induction in pepper (Fig. 1a). Strains 75-3 and 85-10 ectopically expressing avrBs3 also induced hypertrophy in L. esculentum and in the wild tomato species L. pennellii (S. Schornack and U. Bonas, unpublished data). Microscopic analysis revealed that pustules are the consequence of cell hypertrophy in the spongy mesophyll (Fig. 1c, d, e). Hypertrophy appears 4 days after bacterial inoculation and persists until the tissue collapses, which occurs from the center of the infiltrated area toward its margin, where the largest pustules are found. Reproducible observation of the avrBs3-induced hypertrophy in pepper and tomato is best with X. campestris pv. vesicatoria strains growing slowly in vitro and in planta, such as the 85-10 derivative I74A, which was used in this study. Fast-growing X. campestris pv. vesicatoria strains inoculated in laboratory conditions often caused tissue watersoaking and collapse before hypertrophy could develop. Induction of pustules by transient expression of avrBs3 within plant cells. Agrobacterium-mediated transient transformation of leaves has been a powerful tool to study the effect of individual pathogen proteins in plants (Bonas and Van den Ackerveken, 1999). Transient expression of avrBs3 under the control of the 35S Cauliflower mosaic virus promoter in resistant Bs3 pepper leaves resulted in the HR (Van den Ackerveken et al. 1996), indicating that AvrBs3 acts inside host cells. Transient expression of the same avrBs3 construct in Nicotiana clevelandii (Fig. 1b), N. benthamiana, N. tabacum, and in potato (Solanum tuberosum) induced pustules 4 to 5 days postinoculation (dpi). Agrobacterium strains carrying an empty vector did not cause any visible change in Nicotiana spp. and potato leaves (not shown), whereas susceptible pepper and tomato leaves reacted with chlorosis and necrosis 4 to 5 dpi against Agrobacterium spp., which inhibited avrBs3-dependent effects in susceptible host plants. The pustules resulting from avrBs3 expression in the nonhost plants were macroscopically similar to those triggered by X. campestris pv. vesicatoria–delivered AvrBs3 in pepper and tomato leaves. Microscopic examination of the N. clevelandii tissue expressing avrBs3 revealed enlarged mesophyll cells (Fig. 1f, g). Unlike pepper tissue infected with X. campestris pv. vesicatoria, the hypertrophied N. clevelandii tissue never became necrotic. At later time points (Fig. 1h, i) the number of cells in the affected tissue increased. This phenomenon was not observed in the case of type III-delivered AvrBs3 into pepper leaves and might be due to overexpression of avrBs3 under the control of the 35S promoter or to the longer survival time of Agrobacterium-infected Nicotiana cells, allowing them to develop novel AvrBs3-induced phenotypes. Different Nicotiana spp. (N. tabacum, N. benthamiana, and N. clevelandii) developed avrBs3-induced pustules to various extents, the largest being obtained in N. clevelandii (Fig. 1g, i). Transient expression of avrBs3 in Arabidopsis spp. did not induce any visible cellular changes, suggesting restriction of AvrBs3-specific effects to solanaceous plants. Table 1. Induction of hypertrophy and hypersensitive response (HR) by different Xanthomonas campestris pv. vesicatoria strains Straina 85-10 85-10 85-10 75-3 75-3 82-8 82-8DavrBs3 82-8DavrBs2e I74A I74A I74A I74A I74A I74A I74A Presence of avrBs3 or derivative thereofb … avrBs3 avrBs4 … avrBs3 avrBs3, avrBs4 avrBs4 avrBs3, avrBs4 … avrBs3 avrBs3Drep16 avrBs3D1-3f avrBs3D1-3SV40f avrBs3DAAD avrBs3DAAD::VP16 Hypertrophyc HRd – – + + + + + + + + – – – – – – + – – – – – – + – + – a 85-10, 82-8, and 75-3 are field isolates of X. campestris pv. vesicatoria. I74A is a derivative of strain 85-10. b Strains not endogenously expressing avrBs3 or a derivative thereof were transformed with a pDSK602 derivative containing the gene. c Hypertrophy was visually scored on susceptible Early Cal Wonder (ECW) pepper leaves, except for strain 75-3 (on Lycopersicon pennellii); indicates occasional hypertrophy. d HR was recorded on Bs3 pepper line ECW-30R, or on bs3 line ECW in the case of Drep16. + and indicate a fully developed and a partial or delayed HR, respectively. e avrBs2 is an avirulence gene unrelated to avrBs3, the deletion of which causes a fitness penalty (Kearney and Staskawicz 1990). f avrBs3D1-3 is a nuclear localization signal (NLS) deletion mutant, and avrBs3D1-3SV40 is the same mutant complemented by introduction of the SV40 NLS (Van den Ackerveken et al. 1996). Fig. 2. Identification of differentially expressed cDNAs by amplified fragment length polymorphism (AFLP). Autoradiograph of an AFLP gel showing a fraction of the transcription profile for one primer pair. cDNA profiles in leaves of pepper cultivar ECW infected with Xanthomonas campestris pv. vesicatoria strain 85-10 expressing avrBs3DAAD (D) or wild-type avrBs3 (wt) are compared, 9 and 20 h postinoculation (hpi). The differential band corresponds to upa7a. Vol. 15, No. 7, 2002 / 639 The fact that transient avrBs3 expression in plant cells could mimic the phenotype induced by AvrBs3 delivered by X. campestris pv. vesicatoria indicates that hypertrophy induction does not require X. campestris pv. vesicatoria effectors other than AvrBs3. Induction of hypertrophy requires the AvrBs3 repeat region, NLSs, and AAD. A collection of avrBs3 mutant derivatives expressed by the virulent X. campestris pv. vesicatoria strain I74A was tested for hypertrophy induction in pepper (Table 1). We first tested whether the repeat deletion derivative avrBs3Drep16, which renders X. campestris pv. vesicatoria avirulent in the bs3 pepper line Early Cal Wonder (ECW) but virulent in Bs3 lines, induces hypertrophy in Bs3 plants. Neither avrBs3Drep16 nor other avrBs3 repeat deletion derivatives (Herbers et al. 1992) were able to induce hypertrophy, indicating that hypertrophy depends on the wild-type AvrBs3 repeat region. Similarly, the AvrBs4 protein (previously designated AvrBs3-2; Bonas et al. 1993), which is 97% identical to AvrBs3 and differs from it mainly in the repeat region, did not induce hypertrophy in pepper, tomato, or when transiently expressed in N. clevelandii leaves (Fig. 1j). We reported previously that AvrBs3 contains two functional NLSs (Van den Ackerveken et al. 1996). Induction of hypertrophy in susceptible pepper leaves had requirements for the NLSs identical to those for the HR (i.e. hypertrophy developed only when NLS2 or NLS3 were present) (Table 1). A mutant carrying an 83-aa deletion of the NLS region, DNLS1-3 (Van den Ackerveken et al. 1996) could be complemented for hypertrophy induction by the heterologous, 8-aa NLS from the large T-antigen of simian virus SV40. In addition, the deletion derivative of AvrBs3 deleted in the acidic activation domain (AvrBs3DAAD) (Szurek et al. 2001) was unable to induce hypertrophy. Introduction of the heterologous AAD from the Herpes simplex protein VP16 did not restore hypertrophy (Table 1), although HR in resistant plants was partially restored (Szurek et al. 2001). This is probably due to the lower activity of the VP16 constructs already evidenced by a weak complementation for HR induction (Szurek et al. 2001). Transient expression of avrBs3 mutant derivatives in Nicotiana spp. showed that pustule induction had identical requirements for the AvrBs3 motifs as in X. campestris pv. vesicatoria infection experiments (data not shown). Taken together, these data indicate that all the AvrBs3 motifs found earlier to be needed for HR induction in Bs3 pepper plants also are essential for hypertrophy induction. AvrBs3-induced gene expression in pepper. Our previous results suggest that AvrBs3 is transported from X. campestris pv. vesicatoria into the plant nucleus where it may function in modulating transcription. To identify plant genes whose expression is affected by the AAD of AvrBs3, we performed a cDNA-AFLP analysis (discussed below). PolyA+ mRNA from susceptible pepper plants infected with X. campestris pv. vesicatoria strain 85-10 expressing AvrBs3 or AvrBs3DAAD was isolated 9 and 20 h postinoculation (hpi) and used in the cDNA-AFLP procedure. All 256 possible primer combinations corresponding to the ApoI/TaqI enzyme pair were employed, and approximately 21,800 different cDNA fragments (on average, 85 per primer combination) were inspected. Thirty induced (Fig. 2) and two repressed transcripts were identified. AFLP fragments were 60 to 326 bp in length and were extended by polymerase chain reaction (PCR) using a cDNA library as template (discussed below). Sequence analysis revealed that the fragments corresponded to 22 different induced and 2 repressed genes, because the same ApoI/TaqI fragments sometimes were amplified by multiple primer combinations in spite of a mismatch at the second-butlast 3 nucleotide position of a primer. In one case, two different, noncontiguous ApoI/TaqI fragments belonged to the same gene. The majority of differential fragments could already be detected at 9 hpi, but generally were more abundant at 20 hpi. Out of the 24 differentially expressed genes identified by the AFLP screen, 13 could be reproducibly confirmed to be induced by X. campestris pv. vesicatoria expressing avrBs3 (discussed below) using Reverse transcriptase-polymerase chain reaction (RT-PCR) and were studied further. These genes were designated upa1 to upa13 (upregulated by AvrBs3). The 11 other genes from the AFLP screen had highly variable or constitutive expression levels or proved difficult to amplify, and were not studied further. Sequence analysis of AvrBs3-induced genes. Five AvrBs3-induced genes (upa1 to 5) are homologous to members of a family of auxin-induced genes, the SAUR family (small auxin up RNA) (McClure and Guilfoyle 1987) (Table 2). Three upa genes show high homology to a-expansin genes. Two of these, upa7a and upa7b, are 100% identical over 191 bp corresponding to the smaller AFLP fragment, containing an extra ApoI site which allowed its separate isolation. Screening a pepper cDNA library, we obtained a full-length cDNA clone corresponding to the larger fragment upa7a. Its deduced amino acid sequence is 89% identical to NtEXP1, an expansin from tobacco (Link and Cosgrove 1998). RT-PCR primers may not discriminate between the transcripts corresponding to upa7a Table 2. AvrBs3-induced genes identified by cDNA-amplified fragment length polymorphism and confirmed by reverse transcriptase-polymerase chain reaction cDNAa upa1 upa2 upa3 upa4 upa5 upa6 upa7a, 7b upa8 upa9 upa10 upa11 Homology to Most homologous sequence (plant) Protein identity; similarity (%) NAA inducibility Cycloheximide sensitivityb Auxin-induced protein (SAUR family) Auxin-induced protein (SAUR family) Auxin-induced protein (SAUR family) Auxin-induced protein (SAUR family) Auxin-induced protein (SAUR family) a-expansins a-expansins Pectate lyases Hypothetical protein Hypothetical protein Anthocyanidin rhamnosyl transferases pir||T17020 (apple tree) pir||JQ1096 (soybean) pir||T07798 (radish) TGSAUR22 (tulip) pir||JQ1098 (soybean) gb|AAD13633.1| (tomato) gb|AAC96077.1| (tobacco) F22K18.20 (Arabidopsis) AL132959 (Arabidopsis) F22K18.80 (Arabidopsis) Q43716 (Petunia) 73; 86 71; 84 52; 59 59; 73 32; 52 96; 96 89; 91 85; 92 63; 79 49; 66 28; 45 No Yes Yes Yes Yes Yes No No No No No Yes Yes Yes Yes Yes Yes Yes Yes Yes No No a All cDNAs sequences contain a complete open reading frame with the exception of upa3 (370 bp), upa8 (1,196 bp), and upa11 (790 bp), for which only part of the cDNA sequence is known. b Pepper leaves were inoculated with Xanthomonas campestris pv. vesicatoria expressing avrBs3DAAD or wild-type avrBs3 from plasmids pDSF341 and pDSF340, respectively, without (–) or with (+) 50 µM cycloheximide. Samples for RNA isolation were collected 12 h postinoculation. Two samples of each inoculation are shown. The experiment was repeated three times with similar results. 640 / Molecular Plant-Microbe Interactions and upa7b; therefore, the RT-PCR product designated upa7 (Fig. 3) might represent several homologous expansinlike transcripts. The deduced amino acid sequence of upa6 is 96% identical to tomato a-expansin Exp5, expressed in developing fruit (Brummell et al. 1999) and is 62% identical to Upa7a. Transient expression of upa6 or upa7a under the control of the 35S promoter in N. clevelandii (Fig. 1k) or in pepper (data not shown) was not sufficient to trigger tissue alterations such as hypertrophy. The deduced amino acid sequence of upa8 shows homology to plant pectate lyases. Transcript upa9 contains a 67-aa open reading frame that exhibits weak homology (GAP: 30% aa identity) to a 65-aa protein of cucumber (CRG16), whose mRNA levels are gibberellin responsive and increase during cucumber hypocotyl elongation (Chono et al. 1996). The upa10-encoded protein shows no significant homology to any known protein. Computer-based analysis indicated a probable membrane localization (PSORT, 70%), and two polygalacturonase motifs (BLOCKS) that might indicate involvement of Upa10 in the metabolism of cell wall polymers. The gene upa11 is 45% similar on the amino acid level to anthocyanidin glucoside rhamnosyl transferases involved in the formation of purple pigmentation of flowers and stressed tissue (Brugliera et al. 1994). Both upa12 and upa13 encode putative transcription factors that will be described elsewhere. Kinetics and specificity of gene induction. Using RT-PCR, we studied the time-course of upa induction, comparing the mRNA patterns in tissue infiltrated with X. campestris pv. vesicatoria expressing avrBs3, avrBs3DAAD, or 10 mM MgCl2, from 4 to 20 hpi (Fig. 3). Although most genes were induced by wild-type AvrBs3 as early as 6 hpi, the five SAUR-like transcripts (upa1-5) only began to accumulate at 9 hpi, a delay suggesting an AvrBs3-dependent activation cascade. AvrBs3DAAD also induced some of the identified genes (Figs. 3 and 4), albeit more weakly than AvrBs3. Induction of some genes (e.g., upa11) was absolutely dependent on the C-terminal AAD. Previous results in yeast suggested that the N-terminal region in AvrBs3 also bears transcriptional activation activity (Szurek et al. 2001; Zhu et al. 1998), which might be responsible for the residual activation of other genes (e.g., upa10) by AvrBs3DAAD (Figs. 3 and 4). We also tested whether plant gene activation by AvrBs3 is repeat-region specific using X. campestris pv. vesicatoria strains delivering AvrBs3Drep16 or the AvrBs3 homologue AvrBs4 (Fig. 4). Both strains induced some but not all upa genes, and to different levels. For example, AvrBs4 activated upa6 and upa7, but not upa9, upa10, or upa11, while AvrBs3Drep16 activated upa9, but not upa10 and upa11. Induction of the expansinlike gene upa7 by AvrBs4 varied from experiment to experiment (Fig. 4A and B), but upa7 transcript levels usually were markedly higher with AvrBs3 than with AvrBs4. Environmental or developmental cues probably modulate the responsiveness of the upa7 promoter, and AvrBs3 appears to overcome potential repressors more efficiently than AvrBs4. Taken together, these results suggest a complex and specific host-gene activation spectrum for each AvrBs3-like protein. Dependence of upa induction on de novo protein synthesis. To investigate whether any of the upa genes might be directly induced by AvrBs3, we infiltrated cycloheximide together with the bacterial suspension to block plant protein synthesis, and analyzed upa profiles at 12 hpi by RT-PCR (Fig. 5; Table 2). Expression of ubiquitin, used as a control, was not affected by cycloheximide treatment in this time frame. In contrast, cycloheximide prevented induction of most AvrBs3- induced genes. Induction of these genes, therefore, requires synthesis of additional proteinaceous components and may not be directly induced by AvrBs3. However, upa10 and upa11 still were induced in the presence of cycloheximide, indicating either direct activation by AvrBs3 or via an available transcription factor, in an AvrBs3 AAD-dependent manner. AvrBs3-mediated upa11 transcript accumulation was markedly increased by cycloheximide treatment (Fig. 5), implying that a negative feedback loop involving protein synthesis normally limits accumulation of this transcript. This was also true, to a lesser extent, for upa10. Cycloheximide-treated tissue died 48 h after treatment; therefore, the dependency of hypertrophy induction on de novo protein synthesis could not be assessed. In summary, cycloheximide treatment revealed that AvrBs3 induces only a few transcripts directly and most upa genes indirectly. Auxin treatment induces some of the upa genes. To test whether the SAUR-like genes and other upas are induced upon auxin treatment, a solution of the synthetic auxin naphthalene acetic acid (NAA) (10 µg/ml) was infiltrated into pepper leaves. RT-PCR (Fig. 6; Table 2) revealed that four of the five SAUR-like genes were induced by NAA, indicating that they are probably the pepper equivalent of known SAURs. Interestingly, upa6 (expansinlike gene) also appeared to be induced upon NAA infiltration. Genes upa7 to upa11 did not show induction after NAA treatment (Fig. 6), even at later time points (data not shown). These results suggest that the AvrBs3induced genes fall into several classes distinguished by their auxin responsiveness. DISCUSSION AvrBs3 affects host cell morphology. Delivery of AvrBs3 by X. campestris pv. vesicatoria into susceptible pepper and tomato plants induces hypertrophy. Fig. 3. Time course of pepper gene induction. The kinetics of upa (upregulated by AvrBs3) expression after infection with Xanthomonas campestris pv. vesicatoria was analyzed by reverse transcriptasepolymerase chain reaction (RT-PCR). Pepper cultivar ECW leaves were inoculated with 10 mM MgCl2 (mock), X. campestris pv. vesicatoria strain 85-10 expressing avrBs3 (from pDSF340), or avrBs3DAAD (from pDSF341). Tissue samples for RNA isolation were taken at 4, 6, 9, 12 and 20 h postinoculation. UBI: ubiquitin was used as control for a constitutively expressed gene (PCR yields two ubiquitin-specific bands). Sequence homologies are shown in Table 2. Vol. 15, No. 7, 2002 / 641 This phenomenon is reminiscent of various pustule and canker symptoms induced by several Xanthomonas spp. in their hosts, including X. campestris pv. glycines in soybean, X. citri in citrus trees, and X. populi in poplar (Swings and Civerolo 1993). Pustules induced in soybean by X. campestris pv. glycines have been associated with mesophyll cell hypertrophy (Jones and Fett 1987), but the bacterial effectors responsible for this symptom have not been studied. Citrus canker is due to cell proliferation caused by the X. citri pthA gene (Swarup et al. 1992), which encodes a protein 96% identical to AvrBs3. When transiently expressed in Citrus spp. using particle bombardment or Agrobacterium spp., pthA triggered cell hypertrophy and proliferation (Duan et al. 1999). Transient expression of avrBs3 in leaves of Nicotiana spp. and potato plants also resulted in cell hypertrophy and, later, in cell division. However, pthA expression in Citrus spp. leads to eruption of the mesophyll tissue through the abaxial leaf epidermis and to cell death (Duan et al. 1999), which was not observed with avrBs3 in solanaceous plants. As the hypertrophied Nicotiana spp. tissue ages, cell division also occurs, which was not observed in pepper. Possible explanations for this difference include: (i) AvrBs3 sharing with PthA a capability to induce cell division if the infected cells survive long enough, which is not the case in X. campestris pv. vesicatoria-infected pepper tissue; (ii) dif- ferent reactions to AvrBs3 depending on the plant species; and (iii) cell division resulting from AvrBs3 overexpression under the control of the 35S promoter. For growth of X. campestris pv. vesicatoria in pepper leaves in laboratory conditions, hypertrophy is not a prerequisite. This is in contrast to the PthA-induced canker on citrus trees, which appears to provide an ecological niche necessary for bacterial growth (Swarup et al. 1991). Hypertrophy also could play a role in bacterial dispersal by decreasing the intercellular space volume at the end of the bacterial growth phase, resulting in a pressure that might expulse the bacteria out of the leaves, a dissemination mechanism proposed for PthA (Gabriel 1999). Another member of the avrBs3 family, avrb6 from X. campestris pv. malvacearum, was shown to enhance watersoaking of the infected tissue and promote release of the pathogen to the surface of cotton leaves (Yang et al. 1994). In field conditions, these effects might also apply for AvrBs3. Testing avrBs3 repeat-, NLS-, and AAD-mutant derivatives for hypertrophy-inducing activity largely confirmed the information on the functional regions in AvrBs3 based on HR tests. The requirement for these regions in both HR and hypertrophy induction implies that the two pathways share a similar beginning, probably up to gene activation. upa genes indeed are activated in resistant Bs3 plants before the onset of the HR Fig. 4. Effect of AvrBs3, AvrBs3Drep16, and AvrBs4 on upa induction. A, Leaves from pepper cultivar ECW were infiltrated with Xanthomonas campestris pv. vesicatoria strain 85-10 expressing avrBs3DAAD, avrBs3, avrBs3Drep16, and avrBs4 from plasmids pDSF341, pDSF340, pDSF316, and pDSF200, respectively. Pepper cultivar ECW leaves were inoculated with 10 mM MgCl2 (mock), X. campestris pv. vesicatoria strain 85-10 expressing avrBs3 (from pDSF340), or avrBs3DAAD (from pDSF341). Tissue samples for RNA isolation and reverse transcriptase-polymerase chain reaction analysis were taken at 4, 6, 9, 12 and 20 h postinoculation. Note that for avrBs3Drep16, the 22 h time point corresponds to the onset of the hypersensitive response. In this experiment, upa1 could not be amplified. B, Independent repeat of this experiment showing the expression pattern of upa7. 642 / Molecular Plant-Microbe Interactions (data not shown). Interestingly, none of the repeat deletion derivatives of AvrBs3 that retained or gained avirulence activity on different pepper and tomato lines (Herbers et al. 1992) displays hypertrophy-inducing activity. In contrast, a number of repeat deletion derivatives of PthA could still induce canker (Yang and Gabriel 1995a). Although the AvrBs3 homologue AvrBs4 induced some upa genes (e.g., the expansinlike transcripts), we never observed any AvrBs4-induced hypertrophy. AvrBs4 probably fails to induce hypertrophy because genes are not induced to a sufficient level or induction of key hypertrophy-related transcripts is missing. Consistent with the idea that hypertrophy may result from the cooperative action of many induced genes, overexpression of the upa7a expansinlike cDNA alone did not induce hypertrophy in pepper or N. clevelandii (Fig. 1k and data not shown). cell expansion (Carpita and Gibeaut 1993; Domingo et al. 1998; Inouhe and Nevins 1991), whereby they might function in synergy with expansins that promote “polymer creep” (Cosgrove 2000). More puzzling is the AvrBs3-induced expression of upa11, 45% similar on the amino acid level to plant anthocyanidin glucoside rhamnosyl transferases. Members of this gene family are involved in anthocyanin synthesis in flowers and stressed tissue. The fact that cycloheximide does not block upa11 induction suggests that the upa11 promoter might contain a consensus sequence required for AvrBs3 induction. Mechanism of AvrBs3-mediated gene induction. Our results suggest that different AvrBs3 family members activate distinct sets of plant genes. In support of this hypothesis, AvrBs3 induces host genes in an AAD-dependent manner. In the experimental design of our cDNA-AFLP screen, differences in gene expression are due only to a 36-aa deletion in the AAD of the X. campestris pv. vesicatoria effector AvrBs3. In this case, the number of differential genes is expected to be lower than when compatible and incompatible interactions are compared (Durrant et al. 2000) and the identified genes should reflect the specific activity of AvrBs3 in susceptible plants. The ApoI–TaqI enzyme pair was chosen to maximize the number of appropriate cDNA fragments for the AFLP technique. Based on the restriction site frequency in 33 known pepper cDNAs from the GenBank database, our screen is estimated to cover 30 to 40% of all expressed pepper genes, hence of the total number of AvrBs3-induced genes. The success in identifying upa genes whose induction is dependent on the AvrBs3 C-terminal activation domain is a strong argument in favor of the hypothesis that AvrBs3 acts as a transcription factor within the plant cell. This makes AvrBs3 the first known bacterial type III effector reported to target directly the host’s genome. The results of cycloheximide treatment experiments suggest a model in which AvrBs3 induces a few genes directly, whereas most upa genes are probably activated subsequently by AvrBs3-induced transcription factors. Presumed role of the induced genes. It is striking that many of the AvrBs3-induced genes show homology to genes involved in cell expansion, such as the expansinlike transcripts upa6, upa7a, and upa7b. The association of expansins with cell enlargement is well documented (Cosgrove 2000) and several expansin transcripts have been identified as auxin-induced (Catala et al. 2000; Civello et al. 1999; Hutchison et al. 1999), which was observed as well for upa6 (Fig. 6). The presence of SAUR transcripts shortly before cell enlargement has been reported (McClure and Guilfoyle 1989). Rapid induction of SAUR transcripts (McClure and Guilfoyle 1987) also was observed for upa2 to upa5. The small SAUR-encoded proteins, recently found to have calmodulinbinding activity, are hypothesized to be involved in the auxin signal transduction pathway (Yang and Poovaiah 2000). Several families of early auxin-induced genes are known (Abel and Theologis 1996). Among these, our cDNA-AFLP screen identified only SAUR genes, which suggests action of AvrBs3 downstream of auxin rather than in stimulating plant auxin synthesis. In preliminary experiments to address whether AvrBs3 stimulates the synthesis of auxin, auxin concentration in X. campestris pv. vesicatoria-infected pepper leaves appeared not to be correlated with presence or absence of avrBs3 (data not shown). Consistent with the AvrBs3-induced cellular phenotype, upa8 encodes a putative pectate-lyase. Hydrolysis of wall polymers by such enzymes has been hypothesized to facilitate Fig. 5. Effect of cycloheximide treatment on upa induction. Reverse transcriptase-polymerase chain reaction analysis of upa gene induction in the presence of the eukaryotic protein synthesis inhibitor cycloheximide. Pepper leaves were inoculated with Xanthomonas campestris pv. vesicatoria expressing avrBs3DAAD or wild-type avrBs3 from plasmids pDSF341 and pDSF340, respectively, without (–) or with (+) 50 µM cycloheximide. Samples for RNA isolation were collected 12 h postinoculation. Two samples of each inoculation are shown. The experiment was repeated three times with similar results. Vol. 15, No. 7, 2002 / 643 the many AvrBs3 family members present simultaneously in certain Xanthomonas strains contribute to virulence symptoms through distinct pathways rather than in an additive manner (Bai et al. 2000; Yang et al. 1996). It is not yet understood how subtle amino acid differences between the repeat regions of different AvrBs3 family members account for their different specificities. A central question concerning the likely role of AvrBs3 as a transcription factor is whether it binds to target promoter sequences or to host nuclear proteins which, themselves, make contact with regulatory DNA sequences. Our study provides the basis for the identification of AvrBs3-responsive sequences in the plant that will allow to test this hypothesis. MATERIALS AND METHODS Bacterial strains and plasmids, plant inoculations. Most bacterial strains and plasmids were described earlier (Szurek et al. 2001; Van den Ackerveken et al. 1996). Stability of all AvrBs3 derivatives was verified by immunoblotting. Strain I74A is a derivative of strain 85-10 with a wild-type Hrp phenotype but slower growth than the wild type in culture medium and in planta. For plant inoculations, X. campestris pv. vesicatoria was resuspended at an optical density at 600 nm (OD600) of 0.4 (5 108 CFU/ml) in 10 mM MgCl2 and inoculated with a needleless syringe into the intercellular space of the abaxial leaf surface. HR was scored 24 to 48 hpi, watersoaking or hypertrophy 4 to 6 dpi. Agrobacterium-mediated transient expression assays were performed with A. tumefaciens strain GV3101 as described (Van den Ackerveken et al. 1996) with the following modifications: incubation of bacteria in induction medium for 5 to 7 h and inoculation at an OD600 of 0.5 to 1 in infiltration medium (10 mM MgCl2, 5 mM MES, pH 5.3, 150 µM acetosyringone). Binary vector constructs have been described (Van den Ackerveken et al. 1996). For cycloheximide treatment, leaf tissue was inoculated with a bacterial suspension as above, containing 50 µM cycloheximide. Bacterial suspensions without cycloheximide were used as controls. Plant material. Pepper (Capsicum annuum) plants of cultivar ECW and the near-isogenic line ECW-30R containing the resistance gene Bs3 (Minsavage et al. 1990), tomato cultivar MoneyMaker, and Nicotiana spp. were grown in greenhouse conditions. Sixweek old plants were used for bacterial inoculations. Inoculated plants were transferred to a Percival growth chamber (Percival Scientific, Perry, IA, U.S.A.) at 28°C/24°C, 16 h of light; Nicotiana and tomato plants were transferred to a walkin chamber (Vötsch Industrietechnik, Balingen-Frommern, Germany) at 25°C/22°C and 16 h of light. Pepper cDNA library. A cDNA library was constructed from pepper line ECW using the lZAP cDNA kit (Stratagene, La Jolla, CA, U.S.A.). RNA was isolated from a mix of healthy leaves and leaves infected with avrBs3-containing X. campestris pv. vesicatoria strain 85-10 (pDSF340) collected 6, 9, and 20 hpi. Fig. 6. Effect of auxin on upa induction. Reverse transcriptase-polymerase chain reaction analysis of upa gene expression was performed after infiltration of 10 µg of the synthetic auxin naphthalene acetic acid (in 2 mM NaOH) or 2 mM NaOH alone (mock) per ml into pepper leaves. RNA was isolated from tissue collected 10, 30, and 60 min after infiltration. 644 / Molecular Plant-Microbe Interactions cDNA-AFLP analysis. cDNA-AFLP was performed as described (Bachem et al. 1996; Durrant et al. 2000). To minimize the identification of false positive transcripts due to leaf-to-leaf variability, one half of each leaf was infected with one strain and the other half with the second strain. For each time point, nine infected leaf halves from three different ECW plants were pooled for RNA extraction using standard protocols. PolyA+ RNA was isolated using Oligo(dT)-coupled DynaBeads (DYNAL, Oslo, Norway), and cDNA was produced using Expand Reverse-Transcriptase (Roche, Mannheim, Germany) according to the manufacturer’s instructions. AFLP reactions were carried out with 33P-labeled ApoI primers and resolved on 5% sequencing gels which were dried and exposed to X-ray film (Eastman Kodak, Rochester, NY, U.S.A.). After visual inspection of the autoradiographs, differential fragments were excised from the gel, eluted for 16 h in water, reamplified by PCR, and sequenced using an ABIPrism 377 DNA sequencer (Applied Biosystems, Foster City, CA, U.S.A.). For each primer combination yielding a differential fragment in the first AFLP screening, the AFLP procedure was repeated three times with cDNA isolated from independent inoculation experiments. The full-length sequence of most identified cDNAs was obtained by carrying out PCR reactions on the pepper cDNA library described above using primers internal to the AFLP fragment, in combination with M13 primers binding in the vector. Homology searches were performed with the BLAST and BLASTX programs (Altschul et al. 1990). Semiquantitative RT-PCR experiments. RT-PCR experiments were performed using primers yielding a 350- to 450-bp product for each upa gene. RT-PCR tem- plates were produced as follows: four leaf discs (1.3-cm diameter) from different pepper plants infected with the tested strains were pooled for RNA isolation. First-strand cDNA was synthesized from 4 µg of total RNA, with 200 pmol oligo(dT)20 and 200 units MMuLV-Reverse Transcriptase (Roche), according to the manufacturer’s instructions. Reactions were diluted 10 times and used as a template for PCR. Specific primers amplifying the ubiquitin transcript were used as control. Appropriate PCR cycle numbers specific to each gene and to the primer combination were determined by testing a wide range of cycle numbers and choosing one for which DNA amplification was still in the exponential phase. PCR conditions appropriate for each gene and the corresponding primer pair sequences are available upon request. Microscopy. Sections of plant leaf material (Fig. 1c) were made in agarose-embedded tissue maintained in styrofoam using a vibratome. For semithin sections (Fig. 1d–k), leaf segments were fixed for 3 h with 3% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) and dehydrated in a graded ethanol series. Ethanol was substituted by epoxy resin (Spurr 1969) and samples were polymerized at 70°C. Sections (1 µm) were made with a Ultracut-S ultramicrotome (LEICA, Reichert, Vienna) and stained with toluidine blue. ACKNOWLEDGMENTS We thank G. Hause for his help with microscopy and B. Szurek and L. Noël for fruitful discussions; B. Sotta and E. Miginiac (University of Paris 6, France) as well as A. Müller and E. Weiler (University of Bochum, Germany) for measuring auxin levels in pepper leaves and for stimulating discussions; A. Varet for suggesting the cycloheximide experiment; and T. Lahaye for critically reading of the manuscript. This work was supported by an E.U. Marie-Curie fellowship to E. Marois. LITERATURE CITED Abel, S., and Theologis, A. 1996. Early genes and auxin action. Plant Physiol. 111:9-17. Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. Bachem, C. W. B., van der Hoeven, R. S., de Bruijn, S. M., Vreugdenhil, D., Zabeau, M., and Visser, R. G. F. 1996. Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: Analysis of gene expression during potato tuber development. Plant J. 9:745-753. Bai, J., Choi, S.-H., Ponciano, G., Leung, H., and Leach, J. E. 2000. Xanthomonas oryzae pv. oryzae avirulence genes contribute differently and specifically to pathogen aggressiveness. Mol. Plant-Microbe Interact. 13:1322-1329. Ballvora, A., Pierre, M., van den Ackerveken, G., Schornack, S., Rossier, O., Ganal, M., Lahaye, T., and Bonas, U. 2001. Genetic mapping and functional analysis of the tomato Bs4 locus governing recognition of the Xanthomonas campestris pv. vesicatoria AvrBs4 protein. Mol. PlantMicrobe Interact. 14:629-638. Bonas, U., Conrads-Strauch, J., and Balbo, I. 1993. Resistance in tomato to Xanthomonas campestris pv. vesicatoria is determined by alleles of the pepper-specific avirulence gene avrBs3. Mol. Gen. Genet. 238:261269. Bonas, U., Schulte, R., Fenselau, S., Minsavage, G. V., Staskawicz, B. J., and Stall, R. E. 1991. Isolation of a gene cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and the hypersensitive response on pepper and tomato. Mol. Plant-Microbe Interact. 4:81-88. Bonas, U., Stall, R. E., and Staskawicz, B. 1989. Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol. Gen. Genet. 218:127-136. Bonas, U., and Van den Ackerveken, G. 1999. Gene-for-gene interactions: Bacterial avirulence proteins specify plant disease resistance. Curr. Opin. Microbiol. 2:94-98. Brugliera, F., Holton, T. A., Stevenson, T. W., Farcy, E., Lu, C. Y., and Cornish, E. C. 1994. Isolation and characterization of a cDNA clone corresponding to the Rt locus of Petunia hybrida. Plant J. 5:81-92. Brummell, D. A., Harpster, M. H., and Dunsmuir, P. 1999. Differential expression of expansin gene family members during growth and ripening of tomato fruit. Plant Mol. Biol. 39:161-169. Carpita, N. C., and Gibeaut, D. M. 1993. Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3:1-30. Catala, C., Rose, J. K. C., and Bennett, A. B. 2000. Auxin-regulated genes encoding cell wall-modifying proteins are expressed during early tomato fruit growth. Plant Physiol. 122:527-534. Chen, Z., Kloek, A. P., Boch, J., Katagiri, F., and Kunkel, B. N. 2000. The Pseudomonas syringae avrRpt2 gene product promotes pathogen virulence from inside plant cells. Mol. Plant-Microbe Interact. 13:13121321. Chono, M., Yamauchi, T., Yamaguchi, S., Yamane, H., and Murofushi, N. 1996. cDNA cloning and characterization of a gibberellin-responsive gene in hypocotyls of Cucumis sativus L. Plant Cell Physiol. 37:686691. Civello, P. M., Powell, A. L., Sabehat, A., and Bennett, A. B. 1999. An expansin gene expressed in ripening strawberry fruit. Plant Physiol. 121:1273-1280. Cornelis, G. R., and van Gijsegem, F. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. Cosgrove, D. J. 2000. Loosening of plant cell walls by expansins. Nature 407:321-326. Domingo, C., Roberts, K., Stacey, N. J., Connerton, I., Ruíz-Teran, F., and McCann, M. C. 1998. A pectate lyase from Zinnia elegans is auxin inducible. Plant J. 13:17-28. Duan, Y. P., Castañeda, A., Zhao, G., Erdos, G., and Gabriel, D. W. 1999. Expression of a single, host-specific, bacterial pathogenicity gene in plant cells elicits division, enlargement, and cell death. Mol. PlantMicrobe Interact. 12:556-560. Durrant, W. E., Rowland, O., Piedras, P., Hammond-Kosack, K. E., and Jones, J. D. 2000. cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell 12:963-977. Gabriel, D. W. 1999. The Xanthomonas avr/pth gene family. Pages 39-55 in: Plant-Microbe Interactions. Vol. 4. G. Stacey and N. T. Keen, eds. American Phytopathological Press, St. Paul, MN, U.S.A. Herbers, K., Conrads-Strauch, J., and Bonas, U. 1992. Race-specificity of plant resistance to bacterial spot disease determined by repetitive motifs in a bacterial avirulence protein. Nature 356:172-174. Hutchison, K. W., Singer, P. B., McInnis, S., Diaz-Sala, C., and Greenwood, M. S. 1999. Expansins are conserved in conifers and expressed in hypocotyls in response to exogenous auxin. Plant Physiol. 120:827-832. Inouhe, M., and Nevins, D. J. 1991. Inhibition of auxin-induced cell elongation of maize coleoptiles by antibodies specific for cell wall glucanases. Plant Physiol. 96:426-431. Jones, J. B., Stall, R. E., and Bouzar, H. 1998. Diversity among xanthomonads pathogenic on pepper and tomato. Annu. Rev. Phytopathol. 36:4158. Jones, S. B., and Fett, W. F. 1987. Bacterial pustule disease of soybean: Microscopy of pustule development in a susceptible cultivar. Phytopathology 77:266-274. Kearney, B., and Staskawicz, B. J. 1990. Widespread distribution and fitness contribution of Xanthomonas campestris avirulence gene avrBs2. Nature 346:385-386. Kjemtrup, S., Nimchuk, Z., and Dangl, J. L. 2000. Effector proteins of pathogenic bacteria: Bifunctional signals in virulence and host recognition. Curr. Opin. Microbiol. 3:73-78 Link, B. M., and Cosgrove, D. J. 1998. Acid growth response and a-expansins in suspension cultures of bright yellow 2 tobacco. Plant Physiol. 118:907-916. Lorang, J. M., Shen, H., Kobayashi, D., Cooksey, D., and Keen, N. T. 1994. avrA and avrE in Pseudomonas syringae pv. tomato PT23 play a role in virulence on tomato plants. Mol. Plant-Microbe Interact. 7:508515. McClure, B. A., and Guilfoyle, T. J. 1987. Characterization of a class of small auxin-induced soybean polyadenylated RNAs. Plant Mol. Biol. 9:611-623. McClure, B. A., and Guilfoyle, T. J. 1989. Rapid redistribution of auxinregulated RNAs during gravitropism. Science 243:91-93. Mindrinos, M., Katagiri, F., Yu, G. L., and Ausubel, F. M. 1994. The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78:1089-1099. Minsavage, G. V., Dahlbeck, D., Whalen, M. C., Kearney, B., Bonas, U., Staskawicz, B. J., and Stall, R. E. 1990. Gene-for-gene relationships specifying disease resistance in Xanthomonas campestris pv. vesicatoria - pepper interactions. Mol. Plant-Microbe Interact. 3:41-47. Morel, J. B., and Dangl, J. L. 1997. The hypersensitive response and the Vol. 15, No. 7, 2002 / 645 induction of cell death in plants. Cell Death Differ. 4:671-683. Ritter, C., and Dangl, J. L. 1995. The avrRpm1 gene of Pseudomonas syringae pv. maculicola is required for virulence on Arabidopsis. Mol. Plant-Microbe Interact. 8:444-453. Rossier, O., Wengelnik, K., Hahn, K., and Bonas, U. 1999. The Xanthomonas Hrp type III system secretes proteins from plant and mammalian pathogens. Proc. Natl. Acad. Sci. U.S.A. 96:9368-9373. Shan, L., He, P., Zhou, J.-M., and Tang, X. 2000. A cluster of mutations disrupt the avirulence but not the virulence function of AvrPto. Mol. Plant-Microbe Interact. 13:592-598. Spurr, A. R. 1969. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26:31-43. Swarup, S., De Feyter, R., Brlansky, R. H., and Gabriel, D. W. 1991. A pathogenicity locus from Xanthomonas citri enables strains from several pathovars of Xanthomonas campestris to elicit cankerlike lesions on citrus. Phytopathology 81:802-809. Swarup, S., Yang, Y., Kingsley, M. T., and Gabriel, D. W. 1992. A Xanthomonas citri pathogenicity gene, pthA, pleiotropically encodes gratuitous avirulence on nonhosts. Mol. Plant-Microbe Interact. 5:204-213. Swings, J. G., and Civerolo, E. L. 1993. Pages 44-47, 48-50, and 64-68 in: Xanthomonas. Chapman and Hall, London. Szurek, B., Marois, E., Bonas, U., and Van den Ackerveken, G. 2001. Eukaryotic features of the Xanthomonas type III effector AvrBs3: protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. Plant J. 26:523-534 Tsiamis, G., Mansfield, J. W., Hockenhull, R., Jackson, R. W., Sesma, A., Athanassopoulos, E., Bennett, M. A., Stevens, C., Vivian, A., Taylor, J. D., and Murillo, J. 2000. Cultivar-specific avirulence and virulence functions assigned to avrPphF in Pseudomonas syringae pv. phaseolicola, the cause of bean halo-blight disease. EMBO (Eur. Mol. Biol. Organ.) J. 19:3204-3214. Vancanneyt, G., Schmidt, R., O’Connor-Sanchez, A., Willmitzer, L., and Rocha-Sosa, M. 1990. Construction of an intron-containing marker 646 / Molecular Plant-Microbe Interactions gene: Splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol. Gen. Genet. 220:245–250. Van den Ackerveken, G., Marois, E., and Bonas, U. 1996. Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell 87:1307-1316. Vivian, A., and Arnold, D. L. 2000. Bacterial effector genes and their role in host-pathogen interactions. J. Plant Pathol. 82:163-178. Yang, B., Zhu, W., Johnson, L. B., and White, F. F. 2000. The virulence factor AvrXa7 of Xanthomonas oryzae pv. oryzae is a type III secretion pathway-dependent nuclear-localized double-stranded DNA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 97:9807-9812. Yang, T. B., and Poovaiah, B. W. 2000. Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action. J. Biol. Chem. 275:3137-3143. Yang, Y., and Gabriel, D. W. 1995a. Intragenic recombination of a single plant pathogen gene provides a mechanism for the evolution of new host specificities. J. Bacteriol. 177:4963-4968. Yang, Y., and Gabriel, D. W. 1995b. Xanthomonas avirulence/pathogenicity gene family encodes functional plant nuclear targeting signals. Mol. Plant-Microbe Interact. 8:627-631. Yang, Y., Defeyter, R., and Gabriel, D. W. 1994. Host-specific symptoms and increased release of Xanthomonas citri and X. campestris pv. malvacearum from leaves are determined by the 102-bp tandem repeats of pthA and avrb6, respectively. Mol. Plant-Microbe Interact. 7:345-355. Yang, Y., Yuan, Q., and Gabriel, D. W. 1996. Watersoaking function(s) of XcmH1005 are redundantly encoded by members of the Xanthomonas avr/pth gene family. Mol. Plant-Microbe Interact. 9:105-113. Zhu, W., Yang, B., Chittoor, J. M., Johnson, L. B., and White, F. F. 1998. AvrXa10 contains an acidic transcriptional activation domain in the functionally conserved C terminus. Mol. Plant-Microbe Interact. 11:824-832. Article 3: 2005 Nature Lipoprotein particles carry lipid-­linked proteins and are required for long-­range Hedgehog and Wingless signalling. Panáková D., Sprong H., Marois E., Thiele C., Eaton S. articles Lipoprotein particles are required for Hedgehog and Wingless signalling Daniela Panáková*, Hein Sprong*†, Eric Marois, Christoph Thiele & Suzanne Eaton Max Planck Institute of Molecular Cell Biology and Genetics, Pfotenhauerstrasse-108, 01307 Dresden, Germany * These authors contributed equally to this work † Present address: Department of Membrane Enzymology, Institute of Biomembranes, Utrecht University, Padualaan 8, 3584 CH Utrecht, The Netherlands ........................................................................................................................................................................................................................... Wnt and Hedgehog family proteins are secreted signalling molecules (morphogens) that act at both long and short range to control growth and patterning during development. Both proteins are covalently modified by lipid, and the mechanism by which such hydrophobic molecules might spread over long distances is unknown. Here we show that Wingless, Hedgehog and glycophosphatidylinositol-linked proteins copurify with lipoprotein particles, and co-localize with them in the developing wing epithelium of Drosophila. In larvae with reduced lipoprotein levels, Hedgehog accumulates near its site of production, and fails to signal over its normal range. Similarly, the range of Wingless signalling is narrowed. We propose a novel function for lipoprotein particles, in which they act as vehicles for the movement of lipid-linked morphogens and glycophosphatidylinositol-linked proteins. In the developing wing of Drosophila, Hedgehog activates shortrange target gene expression up to five cells away from its source of production, and longer-range targets over more than twelve cell diameters1. Wingless can signal through a range of over 30 cell diameters2. These morphogens are anchored to the membrane via covalent lipid modification3–9. The mechanisms that allow longrange movement of molecules with such strong membrane affinity are unclear. Like Wingless and Hedgehog, glycophosphatidylinositol (gpi)linked proteins transfer between cells with their lipid anchor intact10–12. We observed that gpi-linked green fluorescent protein (GFP) expressed in Wingless-producing cells spreads into receiving tissue at the same rate as Wingless, where it co-localizes with Wingless in endosomes. Thus, we proposed that these proteins travel together on a membranous particle, which we called an argosome13. How might argosomes form? One possibility is that argosomes are membranous exovesicles. Such particles could be generated by plasma membrane vesiculation, or by an exosomerelated mechanism14. Alternatively, argosomes might resemble lipoprotein particles like low-density lipoprotein (LDL). Vertebrate lipoprotein particles are scaffolded by apolipoproteins and comprise a phospholipid monolayer surrounding a core of esterified cholesterol and triglyceride. Insects construct similar particles called lipophorins15,16. Lipid-modified proteins of the exoplasmic face of the membrane (such as GFPgpi, Wingless or Hedgehog) might insert into the outer phospholipid monolayer of such a particle via their attached lipid moieties. Here, we use biochemical fractionation to determine the sort of particle with which lipid-linked proteins associate, and genetic means to address its function. Lipid-linked proteins copurify with lipophorin We compared sedimentation of Wingless, Hedgehog and gpi-linked proteins to that of transmembrane proteins, exosomes and lipophorin particles. To mark exosomes, we used flies expressing a vertebrate CD63:GFP fusion construct. CD63 is a tetraspanin that localizes to internal vesicles of multivesicular endosomes, and is released on exosomes17,18. In Drosophila imaginal discs, CD63:GFP localizes to late endosomes in producing cells, consistent with vertebrate studies (Supplementary Fig. S1A–D). It is released and endocytosed by neighbouring cells between one and three cell diameters away (Supplementary Fig. S1A,E), indicating that it is present on exosomes. To mark lipoprotein particles, we made antibodies to Drosophila 58 apolipophorins I and II (ApoLI and ApoLII); these proteins are generated by cleavage of the precursor pro-Apolipophorin19,20. Lipophorin is produced in the fat body20; consistent with this, we cannot detect apolipophorin transcripts in imaginal discs (data not shown). Nevertheless, the ApoLI and ApoLII proteins are as abundant in discs as in the fat body (Supplementary Fig. S1F and G). Plasma membrane and exosomal markers are completely pelleted after centrifugation for 3 h at 120,000g, whereas most ApoLII remains in the supernatant (Fig. 1a). Most Wingless:GFP and Hedgehog is present in the pellet, as are the gpi-linked proteins Fasciclin I21, Connectin22, Klingon23 and Acetylcholineasterase24 (Fig. 1b); this is not unexpected, because these proteins localize to the plasma membrane and internal membrane compartments. Surprisingly, however, some Wingless:GFP (6%), Hedgehog (2%) and gpi-linked proteins (14–22%) remain in the supernatant (Fig. 1b). The 120,000g supernatant (S120) contains both free soluble proteins and lipoprotein particles. To separate them, we performed isopycnic density centrifugation. In these gradients, lipophorin moves to the top low-density fraction whereas soluble proteins are present in higher-density fractions (top two panels of Fig. 1c). Gpi-linked proteins are found almost entirely in the top fraction with lipophorin. Treating the S120 with Phosphatidylinositolspecific phospholipase C (PI-PLC) before density centrifugation shifts their migration to higher-density fractions (Fig. 1c). This suggests that gpi-linked proteins associate with low-density particles via their gpi anchor. Similarly, when S120s from larvae that express Wingless:GFP or Hedgehog:HA in imaginal discs are subjected to isopycnic density centrifugation, these proteins are found in the lowest-density fraction with ApoLII, as is endogenous Hedgehog (Fig. 1d). Antibodies to endogenous Wingless detect a doublet in the top fraction and a band of somewhat higher mobility in high-density fractions. These data indicate that non-membrane-bound Wingless and Hedgehog associate with low-density particles in imaginal discs in vivo; other larval tissues may secrete Wingless in a non-lipophorin-associated form. We worried that lipophorin particles in the haemolymph might extract proteins from discs during larval homogenization, so we repeated these experiments using dissected discs. All Wingless, Hedgehog and ApoLII in the imaginal disc S120s are present on low-density particles (Fig. 1e), suggesting that their association is not an artefact of homogenization. Consistent with this, incubating © 2005 Nature Publishing Group NATURE | VOL 435 | 5 MAY 2005 | www.nature.com/nature articles pelleted imaginal disc membranes with an excess of purified lipoprotein particles does not extract Hedgehog:HA from membranes under the conditions used for homogenization (Fig. 1f). This suggests that association of lipid-linked morphogens with lipophorin depends on active cellular processes and does not occur during extract preparation. To ask whether lipid-linked proteins associated with lipophorin, or with some other low-density particle, we immunoprecipitated ApoLII from larval S120s and probed precipitates for Wingless, Hedgehog or GFPgpi. These proteins are immunoprecipitated by anti-ApoLII, but not pre-immune serum (Fig. 1g). Furthermore, anti-ApoLII is unable to precipitate secreted GFP that does not contain a gpi anchor (Fig. 1h). Hedgehog and Fas-1 also immunoprecipitated with ApoLII from the more purified top fraction of KBr gradients (Supplementary Fig. S1). Thus, lipid-linked morphogens and gpi-linked proteins associate directly with lipophorin particles. these morphogens are abundant; lipophorin has a nutritional function as well and many potential receptors are encoded in the genome27. Strong co-localization between lipophorin and lipidlinked morphogens is predicted if Wingless and Hedgehog are endocytosed with lipophorin. Nevertheless, we cannot exclude the possibility that these proteins were internalized separately and converged in the same endosomes. Lipophorin–RNAi perturbs lipid transport These experiments do not exclude the possibility that some Wingless or Hedgehog in the 120,000g pellet (P120) might be present on exosomes. To investigate this, we expressed CD63:GFP in either Wingless- or Hedgehog- producing cells and looked for colocalization with CD63:GFP-labelled exosomes in receiving tissue. No significant co-localization is detected (Fig. 2d–f, j–l). Thus, it seems unlikely that imaginal disc cells release Wingless or Hedgehog on exosomes, although the mechanism remains a possibility for transmembrane ligands such as Boss or Notch25,26. To test whether Wingless or Hedgehog co-localized with lipoprotein particles, we incubated imaginal discs with purified lipophorin particles fluorescently labelled with Alexa488; although they work for western blotting, neither anti-ApoLI nor ApoLII antibodies detect endogenous lipophorin by immunofluorescence. Immunostaining reveals that Wingless and Hedgehog are found in the same endosomes as Alexa488lipophorin (Fig. 2a–c, g–i). Unsurprisingly, lipophorin uptake is not limited to areas where To assess the role of lipophorin in larval growth and development, we reduced the levels of ApoLI and II by RNA interference directed against two different regions of the apolipophorin messenger RNA. Similar phenotypes were produced by each construct. To express double-stranded (ds)RNA, we used a modified GAL4:UAS system in which expression of inverted repeats can be temporally controlled by heat-shock-dependent excision of an intervening HcRed cassette by the flippase (FLP) recombinase. We tested extracts from wildtype larvae or larvae harbouring hs-flp, GAL4 driver and UAS dsRNA constructs at various times after heat shock to see how fast lipophorin levels were reduced (Fig. 3a). Larvae of the latter genotype made only 50% of the wild-type level of ApoLII, even in the absence of heat shock; basal activity of the heat-shock promoter in the fat body causes HcRed excision in approximately 50% of fat-body cells, although excision strictly depends on heat shock in other larval tissues (data not shown). Although they survive less frequently, these flies have no obvious phenotype. After heat shock, all fat-body cells excise the HcRed cassette and ApoLII levels decrease further. After four days, ApoLII is reduced to 5% of wild-type levels. ApoLI levels are reduced with similar kinetics (Supplementary Fig. S3). These animals prolong the third larval instar and rarely pupariate. We performed all the experiments described below on third-instar larvae 4–6 days after heat shock. To investigate the requirement for lipophorin in lipid transport, we assessed the accumulation of neutral lipids in larval tissues by staining them with Nile Red28. Cells of the posterior midgut Figure 1 Lipid-linked proteins co-fractionate with lipophorin. Western blots of fractionated extracts probed with antibodies to indicated proteins. a, b, Larval S120s and indicated proportions of larval P120s. AchE, acetylcholinesterase. c–e, KBr isopycnic density gradient fractions made from larval (c, d) or disc (e) S120s. Top fraction, 1.14 g cm23; bottom fraction, 1.4 g cm23. þ, PI-PLC-treated, 2, mock-treated. f, Top panel: P120 and S120 from Hedgehog:haemaglutinin (HA)-expressing discs, probed with anti-HA. Lower panel: P120 and S120 after incubating P120 at 4 8C with fivefold excess of purified lipoprotein particles and recentrifuging. g, h, S120s from wild-type or fusionprotein-expressing larvae immunoprecipitated with pre-immune, anti-ApoLII or anti-GFP serum. Morphogens colocalize with lipophorin NATURE | VOL 435 | 5 MAY 2005 | www.nature.com/nature © 2005 Nature Publishing Group 59 articles normally contain many small lipid droplets (Fig. 3b). Lipophorin reduction causes a dramatic expansion of these droplets (Fig. 3c), suggesting that lipophorin is required for the efficient extraction of lipid from the midgut. The wild-type fat body contains both small and large lipid droplets (Fig. 3d). Fat bodies of lipophorin–RNAi larvae are reduced in size and have fewer small lipid droplets (Fig. 3e), although larger droplets appeared normal. These data suggest that lipophorin delivers lipid to the fat body. Lipid droplets in discs from lipophorin–RNAi larvae are fewer and smaller than in the wild type (compare Fig. 3f and g). Their discs are also reduced in size, particularly in the wing pouch (data not shown). Thus, discs require lipophorin for accumulation of lipid droplets and for growth. Neither Caspase3 activation nor membrane phosphatidylinositol 3,4,5-phosphate (PIP3) accumulation is altered in lipophorin–RNAi discs (Supplementary Fig. S4), suggesting that their small size is not due to cell death or reduced insulin signalling29. Hedgehog function requires lipophorin To test whether lipophorin association was required for Hedgehog Figure 2 Wingless and Hedgehog co-localize with Alexa488lipophorin. Scale bars ¼ 10 mm. Blue lines indicate AP boundaries. a–c, Disc stained with anti-Wingless (a, and red in b) after 20-min incubation with Alexa488lipophorin (green in b, and c). d–f, Disc expressing CD63:GFP (green in b, and f) in Wingless-producing cells stained with anti-Wingless (d, and red in e). g–i, Disc stained with anti-Hedgehog (g, and red in h) after 20-min incubation with Alexa488lipophorin (green in h, and i). j–l, Disc expressing CD63:GFP (green in k, and l) in Hedgehog-producing cells stained with anti-Hedgehog (j, and red in k). 60 function, we examined Hedgeghog distribution and signalling in lipophorin–RNAi larval discs. In wild-type discs, Hedgehog expressed in the posterior compartment moves across the anterior–posterior (AP) compartment boundary and activates transcription of short and long-range target genes. Cells closest to the source respond by activating the transcription of collier (Fig. 4b, c) and patched (Fig. 4h, i). Further away, Hedgehog activates transcription of decapentaplegic (Fig. 4a, b)1,30,31. We monitored levels of Collier and a decapentaplegic reporter construct (dpplacZ) in wild-type and lipophorin–RNAi discs stained in parallel and imaged under identical conditions. Discs from lipophorin–RNAi larvae activate collier at least as efficiently as those of the wild type (compare Fig. 4c and f). In contrast, the range of activation of dppLacZ is significantly narrowed in lipophorin RNAi discs. dppLacZ is expressed up to 11 cells away from the AP boundary in wild-type discs (Fig. 4a, b), but only up to six cells away in lipophorin–RNAi larvae (Fig. 4d, e, m). These data suggest that lipophorin knockdown decreases the range of Hedgehog signalling. To discover whether Hedgehog trafficking was altered, we stained discs for Hedgehog and Patched. In wild-type discs, Hedgehog moves into the anterior compartment, where it is found in endosomes, often with Patched32,33 (Fig. 4g–i). Patched-mediated endocytosis is thought to sequester Hedgehog and limit its spread32,34. Hedgehog is most abundant up to five cell rows away from the AP Figure 3 Lipophorin–RNAi perturbs lipid transport. a, Extracts from equal numbers of wild-type (WT) or hs-flippase/þ; UAS dsRNA/Tubulin:GAL4 larvae probed for ApoLII at indicated hours after heat shock. b–g, Posterior midgut (b, c), fat body (d, e) or imaginal discs (f, g) from wild-type larvae (b, d, f) or hs-flippase/þ; Adh:GAL4/þ; UAS dsRNA/þ larvae (c, e, g) 5 days after heat shock. Yellow, neutral lipid; red, plasma membrane. Scale bar ¼ 40 mm (b, c) or 10 mm (d–g). © 2005 Nature Publishing Group NATURE | VOL 435 | 5 MAY 2005 | www.nature.com/nature articles boundary; although Hedgehog signals over a wider range, specific staining there cannot be distinguished from background. In lipophorin–RNAi discs, Hedgehog (Fig. 4j, k) accumulates to abnormally high levels in the first five rows of anterior cells. We counted 380 Hedgehog spots in the most apical 10 mm of the wild-type disc shown in Fig. 4g. The lipophorin–RNAi disc shown in Fig. 4j contains 1,208 Hedgehog spots in the same region. Most accumulated Hedgehog colocalizes with Patched (Fig. 4k, l) in endosomes (Supplementary Fig. S5). Furthermore, Patched co-accumulates more extensively with Hedgehog in endosomes than it does in wild-type (Fig. 4h, k). These data indicate that lipophorin RNAi either increases the susceptibility of Hedgehog to Patched-mediated endocytosis, or prevents subsequent degradation of the protein. We wondered whether lipophorin depletion might affect Hedgehog trafficking indirectly by preventing release of a needed co-factor from some other larval tissue. To investigate this, we added purified lipophorin particles to explanted lipophorin–RNAi discs and examined Hedgehog and Patched distribution. Abnormal Hedgehog and Patched accumulation was strongly reduced by a two-hour incubation of dissected discs with lipophorin particles (Supplementary Fig. S7). Thus lipophorin acts directly in imaginal discs to control Hedgehog trafficking, although it is still possible that its effects on signalling are indirect. Drosophila cannot synthesize sterols and relies on dietary sources. To assess whether reduced uptake of sterols or other lipids might cause the changes we see, we explored the effects of lipid deprivation on larval development. Larvae were allowed to hatch and feed on sucrose/agarose plates supplemented with yeast for 2–3 days, then transferred to plates containing chloroform-extracted yeast autolysate, rather than yeast. These larvae are developmentally delayed; after 7 days of lipid deprivation, their discs are much smaller than those of younger late-third-instar larvae (compare Fig. 5a, b). In contrast, yeast-fed siblings pupariate and begin to eclose by this time. Those flies that infrequently eclose after larval lipid depletion are small (35–60% of normal body weight) but normally patterned (Fig. 5c, d). Thus, lipid depletion stalls imaginal growth. To discover whether lipid starvation affected Hedgehog trafficking or signalling, we deprived larvae of lipid 2 days after hatching and stained their discs 6 days later (Fig. 5h–j). No changes in Hedgehog or Patched distribution are apparent in these discs compared with younger yeast-fed discs of similar size (Fig. 5e–g). Furthermore, the range of dpp and collier expression does not differ in lipid-starved and yeast-fed discs (Fig. 5k–p). Thus, lipid starvation does not mimic the effects of lipophorin knockdown. We speculate that lipid-starvation-induced growth arrest prevents membrane sterol from dropping to levels that would interfere with the Hedgehog pathway. Thus, lipophorin does not indirectly affect the Hedgehog pathway via lipid deprivation. Figure 4 Lipophorin–RNAi alters Hedgehog distribution and signalling. Blue lines ¼ AP boundary. Scale bar ¼ 10 mm. a–c, dpplacZ/þ disc 4 days after heat shock, stained for LacZ (a, and red in b) and Collier (green in b, and c). d–f, hs-flippase/þ;dpplacZ/þ; Tubulin:GAL4/UAS:dsRNA disc 4 days after heat shock, stained for LacZ (d, and red in e) and Collier (green in e, and f). g–i, Wild-type disc 4 days after heat shock, stained for Hedgehog (g, and red in h) and Patched (green in h, and i). j–l, hs-flippase/ þ;UAS:dsRNA/Tubulin:GAL4 wing disc 4 days after heat shock, stained for Hedgehog (j, and red in k) and Patched (green in k, and l). m, Average Collier and DppLacZ staining intensities for four wild-type and four lipophorin–RNAi discs. Blue line indicates AP boundary. Average distance from AP boundary of peak LacZ staining was 16.6 ^ 2.7 mm for the wild type, and 11.1 ^ 1.5 mm for lipophorin–RNAi. NATURE | VOL 435 | 5 MAY 2005 | www.nature.com/nature Wingless function requires lipophorin To discover whether lipophorin RNAi perturbed Wingless trafficking, we examined Wingless distribution. In lipophorin–RNAi discs, extracellular Wingless is less abundant on both the apical and basolateral epithelial surfaces and spreads over shorter distances © 2005 Nature Publishing Group 61 articles (Fig. 6a–d). However, no consistent alterations in intracellular Wingless are detected (not shown). Thus, lipophorin promotes accumulation of extracellular Wingless. To investigate whether Wingless signalling required lipophorin, we examined the activation of two target genes. Senseless is produced only in cells near the Wingless source and its expression is unaffected by lipophorin RNAi (not shown). Distalless is normally produced in a gradient throughout most of the wing pouch. In lipophorin–RNAi discs, the Distalless gradient is abnormally narrow (Fig. 6e–g). This suggests that lipophorin knockdown specifically perturbs long-range Wingless signalling. Here, we establish the principle that lipid-linked proteins of the exoplasmic face of the membrane associate with lipoproteins. These include many gpi-linked proteins with diverse functions, as well as the lipid-linked morphogens Wingless and Hedgehog. The mechanism allowing long-range dispersal of lipid-linked proteins is not yet understood. The finding that these proteins exist in both membrane-associated and lipoprotein-associated forms suggests reversible binding to lipoprotein particles as a plausible mechanism for intercellular transfer, and the consequences of lowering lipoprotein levels in Drosophila larvae supports this idea. Lipophorin knockdown narrows the range of both Wingless and Hedgehog signalling. Hedgehog accumulates to an abnormally high level in cells near the source of production and long-range signalling is inhibited; short-range target genes, however, are expressed normally. These data suggest that Hedgehog does not move as far when lipophorin levels are low. The range over which Hedgehog moves is normally restricted by Patched-mediated endocytosis. In discs from lipophorin RNAi larvae, accumulated Hedgehog co-localizes with Patched in endosomes, suggesting that it is more efficiently sequestered by Patched. How might lipophorin antagonize Patched-mediated sequestration and promote long-range movement? Our data are consistent with the idea that lipophorin is continuously needed for movement, rather than required only for the release of morphogens. If lipophorin were important only for Hedgehog secretion, we would expect lipophorin RNAi to decrease the amount of Hedgehog found in receiving tissue; this seems not to Figure 5 Hedgehog signalling is unaffected by lipid-depletion. a, b, Discs from fully fed (a) or lipid-starved (b) larvae. c, d, Wings from fully fed (c) or lipid-starved (d) flies. Scale bar ¼ 250 mm. e–j, Discs from fully fed (e–g) or lipid-starved (h–j) larvae stained for Hedgehog (e, h, and red in f, i) and Patched (green in f, i; and g, j). Scale bar ¼ 30 mm. k–p, Disc from fully fed (k–m) or lipid-starved (n–p) dpplacZ/þ larvae stained for LacZ (k, n, and green in l, o) and Collier (red in l, o; and m, p). Blue lines indicate AP boundary. Scale bars ¼ 10 mm. 62 © 2005 Nature Publishing Group NATURE | VOL 435 | 5 MAY 2005 | www.nature.com/nature articles Figure 6 Lipophorin–RNAi narrows the range of Wingless signalling. a–d, Apical (a, b) and basolateral (c, d) sections of wild-type (a, c) and Adh:GAL4/þ; UASdsRNA/þ (b, d) wing discs 5 days after heat shock, stained for extracellular Wingless. e, f, Distalless protein accumulation in wild-type (e) and hs-flippase/þ;UAS dsRNA/ TubulinGAL4 (f) wing discs 5 days after heat shock. g, Average Distalless staining intensity with distance from dorsal–ventral boundary of five wild-type (pink) and five hs-flippase/þ;UAS dsRNA/ TubulinGAL4 (blue) wing discs. For plots of individual discs, see Supplementary Fig. S6. be the case. Furthermore, altered Hedgehog trafficking in receiving tissue is consistent with a model in which lipophorin is required at each step of intercellular transfer. We favour the idea that reversible association of Hedgehog with lipophorin particles facilitates its transfer from the plasma membrane of one cell to that of the next. This model predicts that lowering lipophorin levels should increase the length of time that Hedgehog spends in the plasma membrane before becoming associated with lipophorin. This would slow its rate of transfer and increase the probability of Patched endocytosing Hedgehog before it moved to the next cell. Hedgehog would then signal efficiently in the short range, but be so efficiently sequestered by Patched that very little protein would travel far enough to activate long-range target genes. These predictions are completely consistent with our observations. This model differs significantly from our original concept of argosome function. We initially speculated that argosomes were exosome-like particles with an intact membrane bilayer, and that lipid-linked morphogens needed to be assembled on these particles to be secreted by producing cells. Instead, we find that argosomes are exogenously derived lipoproteins that facilitate the movement of morphogens through the epithelium. Many questions remain as to how morphogens become associated with argosomes, and how the spread and cell-interactions of these particles are regulated. Clearly, heparan sulphate proteoglycans are essential for the movement of Hedgehog and Wingless into receiving tissue35,36. Because heparan sulphate binds to vertebrate lipoprotein particles37,38, one might speculate that heparan sulphate proteoglygans (HSPGs) facilitate morphogen movement through lipoprotein binding. Conversely, we find many gpi-linked proteins, including the HSPG’s Dally and Dally-like (unpublished data), on lipoprotein particles themselves. These associated proteins have the potential to modulate the cellular affinities or trafficking properties of lipoproteins and the morphogens they carry. Our data suggest that lipophorin particles not only mediate intercellular transfer of Hedgehog, but may also be endocytosed together with the morphogen. Interestingly, LDL-receptor-related proteins Arrow and Megalin have demonstrated roles in Wingless signalling and Hedgehog endocytosis, respectively39–41. It is intriguing to speculate that these receptors might be important for interaction with the lipoprotein-associated form of the morphogen. Cholesterol has the potential to modulate the activity of the Hedgehog pathway at many different points3,42–44. Whether changes in the level of cellular cholesterol normally play a role in regulating the activity of the pathway is unclear. Here we show that Hedgehog interacts with the particle that delivers sterol to cells. This observation raises the possibility that internalization of Hedgehog is linked to sterol uptake, and suggests new mechanisms to link nutrition, growth and signalling during development. A NATURE | VOL 435 | 5 MAY 2005 | www.nature.com/nature Methods Fractionation Five millilitres of larvae were homogenized with 5 ml of 150 mM NaCl, 50 mM Tris-Cl pH 7.4, 2 mM EGTA plus protease inhibitors on ice. The 1,000g supernatant was centrifuged for 3 h at 33,600 r.p.m. (120,000g) at 4 8C in a SW40Ti rotor generating a pellet (P120) and supernatant (S120). For isopycnic density centrifugation, we added 0.33 g ml21 KBr to the S120, and centrifuged for 2 days at 40,000 r.p.m. (285,000g) at 10 8C in an SW40Ti rotor. Immunoprecipitation S120 pre-cleared 2 h with protein A-sephacryl CL4B beads was incubated with beads linked to different sera. Beads were washed with PBS, 1% BSA, then PBS, and eluted using Laemmli sample buffer or 150 mM NaCl, 2 mM EDTA, 100 mM Tris-Cl pH 8.3, 0.5% Nonidet-P40, 0.5% sodiumdeoxycholate and 0.1% SDS. © 2005 Nature Publishing Group 63 articles Antisera Rabbits were immunized with synthetic peptide LEGVIRRDSPKFKDL (Hedgehog amino acids 123–138), conjugated to keyhole limpet haemocyanin (Eurogentec). Antibody was affinity-purified on peptide-conjugated affigel-15 columns (Biorad). DNA encoding amino acids 195–509 or 891–1070 (parts of ApoLII and ApoLI, respectively) was amplified from GH18004 (Resgen) and cloned into pQE30. Histagged fusion proteins (Qiagen) were used to immunize rats or rabbits. Expression construct CD63:EGFP amplified from pEGFP-C1-bos (gift from G. Griffiths) was cloned into pUAST45. RNA interference RNA interference was induced by expressing inverted repeats derived from two different regions of the Pro-apolipophorin cDNA (607 bp ending 47 bp from stop codon, and 500 bp starting at ATG). The first was amplified and inserted into pENTR2B (Invitrogen). Using the Gateway system, we inserted it twice in inverted orientation into pFRIPE. pFRIPE is derived from pUAST; downstream of the UAS are two Gateway insertion sites flanking an FLP cassette containing the HcRed gene and a transcription termination sequence. The second fragment was amplified and cloned as an inverted repeat into pUhr. pUhr was derived from pUAST by inserting an HcRed-containing FLP cassette between the UAS and the multiple cloning site. Flies containing lipophorin–RNAi constructs were crossed with others harbouring heat-shock-inducible FLP and one of several GAL4 drivers. After 5 days at 25 8C, larvae were heat-shocked for 90 min at 37 8C; this causes excision in all cells as determined by HcRed fluorescence. No excision occurs without heat shock in any larval tissue except the fat body (not shown). dsRNA expressed under the control of either TubulinGAL4 (ubiquitous), AdhGAL4 (fat body and part of the gut) or C765GAL4 (disc-specific) was semi-lethal and produced identical larval phenotypes. No phenotype was ever observed when lipophorin dsRNA was expressed in imaginal discs. Immunohistochemistry Imaginal discs were fixed and stained as described13. Antibodies were diluted as follows: anti-Wg46, 1:200; anti-Hh47, 1:500; 1:100; anti-Ptc48 1:50; anti-bgal (Promega Z378A) 1:100; anti-Col31 1:200. To compare wild type and lipophorin–RNAi animals, tissues were stained in parallel and imaged under identical conditions with an LSMZeiss or Leica confocal microscope. Image analysis Hedgehog-positive spots in wild-type and lipophorin–RNAi discs were quantified in ten confocal sections 1 mm apart. The signal threshold was adjusted to 130 and images were despeckled using ImageJ (http://rsb.info.nih.gov/ij/). Grids were overlaid on the processed image and spots were counted manually. We used ImageJ to quantify the Hedgehog signalling range in five projected apical sections of Col- and Dpp-stained discs. For each image, we determined pixel intensity along ten lines centred at the AP boundary using the Plot Profilefunction of ImageJ and averaged them to obtain a plot for each disc. Average plots from four discs of each type were generated using Microsoft Excel. Distalless range in Fig. 6 and Patched staining in Supplementary Fig. S7 were quantified similarly. Lipid starvation Eggs were collected on apple juice/agar plates þ yeast for 24 h, then allowed to develop for 2–3 days on the same yeast-containing plates. Larvae were rinsed with PBS þ 0.05% TritonX100, treated for 10 s with 50% Na hypochlorite, and rinsed with sterile H2O. Larvae were transferred with sterile forceps to 10-cm plates containing 2% chloroform extracted agarose, 2.5% sucrose and 0.15% Nipagen, supplemented with either 0.3 g chloroform extracted yeast autolysate (for lipid starvation), or 0.3 g yeast (for lipid-fed controls). Labelling lipophorin with Alexa488 Lipophorin particles were fluorescently labelled with AlexaFluor 488 (Molecular Probes) according to the manufacturer’s instructions. Conjugate was separated from un-reacted label using Sephadex G-25 PD-10 columns (Amersham Pharmacia Biotech) and eluted with 100-mM Na-phosphate, pH 7.4, 100 mM NaCl, 10% sucrose. Incubation of dissected discs with lipophorin particles For experiments shown in Fig. 2 and Supplementary Fig. 7, imaginal discs were incubated at 29 8C with 50 mg ml21 lipophorin particles for 20 min and 2 h, respectively. On the basis of the starting volume of larvae and the final volume in which lipophorin was eluted, we estimate that this represents approximately 1/10 of the concentration of lipoprotein particles present in the haemolymph. Received 10 November 2004; accepted 28 February 2005; doi:10.1038/nature03504. 1. Strigini, M. & Cohen, S. M. A Hedgehog activity gradient contributes to AP axial patterning of the Drosophila wing. Development 124, 4697–4705 (1997). 2. Neumann, C. J. & Cohen, S. M. Long-range action of Wingless organizes the dorsal-ventral axis of the Drosophila wing. Development 124, 861–870 (1997). 64 3. Porter, J. A. et al. Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell 86, 21–34 (1996). 4. Willert, K. et al. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature 423, 448–452 (2003). 5. Pepinsky, R. B. et al. Identification of a palmitic acid-modified form of human Sonic hedgehog. J. Biol. Chem. 273, 14037–14045 (1998). 6. Williams, K. P. et al. Functional antagonists of sonic hedgehog reveal the importance of the N terminus for activity. J. Cell Sci. 112, 4405–4414 (1999). 7. Chamoun, Z. et al. Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science 293, 2080–2084 (2001). 8. Micchelli, C. A.,, The, I., Selva, E., Mogila, V. & Perrimon, N. Rasp, a putative transmembrane acyltransferase, is required for Hedgehog signaling. Development 129, 843–851 (2002). 9. Lee, J. D. et al. An acylatable residue of Hedgehog is differentially required in Drosophila and mouse limb development. Dev. Biol. 233, 122–136 (2001). 10. Liu, T. et al. Intercellular transfer of the cellular prion protein. J. Biol. Chem. 277, 47671–47678 (2002). 11. Dunn, D. E. et al. A knock-out model of paroxysmal nocturnal hemoglobinuria: Pig-a(-) hematopoiesis is reconstituted following intercellular transfer of GPI-anchored proteins. Proc. Natl Acad. Sci. USA 93, 7938–7943 (1996). 12. Anderson, S. M., Yu, G., Giattina, M. & Miller, J. L. Intercellular transfer of a glycosylphosphatidylinositol (GPI)-linked protein: release and uptake of CD4-GPI from recombinant adeno-associated virus-transduced HeLa cells. Proc. Natl Acad. Sci. USA 93, 5894–5898 (1996). 13. Greco, V., Hannus, M. & Eaton, S. Argosomes: a potential vehicle for the spread of morphogens through epithelia. Cell 106, 633–645 (2001). 14. Denzer, K., Kleijmeer, M. J., Heijnen, H. F., Stoorvogel, W. & Geuze, H. J. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J. Cell Sci. 113, 3365–3374 (2000). 15. Arrese, E. L. et al. Lipid storage and mobilization in insects: current status and future directions. Insect Biochem. Mol. Biol. 31, 7–17 (2001). 16. van der Horst, D. J., van Hoof, D., van Marrewijk, W. J. & Rodenburg, K. W. Alternative lipid mobilization: the insect shuttle system. Mol. Cell. Biochem. 239, 113–119 (2002). 17. Escola, J. M. et al. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 273, 20121–20127 (1998). 18. Wubbolts, R. et al. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J. Biol. Chem. 278, 10963–10972 (2003). 19. Sundermeyer, K., Hendricks, J. K., Prasad, S. V. & Wells, M. A. The precursor protein of the structural apolipoproteins of lipophorin: cDNA and deduced amino acid sequence. Insect Biochem. Mol. Biol. 26, 735–738 (1996). 20. Kutty, R. K. et al. Molecular characterization and developmental expression of a retinoid- and fatty acid-binding glycoprotein from Drosophila. A putative lipophorin. J. Biol. Chem. 271, 20641–20649 (1996). 21. Hortsch, M. & Goodman, C. S. Drosophila fasciclin I, a neural cell adhesion molecule, has a phosphatidylinositol lipid membrane anchor that is developmentally regulated. J. Biol. Chem. 265, 15104–15109 (1990). 22. Nose, A., Mahajan, V. B. & Goodman, C. S. Connectin: a homophilic cell adhesion molecule expressed on a subset of muscles and the motoneurons that innervate them in Drosophila. Cell 70, 553–567 (1992). 23. Butler, S. J., Ray, S. & Hiromi, Y. klingon, a novel member of the Drosophila immunoglobulin superfamily, is required for the development of the R7 photoreceptor neuron. Development 124, 781–792 (1997). 24. Incardona, J. P. & Rosenberry, T. L. Replacement of the glycoinositol phospholipid anchor of Drosophila acetylcholinesterase with a transmembrane domain does not alter sorting in neurons and epithelia but results in behavioral defects. Mol. Biol. Cell 7, 613–630 (1996). 25. Cagan, R. L., Kramer, H., Hart, A. C. & Zipursky, S. L. The bride of sevenless and sevenless interaction: Internalization of a transmembrane ligand. Cell 69, 393–399 (1992). 26. Klueg, K. M. & Muskavitch, M. A. Ligand-receptor interactions and trans-endocytosis of Delta, Serrate and Notch: members of the Notch signalling pathway in Drosophila. J. Cell Sci. 112, 3289–3297 (1999). 27. Culi, J. & Mann, R. S. Boca, an endoplasmic reticulum protein required for wingless signaling and trafficking of LDL receptor family members in Drosophila. Cell 112, 343–354 (2003). 28. Greenspan, P., Mayer, E. P. & Fowler, S. D. Nile red: a selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 100, 965–973 (1985). 29. Britton, J. S., Lockwood, W. K., Li, L., Cohen, S. M. & Edgar, B. A. Drosophila’s insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2, 239–249 (2002). 30. Vervoort, M. hedgehog and wing development in Drosophila: a morphogen at work? Bioessays 22, 460–468 (2000). 31. Vervoort, M., Crozatier, M., Valle, D. & Vincent, A. The COE transcription factor Collier is a mediator of short-range Hedgehog-induced patterning of the Drosophila wing. Curr. Biol. 9, 632–639 (1999). 32. Torroja, C., Gorfinkiel, N. & Guerrero, I. Patched controls the Hedgehog gradient by endocytosis in a dynamin-dependent manner, but this internalization does not play a major role in signal transduction. Development 131, 2395–2408 (2004). 33. Tabata, T. & Kornberg, T. B. Hedgehog is a signaling protein with a key role in patterning Drosophila imaginal discs. Cell 76, 89–102 (1994). 34. Chen, Y. & Struhl, G. Dual roles for patched in sequestering and transducing Hedgehog. Cell 87, 553–563 (1996). 35. The, I., Bellaiche, Y. & Perrimon, N. Hedgehog movement is regulated through tout velu-dependent synthesis of a heparan sulfate proteoglycan. Mol. Cell 4, 633–639 (1999). 36. Takei, Y., Ozawa, Y., Sato, M., Watanabe, A. & Tabata, T. Three Drosophila EXT genes shape morphogen gradients through synthesis of heparan sulfate proteoglycans. Development 131, 73–82 (2004). 37. Mahley, R. W. & Ji, Z. S. Remnant lipoprotein metabolism: key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E. J. Lipid Res. 40, 1–16 (1999). © 2005 Nature Publishing Group NATURE | VOL 435 | 5 MAY 2005 | www.nature.com/nature articles 38. Camejo, G., Hurt-Camejo, E., Wiklund, O. & Bondjers, G. Association of apo B lipoproteins with arterial proteoglycans: pathological significance and molecular basis. Atherosclerosis 139, 205–222 (1998). 39. McCarthy, R. A., Barth, J. L., Chintalapudi, M. R., Knaak, C. & Argraves, W. S. Megalin functions as an endocytic sonic hedgehog receptor. J. Biol. Chem. 277, 25660–25667 (2002). 40. Wehrli, M. et al. arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature 407, 527–530 (2000). 41. Tamai, K. et al. LDL-receptor-related proteins in Wnt signal transduction. Nature 407, 530–535 (2000). 42. Burke, R. et al. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell 99, 803–815 (1999). 43. Nakano, Y. et al. A protein with several possible membrane-spanning domains encoded by the Drosophila segment polarity gene patched. Nature 341, 508–513 (1989). 44. Cooper, M. K. et al. A defective response to Hedgehog signaling in disorders of cholesterol biosynthesis. Nature Genet. 33, 508–513 (2003). 45. Brand, A. H. & Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993). 46. Strigini, M. & Cohen, S. M. Wingless gradient formation in the Drosophila wing. Curr. Biol. 10, 293–300 (2000). 47. Taylor, A. M., Nakano, Y., Mohler, J. & Ingham, P. W. Contrasting distributions of patched and hedgehog proteins in the Drosophila embryo. Mech. Dev. 42, 89–96 (1993). 48. Capdevila, J., Pariente, F., Sanpedro, J., Alonso, J. & Guerrero, I. Subcellular localization of the segment NATURE | VOL 435 | 5 MAY 2005 | www.nature.com/nature polarity protein Patched suggests an interaction with the Wingless reception complex in Drosophila embryos. Development 120, 987–988 (1994). Supplementary Information accompanies the paper on www.nature.com/nature. Acknowledgements We thank T. Kurzchalia for advice regarding lipid depletion. We thank S. Cohen, I. Guerrero, P. Ingham, Y. Hiromi, M. Horscht, John Incardona and R. White for gifts of antibodies, and G. Griffiths for the CD63:GFP fusion construct. We are grateful to A. Mahmoud and S. Bowman for developing CFP:Rab5-expressing flies, and to V. Greco for helping to initiate these studies. We thank D. Backasch for performing embryo injections. K. Simons, M. Zerial, T. Kurzchalia and C. Dahmann provided comments on the manuscript. Author contributions This work has been a collaborative effort between the groups of C. Thiele and S. Eaton, the Thiele laboratory contributing biochemical expertise and the Eaton laboratory expertise in working with Drosophila. First authors appear in alphabetical order. Competing interests statement The authors declare that they have no competing financial interests. Correspondence and requests for materials should be addressed to S.E. (eaton@mpi-cbg.de) or C.T. (thiele@mpi-cbg.de). © 2005 Nature Publishing Group 65 Curriculum vitae Article 4: 2006 Development The endocytic pathway and formation of the Wingless morphogen gradient. Marois E., Mahmoud A., Eaton S. RESEARCH ARTICLE 307 Development 133, 307-317 doi:10.1242/dev.02197 The endocytic pathway and formation of the Wingless morphogen gradient Eric Marois, Ali Mahmoud and Suzanne Eaton* Controlling the spread of morphogens is crucial for pattern formation during development. In the Drosophila wing disc, Wingless secreted at the dorsal-ventral compartment boundary forms a concentration gradient in receiving tissue, where it activates shortand long-range target genes. The glypican Dally-like promotes Wingless spreading by unknown mechanisms, while Dynamindependent endocytosis is thought to restrict Wingless spread. We have utilized short-term expression of dominant negative Rab proteins to examine the polarity of endocytic trafficking of Wingless and its receptors and to determine the relative contributions of endocytosis, degradation and recycling to the establishment of the Wingless gradient. Our results show that Wingless is internalized via two spatially distinct routes: one on the apical, and one on the basal, side of the disc. Both restrict the spread of Wingless, with little contribution from subsequent degradation or recycling. As previously shown for Frizzled receptors, depleting Arrow does not prevent Wingless from entering endosomes. We find that both Frizzled and Arrow are internalized mainly from the apical membrane. Thus, the basal Wingless internalization route must be independent of these proteins. We find that Dally-like is not required for Wingless spread when endocytosis is blocked, and propose that Dally-like promotes the spread of Wingless by directing it to lateral membranes, where its endocytosis is less efficient. Thus, subcellular localization of Wingless along the apicalbasal axis of receiving cells may be instrumental in shaping the Wingless gradient. INTRODUCTION Morphogens are signaling proteins secreted by restricted groups of cells and forming a graded distribution in surrounding tissue (Neumann and Cohen, 1997). This concentration gradient activates different genes as a function of distance from the source, specifying cell identities in developing tissues. Gradient formation requires both a source of morphogen and a sink. Possible sinks include degradation in lysosomes of receiving tissue, degradation by extracellular proteases or release from tissue. The endocytic pathway in receiving tissue is an important regulator of morphogen gradients, although it can have opposing effects on the spread of different morphogens. The TGF homolog Decapentaplegic (Dpp) depends on the functions of Dynamin (Shibire – FlyBase) to move over long distances through the wing disc epithelium of Drosophila (Entchev et al., 2000) (but see also Belenkaya et al., 2004). Dynamin is a GTPase mediating scission of endocytic vesicles (Sever, 2002); thus, Dpp molecules are proposed to be actively passed through cells by planar transcytosis (i.e. successive rounds of internalization and recycling to the surface). By contrast, internalization restricts the range of FGF-8 movement in zebrafish – a process called restrictive clearance (Scholpp and Brand, 2004). Morphogens of the Wingless (Wg) and Hedgehog (Hh) families harbor covalently attached lipid moieties (Pepinsky et al., 1998; Porter et al., 1996; Willert et al., 2003; Zhai et al., 2004). Although lipid confers high membrane affinity on these proteins, they can escape cell membranes by binding to Lipoproteins. In the Drosophila wing disc, Wg and Hh require Lipoproteins to signal over long distances (Panáková et al., 2005). Neither Wg nor Hh Max-Planck Institute for Cell Biology and Genetics, Pfotenhauerstr. 108, 01307 Dresden, Germany. *Author for correspondence (e-mail: eaton@mpi-cbg.de) Accepted 3 November 2005 require Dynamin activity to spread through receiving disc tissue (Strigini and Cohen, 2000; Torroja et al., 2004); therefore, they probably move extracellularly rather than by planar transcytosis in imaginal discs. Loss of Dynamin function does perturb the range of Wg signaling in the embryonic ectoderm, however (Bejsovec and Wieschaus, 1995), suggesting that different transport mechanisms operate in different tissues. Several observations suggest that the endocytic pathway helps shape the Wg gradient. In imaginal discs, Wg accumulates on the cell surface of Dynamin mutant clones (Strigini and Cohen, 2000), suggesting that normal cells internalize and degrade Wg. In embryos, increased Wg lysosomal degradation appears to shorten its signaling range posteriorly (Dubois et al., 2001). Furthermore, recycling Wg back to the extracellular space was proposed to replenish the pool available for movement (Pfeiffer et al., 2002). Nevertheless, the respective contributions of internalization, degradation and recycling in controlling Wg spreading have not been systematically examined. The receptors that restrict the spread of Wg remain unidentified. Removing proteins that mediate Wg internalization and degradation should elevate Wg concentration on cell surfaces. Conversely, overexpressing such proteins might cause excess internalization and deplete extracellular Wg. However, ectopic Frizzled 2 (Fz2) overexpression actually stabilizes extracellular Wg (Cadigan et al., 1998; Baeg et al., 2004), suggesting that this receptor facilitates the spread of Wg. While Wg accumulates on the surface of cells missing both Frizzled receptors Fz1 and Fz2, it is nevertheless still internalized; thus, the role of Fz receptors appears complex (Baeg et al., 2004). The LDL receptor family protein Arrow is thought to act as a Wg co-receptor; its role in Wg trafficking is also unclear. Wg accumulates extracellularly on arrow mutant clones, but this may be an indirect effect; loss of Wg signaling increases the transcription of dally-like (Han et al., 2005), of which the protein product stabilizes Wg at the cell surface (see below). It is not yet known whether Wg internalization depends on Arrow. DEVELOPMENT KEY WORDS: Rab5, Rab7, Rab11, Imaginal disc, Polarized endocytosis, Drosophila RESEARCH ARTICLE Heparan sulfate proteoglycans (HSPGs) play a crucial, if little understood, role in promoting Wg spreading. Wg does not accumulate in tissue that cannot synthesize heparan sulfate (Baeg et al., 2004), and heparinase treatment in vitro causes Wg to disappear from both producing and receiving tissue (Greco et al., 2001). This mainly reflects the activity of the HSPG Dally-like (Dlp), which is required for long-range Wg movement and signaling (Franch-Marro et al., 2005; Han et al., 2005; Kirkpatrick et al., 2004; Kreuger et al., 2004). Dlp overexpression in imaginal discs causes massive accumulation of extracellular Wg (Baeg et al., 2001; Han et al., 2005). Thus, Dlp is thought to promote the interaction of Wg with the cell surface (Baeg et al., 2001; Baeg et al., 2004; Franch-Marro et al., 2005). Cleavage of the Dlp gpi anchor by Notum has also been proposed as a mechanism to promote long-range Wg movement (Kreuger et al., 2004). The possibility that Dlp might influence the endocytic trafficking of Wg has not yet been examined. Development 133 (2) Immunofluorescence Antibodies used were mouse anti-Wg (Strigini and Cohen, 2000), mouse anti-Dlp (Lum et al., 2003), rabbit anti-Dfrizzled2 (Strigini and Cohen, 2000), rat anti-DE-Cadherin2 (Oda et al., 1994), rabbit anti-Caspase (Cell Signalling Technology), recognizing Drosophila Drice and Dcp1 (Yu et al., 2002), rat anti-EGF Receptor (Jékely and Rørth, 2003). To generate Arrow antibodies, guinea pigs were immunized with a fragment of the protein corresponding to amino acids 1222-1450. Antibody specificity was confirmed by staining imaginal discs containing arrow– clones. Antibody stainings were performed as described (Strigini and Cohen, 2000), except that for extracellular protein staining, discs were incubated <3 minutes at room temperature with the Wg antibody before incubation on ice. Confocal images were acquired at 100⫻ magnification on a Zeiss Axiovert 200 microscope. PIPLC treatment MATERIALS AND METHODS To release gpi-linked proteins, wing imaginal discs dissected from third instar larvae were transferred to Grace’s insect medium containing 10 U/ml of PIPLC enzyme (Molecular Probes) and incubated at 25°C with mild agitation for indicated times. Inducible UAS constructs Image analysis pUHR was constructed by inserting an FRT>HcRed>FRT cassette (HcRed and SV40 terminator were PCR-amplified from pHcRed1-N1 [Clontech] with 5⬘ and 3⬘ primers both incorporating a wild-type FRT sequence) between the UAS and the multiple cloning site (MCS) of pUAST (Brand and Perrimon, 1993). When the cassette is present, Gal4 transcribes HcRed and any gene cloned in the MCS is silent. Heat-shock-mediated induction of Flipase excises the FRT cassette and triggers expression of the gene in the MCS. We generated pUHR derivatives containing the following cDNAs: GFPDlp (GFP inserted at a.a. 221); rab5SN subcloned from pUAST-Rab5SN (Entchev et al., 2000); rab7TN; rab11SN; rab4SN (donated by M. GonzálezGaitán). Larvae containing pUHR constructs, a GAL4 driver and hs-Flipase were heated for 1 hour 30 minutes to 37.2°C (causing cassette excision in virtually 100% of the cells) and dissected at indicated times. Inducible tissue-specific RNAi A 406-bp PCR product generated from the dlp coding sequence was cloned as an inverted repeat separated by an intron into pUAST. Transgenic flies were crossed with apterous-Gal4 also expressing ubiquitous Gal80ts, which represses GAL4 at the permissive temperature of 18° (McGuire et al., 2003). Dlp dropped to undetectable levels within 36 hours of shift to 29°. Experiments were performed 40 hours after temperature shift. To induce Dlp RNAi and Rab5SN sequentially, we first inactivated Gal80 (resulting in dlp RNAi) and 2 days later heat-shocked larvae to excise the FRT cassette in pUHR-Rab5SN. For rab7 RNAi, inverted repeats separated by an intron were cloned into pUHR. For rab11RNAi, inverted repeats were cloned without spacer intron into pUHR. For arrow RNAi, we inserted a 575-bp arrow cDNA fragment into pFRIPE. pFRIPE is a pUAST derivative in which inverted repeats can be inserted in one step by Gateway-based recombination (Invitrogen) on either side of an FRT-HcRed-FRT cassette containing a transcription terminator. The FRT cassette separating the two repeats provides inverted repeat stabilization during Escherichia coli growth, and RNAi inducibility in flies by heat-shock dependent cassette excision, which generates an inverted repeat without spacer. FP-Rab constructs To label Rab5, Rab11, Rab4 and Rab7 endosomes, we fused CFP and YFP(Venus) to the N-terminus of Rabs, cloned them into pCasper4 under control of the ubiquitous tubulin promoter and generated transgenic flies. Fluorescence intensity indicates that these constructs drive much weaker expression than the GAL4/UAS system. Dextran uptake Wing imaginal discs were dissected in M3 medium, transferred to medium containing 5 mg/ml Alexa 488-labeled Dextran 10,000 (Molecular Probes) and incubated for 10-15 minutes at 29°C. Discs were washed for 10 minutes in ice-cold PBS and fixed in PBS + 4% formaldehyde. To quantify Wg staining intensity in Fig. S3 (see supplementary material) and Dlp staining intensity in Fig. S6 (see supplementary material), pixel intensity along different lines centered on the dorsal-ventral boundary were quantified using the Plot Profile function of ImageJ and values at each point were averaged. To quantify co-localization between Wg, CFP-Rab5 and YFP-Rab7 in receiving tissue, we used threshold and despeckle functions of Image J to eliminate cytoplasmic signals from FP-Rab proteins. For Wg, this treatment virtually eliminated the dispersed staining outlining cell boundaries. Wg-expressing cells were excluded. We defined a particle as a signal of at least three contiguous pixels and counted particles automatically using the Analyze particles function of Image J. Colocalization was assessed for single confocal sections using the RGB-colocalization plugin and defined as an overlap of at least three pixels. Similar methods were used to quantify co-localization between membrane proteins and Rab5 or 7 endosomes, but the threshold was set higher to reduce signal from the plasma membrane. RESULTS Wingless is found in Rab5 and Rab7 endosomes The Rab5 GTPase regulates endocytic vesicle formation and early endosome fusion (Stenmark et al., 1994; Zerial and McBride, 2001). As Rab5 is replaced by Rab7, these early endosomes acquire degradative capacity and mature into lysosomes (Bucci et al., 2000; Rink et al., 2005). We used transgenic flies ubiquitously expressing moderate levels of CFP-Rab5 and YFP-Rab7 to localize these endosomes in discs. Endosomes positive for Rab5 only were most abundant in the apical- and basal-most regions of disc cells. The fraction of endosomes containing only Rab7 increased in central regions (see Fig. S1A in the supplementary material), suggesting endosomes move centrally as they mature. Consistent with the model that Rab5 is gradually replaced by Rab7, we observed significant co-localization between these two markers (see Fig. S1 in the supplementary material). To test whether Wg was present in these compartments, we examined colocalization between Wg, CFP-Rab5 and YFP-Rab7 in receiving tissue. Between 43 and 58% of Wg spots co-localized with Rab5 and/or Rab7 in the apical half of disc cells (Fig. 1). Basally, between 25 and 29% was found in these compartments (see Fig. S1B in the supplementary material). Thus a significant fraction of Wg was found in Rab5- and Rab7-positive endosomes. Non-colocalizing Wg may be extracellular, or in a different endocytic compartment. DEVELOPMENT 308 Endocytosis and the Wingless gradient RESEARCH ARTICLE 309 Fig. 1. Wingless is found in Rab5 and Rab7 endosomes. Wing imaginal discs ubiquitously expressing moderate levels of CFP-Rab5 and YFP-Rab7 stained with antibodies against Wg. Panels depict the different channels from a single confocal section 4 m below the apical surface (including a small part of the Wg expression domain, at bottom of panel). Yellow arrowheads indicate colocalization between Wg and CFP-Rab5, blue arrowheads colocalization between Wg and YFP-Rab7. Scale bar: 20 m. To identify sites of Wg internalization, we examined the subcellular localization of Wg in Rab5SN-expressing cells. Wg was strongly enriched apical to adherens junctions, and at the very basal side of the epithelium (Fig. 2A-D; quantified in Fig. S3 in the supplementary material), especially when discs were stained specifically for extracellular Wg (Fig. 2C). Polarized accumulation is not an artifact of reduced antibody access to lateral cell membranes, because lateral extracellular Wg accumulation was detected upon Dlp overexpression (Fig. 2E). This suggests that Wg is internalized from apical and basal (but not lateral) surfaces and that internalization normally limits its spread. Rab7-dependent degradation does not modulate the extracellular spread of Wingless To investigate whether the balance of Wg recycling versus degradation affected levels of extracellular Wingless, we perturbed these pathways. In vertebrate cells, expression of dominant negative Rab7 disperses the lysosomal compartment and inhibits degradation of endocytosed proteins (Bucci et al., 2000). In Drosophila disc cells, Rab7TN expression dispersed the punctate distribution of YFP-Rab7, leaving CFP-Rab5-labeled endosomes undisturbed (see Fig. S4A,B in the supplementary material) and caused accumulation of Hedgehog and Patched (Eugster and Eaton, unpublished), suggesting that lysosomal degradation is inhibited. Furthermore, YFP-Rab11-labeled recycling endosomes were more abundant in Rab7TN-expressing cells, suggesting that inhibiting the degradative pathway increased trafficking to recycling endosomes (see Fig. S4E in the supplementary material). Wg-positive endosomes in Rab7TN-expressing tissue were increased in size and number, and were present at a greater distance from the source than in non-expressing control tissue (Fig. 3A). The range of extracellular Wg distribution was unchanged, however (Fig. 3A⬘). Thus, the increased range of detection of Wg in Rab7TNexpressing tissue is not due to an increase in the distance over which extracellular Wg spreads, but rather to enhanced perdurance of internalized Wg protein. Slowing degradation does not increase the pool of Wg available for extracellular movement. Wings of adult flies that expressed Rab7TN were slightly upcurved and showed frequent vein deltas near the margin, but no notching or bristle defects indicative of gain or loss of Wg signaling (not shown). RNAi directed against Rab7 reduced RNA levels and DEVELOPMENT Rab5-dependent endocytosis restricts Wingless spread Previous studies using a temperature-sensitive dynamin mutant to block endocytosis indicated that Wg spreads extracellularly in the wing disc (Strigini and Cohen, 2000). As Dynamin also functions at the Golgi apparatus and Rab11 endosomes (Jones et al., 1998; Pelissier et al., 2003; Sever, 2002; van Dam and Stoorvogel, 2002), we specifically perturbed endocytosis by expressing the dominant negative Rab5SN (Entchev et al., 2000; Stenmark et al., 1994) in different subsets of the wing disc. When Rab5SN expression was induced during the third larval instar (see Materials and methods; see also experimental scheme in Fig. S2A in the supplementary material), endocytosis was inhibited within 2 hours, and cells in the wing pouch began to undergo apoptosis after 6-8 hours (see Fig. S2 in the supplementary material). We therefore limited our observations to discs that had expressed Rab5SN for 2-6 hours. Wg is expressed in a stripe of cells straddling the dorsal-ventral boundary and forms a symmetrical gradient in dorsal and ventral compartments. To ask how Rab5-dependent endocytosis affects Wg distribution, we expressed Rab5SN in the dorsal compartment of third instar discs and compared the shape of the Wg gradient in dorsal and ventral compartments (Fig. 2). One hour after inducing Rab5SN in dorsal cells, no change in Wg distribution was evident (not shown). By 2-4 hours (shortly after the block in Rab5-dependent internalization), Wg puncta disappeared but Wg protein levels were elevated in dorsal tissue nearest the Wg-expressing cells (Fig. 2A). By 5 hours, large amounts of Wg had invaded the entire dorsal wing pouch (Fig. 2B). Elevated Wg levels were not caused by induction of ectopic Wg expression (see Fig. S2H in the supplementary material). The spatial progression of Wg accumulation with time suggests that intervening tissue no longer acts as a sink for Wg when endocytosis is disrupted, and that Rab5-dependent internalization normally limits the range over which Wg spreads. Although Dynamin is needed for Wg secretion from producing cells (Strigini and Cohen, 2000), Rab5-dependent endocytosis is dispensable; targeted Rab5SN expression in the entire wg expression domain had no impact on Wg release even after long-term Rab5SN expression (up to 23 hours; data not shown). 310 RESEARCH ARTICLE Development 133 (2) Fig. 2. Wingless spreads further when Rab5-dependent internalization is blocked. (A-C) Wg staining in single confocal sections of discs that expressed Rab5SN for 4 hours (A) or 5 hours 30 minutes (B,C) in the dorsal compartment. Apical, middle and basal sections are shown. Discs in A,B were stained using a protocol detecting both intra- and extracellular Wg; C shows extracellular Wg. (D) Single xz section perpendicular to the dorsal/ventral boundary of a disc 5 hours 30 minutes after the onset of Rab5SN expression. Dorsal Rab5SN-expressing compartment is to the right. The dorsal-ventral compartment boundary is indicated by a white line. Cadherin (green) marks apical junctions. Elevated Wg (red) accumulates apically at the level of and above junctions. Green arrows indicate junctions within the overlying peripodial membrane. (E) Single confocal section through the middle (8 m below apical surface) of a wing disc that overexpressed GFP-Dlp for 24 hours in the dorsal compartment (indicated by brackets). The section passes through lateral cell membranes. Top: endogenous GFP-Dlp fluorescence. Bottom: extracellular Wg. GFP-Dlp overexpression elevates Wg on the lateral surface. Scale bar: 20 m. produced identical results (see Fig. S4F in the supplementary material). The surprisingly moderate effects of both Rab7 dominant negative expression and RNAi may reflect residual activity of Rab7; alternatively, non-Rab7-dependent degradative pathways may exist in Drosophila. Canonical recycling pathways do not promote Wingless spread To examine whether recycling played a role in Wg gradient formation, we expressed the dominant negative proteins Rab11SN and Rab4SN. Rab11 is required to transport both endocytosed proteins and some de-novo synthesized proteins through recycling endosomes to the plasma membrane (Ang et al., 2004; Lock and Stow, 2005; Maxfield and McGraw, 2004; Satoh et al., 2005). Vertebrate Rab4 directs rapid recycling from early endosomes to the plasma membrane and is essential for recycling certain receptors (de Renzis et al., 2002; Lazzarino et al., 1998; Seachrist et al., 2000; van der Sluijs et al., 1992). Inducing Rab11SN expression in imaginal discs dispersed wildtype YFP-Rab11 within 4 hours (see Fig. S5A in the supplementary material) and inhibited recycling of Delta (Emery et al., 2005), Cadherin (Shotgun – FlyBase) (Classen et al., 2005) and the EGF receptor (see Fig. S5C in the supplementary material). Furthermore, Rab7 endosomes are enlarged by Rab11SN expression, suggesting that inhibiting recycling may increase trafficking to Rab7 endosomes (see Fig. S5B in the supplementary material). Rab4SN expression prevented endosomal localization of wild-type YFPRab4 (see Fig. S5D in the supplementary material), but did not affect the morphology of other endosomes (not shown). Expression of Rab11SN, but not Rab4SN, activated apoptosis within 5 hours (Fig. 3B inset and not shown). Expression of either dominant negative alone, or both Rab11SN and Rab4SN together, did not affect Wg distribution (intra- or extracellular) within 5 hours (Fig. 3B,B⬘ and not shown). Similarly, RNAi directed against Rab11 reduced Rab11 RNA levels (see Fig. DEVELOPMENT Fig. 3. Inhibiting the degradative or recycling pathways does not affect extracellular Wingless distribution. (A,B) Wg conventional staining of wing discs expressing (A) Rab7TN, (B) Rab4SN + Rab11SN (inset shows nuclei positive for activated Caspase). Compare dorsal expressing and ventral non-expressing compartments. (A⬘,B⬘) Wg extracellular staining of the same genotypes. The dorsal-ventral boundary is indicated by a red line. Wg extracellular distribution is not altered by any of these treatments, but note intracellular Wg accumulation in A. Scale bars 20 m. Endocytosis and the Wingless gradient RESEARCH ARTICLE 311 Fig. 4. Arrow knockdown causes intracellular Wg accumulation and loss of Wg signaling. (A) Anti-Arrow staining of wing disc with arrow RNAi induced in the dorsal compartment. Dorsal cells lack Arrow, except in four small clones that failed to excise the HcRed cassette and initiate RNAi (imaged using higher gain than Fig. 5C to maximize residual Arrow detection). (B) Top panel shows a control wing. Middle panel shows a wing from a fly that emerged 6 days after arrow RNAi induction in the posterior compartment. Lower panel shows a wing from a fly that emerged 6 days after induction of GFP-Dlp overexpression in the posterior compartment. (C) Projection of 12 confocal sections 1 m apart from a disc that expressed arrow RNAi in the dorsal compartment for 48 hours, stained for Wg. (D) Single confocal section of the disc shown in (C) showing Wg, CFPRab5 and YFPRab7. Yellow arrowheads show examples of co-localization between Wg and endosomes. Scale bar: 20 m. Arrow is not required for Wingless endocytosis Although it accumulates extracellularly, Wg is still also found in endosomes in Fz1-Fz2 double mutant clones, suggesting that Wg internalization can occur via other receptors (Baeg et al., 2004). Whether Arrow (a Lipoprotein receptor family member required for Wg signaling) internalizes Wg is unknown. To investigate its role in Wg trafficking, we reduced Arrow protein levels by RNAi in the dorsal compartment. Arrow protein became undetectable 2 days after RNAi induction (Fig. 4A), and emerging adults had phenotypes suggesting loss of Wg signaling (Fig. 4B). Wg-positive Rab5 and Rab7 endosomes were more abundant (2.9±0.6-fold, n=3) over a longer range in Arrow RNAi tissue (Fig. 4C,D). These endosomes were also located more apically than those of wild-type cells (see Fig. S6A in the supplementary material). Thus Arrow is not required for Wg internalization, but appears to modulate its trafficking. As Wg is also detected in endosomes over a longer range when Rab7 activity is reduced (Fig. 3A), loss of Arrow may reduce the rate of Wg degradation after endocytosis. Like arrow mutant clones (Han et al., 2005), tissue undergoing arrow RNAi accumulated some extracellular Wg (data not shown); however, this may reflect increased stabilization caused by elevated Dlp protein levels near the dorsal-ventral boundary (see Fig. S6B in the supplementary material), consistent with previous observations (Han et al., 2005). Fz2 and Arrow are internalized apically by Rab5dependent endocytosis Our results suggest that Wg is internalized from both the apical and basal disc surfaces. We expected that receptors mediating Wg internalization should co-accumulate with Wg on these surfaces when endocytosis was blocked. We therefore examined the subcellular localization of Fz1, Fz2 and Arrow after Rab5SN induction. In the steady state, Fz2 is found predominantly on the basal-lateral side of wing epithelial cells (Wu et al., 2004) (Fig. 5C, non-Rab5SN-expressing region). However, after 5 hours of Rab5SN expression, Fz2 accumulated dramatically on the apical side of the epithelium (Fig. 5C,D and Fig. S6C in the supplementary material) and was only slightly elevated on basal and lateral membranes (Fig. 5C). These data suggest that, despite its steady state basal-lateral DEVELOPMENT S5E in the supplementary material), but did not alter Wg distribution (see Fig. S5F,F⬘ in the supplementary material). Thus, it seems unlikely that either Rab11- or Rab4-dependent recycling pathways contributes to the spread of Wg in imaginal discs, although as yet unknown recycling pathways might exist. These data show that Wg gradient formation is controlled by apical and basal internalization, with little contribution from canonical degradative or recycling pathways. RESEARCH ARTICLE localization, Fz2 is delivered to the apical side of the cell but does not accumulate there because of rapid internalization. By contrast, Fz1-GFP localization did not change obviously under these conditions (not shown). As Fz1 and 2 function redundantly, it is possible that Fz1 internalization might be more apparent in the absence of Fz2. Unexpectedly, some Fz2 accumulation may reflect an increase in synthesis, rather than a decrease in endocytosis, because Fz2 mRNA levels rose in response to Rab5SN expression (Fig. 5A). However, the apical shift in Fz2 subcellular distribution Development 133 (2) when endocytosis was blocked indicates that it is normally internalized from the apical surface. As Fz2 is internalized slowly, if at all, from the basal surface, it is unlikely to mediate Wg endocytosis there. Like Fz2, endogenous Arrow protein levels rise with distance from the dorsal-ventral boundary (Fig. 5C, middle). This reflects its transcription (Fig. 5B, left), which, like that of Fz2 (Cadigan et al., 1998), is lowered by Wg signaling (Fig. 5B, middle). Arrow protein is mainly basal-lateral in the steady state (Fig. 5C, non- Fig. 5. Rab5SN expression alters subcellular localization and mRNA levels of Fz2 and Arrow. (A) In-situ hybridization to Fz2 mRNA in a disc after 4 hours 30 minutes Rab5SN expression in the posterior compartment. Inset shows Rab5SN expression domain (green). (B) In-situ hybridizations detecting arrow mRNA in discs of different genotypes. Left: wild type. Middle: disc expressing GFP-Wg in the posterior compartment for 6 hours. Right: disc expressing Rab5SN in the posterior compartment for 4 hours 30 minutes. (C) Discs expressing Rab5SN in the dorsal compartment for 5 hours 30 minutes stained for Arrow, Fz2 and Armadillo to mark apical junctions. Apical, middle sections and the basal region are shown. Note the accumulation of Fz2 and Arrow above the junctions. Large arrows in middle sections indicate the apical cell surface. In the basal region, the wing pouch curves so that the apical-basal axes of the epithelial cells at the edges of the wing pouch lie parallel to the focal plane. Thus, a complete longitudinal section of these cells is visible. Small arrows point to their basal sides. (D) Larger magnification of the areas boxed in C. Armadillo staining reveals the apical junctions of two rows of epithelial cells facing each other. Fz2 and Arrow accumulate above cellular junctions in Rab5SN-expressing tissue. (E) Single confocal section of a wing imaginal disc expressing YFP-Rab7, CFP-Rab5 and stained for Arrow. Arrowheads indicate co-localization between Arrow and Rab5/7 endosomes. Quantifying ten confocal sections corresponding to the apical-most 10 m of the disc shown indicates that 49% of the brightest Arrow spots co-localize with either YFP-Rab7 or CFP-Rab5. Scale bar: 20 m. DEVELOPMENT 312 Endocytosis and the Wingless gradient Rab5SN-expressing cells) and is also present in both Rab5 and Rab7 endosomes (Fig. 5E). In response to Rab5SN expression, punctate Arrow staining disappeared and Arrow accumulated at the apical membrane, co-localizing with Fz2 (Fig. 5C,D and Fig. S6C). Thus Arrow, like Fz2, is internalized apically and is unlikely to mediate basal Wg endocytosis. While these proteins may internalize Wg from the apical surface, an Arrow and Fz2independent Wg internalization pathway must operate at least on the basal surface. We wondered whether arrow mRNA levels, like those of Fz2, might rise in response to Rab5SN expression. Surprisingly, the opposite was true. Arrow mRNA plunged to undetectable levels by 4 hours after Rab5SN induction (Fig. 5B, right). Arrow protein, however, was not reduced within this time. RESEARCH ARTICLE 313 Dlp accumulates upon Rab5SN expression Dlp assists extracellular Wg spreading in discs (Baeg et al., 2001; Franch-Marro et al., 2005; Giraldez et al., 2002; Han et al., 2005) and localizes predominantly to the basal-lateral membrane in the steady state (Baeg et al., 2004) (Fig. 6A,B). As early as 4 hours after inducing Rab5SN expression in the dorsal compartment, endogenous Dlp dramatically accumulated around the apical-lateral region of expressing cells and more modestly on the basal-lateral membrane (Fig. 6A,B and Fig. S6D). dlp mRNA levels rose within 4 hours 30 minutes of initiating Rab5SN expression (Fig. 6C), suggesting that Dlp protein accumulates in part because of increased synthesis. However, the strong apical shift in Dlp subcellular localization suggests that Rab5-dependent internalization normally prevents apical-lateral accumulation of Dlp. We were surprised that Rab5SN expression trapped Dlp at the cell membrane, because gpi-linked proteins are thought to be internalized by a Rab5-independent mechanism (Sabharanjak et al., 2002). To investigate whether Dlp, like Wg and Arrow, was internalized into Rab5 endosomes, we examined co-localization between endogenous Dlp, YFP-Rab7 and CFP-Rab5. Unlike Wg or Arrow, the Dlp signal only rarely overlaps with that of CFP-Rab5 or YFP-Rab7 (Fig. 6D-G). Therefore, apical-lateral enrichment of Dlp in Rab5SN-expressing tissue may occur by an indirect mechanism. For example, Dlp may be recruited there by other proteins accumulating when Rab5 endocytosis is blocked. Alternatively, Dlp may move swiftly through these compartments, failing to accumulate to easily detectable levels. Fig. 6. Rab5SN expression alters subcellular localization and mRNA levels of Dlp. (A,B) Wing disc after 5 hours 30 minutes Rab5SN expression in dorsal compartment, stained for Dlp. Dlp accumulates more strongly apically (A) than basal-laterally (B) in response to Rab5SN expression. (C) In-situ hybridization to dlp mRNA in a wild-type disc (top) and a disc after 4 hours 30 minutes Rab5SN expression in the posterior compartment (to the right). (D-G) Single confocal section of a disc stained for endogenous Dlp and expressing the indicated FP-Rab proteins. Quantifying ten confocal sections corresponding to the apical-most 10 m of this disc indicates that 19% of the brightest Dlp spots co-localize with either YFP-Rab7 or CFP-Rab5. D, dorsal compartment; V, ventral compartment. Wingless spreads independently of Dlp during the endocytosis block Although Dlp is not needed to maintain Wg on the surface of Rab5SN-expressing cells, it might be required earlier to promote Wg spread across Rab5SN-expressing tissue. To address this possibility, we used RNAi to deplete Dlp from the dorsal compartment before Rab5-dependent endocytosis was blocked (see Materials and methods). In discs that did not express Rab5SN, inducing dlp-specific RNAi rendered Dlp protein undetectable within 2 days, and reduced the range of extracellular Wg (Fig. 7E,F), consistent with the effect of dlp null clones (Franch-Marro et al., 2005; Han et al., 2005). dlp RNAi even abolished Dlp detection in Rab5SN-expressing cells (Fig. 7H⬘), which normally have much higher levels of Dlp on their surface (Fig. 6A). Despite the absence of detectable Dlp, blocking Rab5dependent internalization still caused accumulation of extracellular Wg throughout the wing pouch within 5 hours 30 minutes (Fig. DEVELOPMENT Wingless is not recruited by elevated Dlp during the endocytosis block Because Dlp overexpression elevates Wg levels at the plasma membrane (Baeg et al., 2001; Franch-Marro et al., 2005; Giraldez et al., 2002; Han et al., 2005), we wondered whether Wg accumulation on Rab5SN-expressing cells was an indirect effect of increased Dlp. To see whether Wg was bound to these cells via Dlp, we treated discs that had expressed Rab5SN for 5 hours 30 minutes with Phosphatidylinositol-Phospholipase C (PI-PLC) to release gpi-linked proteins. One hour of PI–PLC treatment entirely removed even the high levels of Dlp present on Rab5SNexpressing cells (Fig. 7C). Wg, however, was not released by this treatment (Fig. 7D). Therefore, Wg is not trapped at the surface of Rab5SN-expressing cells by binding to Dlp or any other gpianchored protein. As long as endocytosis is prevented, Wg does not require glypicans to accumulate extracellularly on disc epithelial cells. RESEARCH ARTICLE 7G,H). These data show that Dlp is not needed for the spread of Wg if Rab5-dependent internalization is prevented. Other HSPGs might then ensure the cell-to-cell transfer of Wg. We wondered whether Dlp might normally promote the spreading of extracellular Wg by antagonizing Wg internalization. Dlp overexpression strongly elevates the level of extracellular Wg, Development 133 (2) especially on lateral membranes (Fig. 2E). To ask whether accumulated Wg was accessible to the endocytic pathway, we compared co-localization of Wg with endosomal markers in Dlpoverexpressing versus normal tissue (Fig. 7I-L). Although Dlp overexpression increased the range over which Wg-positive endosomes were found, their frequency was not significantly higher Fig. 7. Removal of Dlp by PI-PLC treatment or by RNAi does not affect Wg accumulation on Rab5SN-expressing cells. (A-D) Imaginal discs after 5 hours 30 minutes Rab5SN expression in the dorsal compartment (dorsal Rab5SN-expressing tissue indicated by brackets). Images are projections of 1-m-spaced confocal sections over 20 m. (A) Disc mock-incubated for 1 hour, stained for Dlp. (B) Disc mock-incubated for 1 hour, stained for Wg. (C) Disc incubated for 1 hour with PI-PLC, stained for Dlp. (D) Disc incubated for 1 hour with PI-PLC, stained for Wg. Wg is not released with Dlp. (E,F) Discs subjected to Dlp RNAi for 40 hours in the dorsal compartment (indicated by brackets) stained for Dlp (E) or extracellular Wg (F). Dlp protein is undetectable and extracellular Wg fails to spread in dorsal cells. (G,H) Apical extracellular Wg staining of discs in which Rab5SN has been expressed for 5 hours in the dorsal compartment (indicated by brackets), either in the presence (G) or absence (H) of Dlp protein. (H⬘) shows depletion of Dlp by RNAi in the dorsal compartment of a disc also expressing Rab5SN. (I-L) Disc ubiquitously expressing YFPRab7, overexpressing Dlp in the dorsal compartment (indicated by bracket) and stained for Wg. A projection of three sections 3 m below the apical membrane is shown. (I) Wg imaged with high gain, emphasizing plasma membrane recruitment. (J) Wg imaged with low gain, emphasizing punctate endosomal staining. (K) YFPRab7-labeled endosomes. (L) Overlay of Wg and YFPRab7. DEVELOPMENT 314 Endocytosis and the Wingless gradient RESEARCH ARTICLE DISCUSSION The mechanisms that promote and inhibit the spread of morphogens play important roles in pattern formation and have been the subject of intense study. Although planar transcytosis (González-Gaitán et al., 1994) has been proposed to explain Wg spread (Bejsovec and Wieschaus, 1995), other data suggest that, at least in imaginal discs, Wg travels extracellularly. Loss of Dynamin activity in cell clones within receiving tissue causes extracellular Wg accumulation (Strigini and Cohen, 2000), suggesting that its range of movement is restricted by a Dynamin-dependent mechanism. Other studies have suggested roles for both degradation and recycling in controlling the spread of Wg (Dubois et al., 2001; Pfeiffer et al., 2002). In this study, we have used dominant negative Rab GTPases to perturb specific branches of the endocytic pathway and address these questions. Our data suggest that the spread of Wg is controlled, like that of zebrafish FGF-8, by restrictive clearance (Scholpp and Brand, 2004). Preventing Rab5-dependent internalization increases the spread of Wg through disc tissue. While this may also elevate extracellular Wg levels by indirect mechanisms (for example by modulating extracellular Wg proteolysis or release from disc tissue), we favor the possibility that the gradient is shaped by internalization of Wg itself, for several reasons. First, Wg is actually found in Rab5and Rab7-positive endosomes. Second, treating living discs with protease inhibitors does not cause Wg to accumulate (E.M and S.E, unpublished). Finally, differential centrifugation experiments suggest that only about 6% of Wg is not membrane-associated (Panáková et al., 2005). Some ligands signal from endocytic compartments after internalization (Bivona and Philips, 2003; González-Gaitán, 2003; Miaczynska et al., 2004). Therefore, we initially wondered whether the high levels of Wg accumulating after Rab5SN expression could increase signal transduction. While Rab5SNexpressing cells both accumulate Armadillo and reduce Senseless expression (not shown), we did not further investigate whether internalization of Wg is required for signaling because of the striking and unexpected transcriptional changes caused by blocking Rab5 activity. For example, transcription of both Fz2 and Wg Dlp degradation apical lateral Wild type degradation degradation degradation spreading basal apical lateral Dlp over-expression Dlp increases and that of Arrow plummets within a few hours of initiating Rab5SN expression – any of these changes by themselves could alter Wg signaling. Although we do not yet understand this phenomenon, one might imagine that coupling transcriptional regulation of endocytic receptors to their actual endocytosis and/or degradation would be an effective homeostatic mechanism. Studies in tissue culture cells have shown that inhibiting Rab7dependent lysosomal degradation can increase recycling of some proteins to the cell surface (Edinger et al., 2003). In imaginal discs, we find that Rab7TN expression increases the abundance of Rab11 recycling endosomes, and that inhibiting recycling via Rab11SN enlarges the Rab7-positive degradative compartment. Thus, the recycling and degradative pathways may compete for some cargo in discs as they do in cultured cells. This raises the possibility that changing the balance of degradation and recycling might affect the pool of extracellular Wingless available for spreading. Indeed, Wg appears to be recycled in embryos (Pfeiffer et al., 2002), and inhibiting lysosomal degradation in the embryonic ectoderm extends its range (Dubois et al., 2001). However, our data suggest that the increase in the range of Wg caused by inhibiting degradation (at least in imaginal discs) is not the result of increased recycling and extracellular spread. Although Wg protein is detected in endosomes over a broader range in imaginal disc tissue expressing Rab7TN, there is no increase in the range of extracellular Wg. Furthermore, neither extracellular nor intracellular Wg distribution is affected by inhibiting the Rab4- or Rab11-dependent recycling pathways. Inhibiting degradation probably extends the apparent range of Wg by stabilizing internalized protein, raising its levels above the threshold of detectability in more distant cells. The idea that apical-basal polarity of epithelial cells might play a role in regulating morphogen trafficking has been suggested by the observation that wg mRNA is enriched apically in the embryonic ectoderm. Changing mRNA localization alters the distribution of Wg protein in receiving tissue (Simmonds et al., 2001), raising the intriguing possibility that Wg might be trafficked differently depending on whether it is secreted apically or basal-laterally. In support of this idea, our data show that Wingless is internalized specifically from the apical and basal (but not lateral) surfaces of the disc epithelium. Indeed, the distribution of Rab5- and Rab7-positive endosomes in general suggests that the apical and basal surfaces are more endocytically active than other regions. The apical and basal internalization mechanisms may be distinct; the known receptors for basal spreading Fig. 8. A model for Wingless gradient formation. Cartoon of Wg trafficking through wild-type tissue (upper panel) and Dlp overexpressing tissue (lower panel). Wg is released from producing cells (red) and moves into receiving tissue, where it is endocytosed from apical and basal surfaces. Dlp overexpression (green in lower panel) biases Wg localization to the lateral surface, where it can diffuse without being endocytosed. This causes both elevation of Wg protein levels and extension of its spreading range (illustrated in flow charts). This model presupposes that Wg moves freely between apical, lateral and basal surfaces. If apical junctions establish a fence preventing diffusion of membrane proteins between apical and basal-lateral domains, Wg movement would have to occur via apical/basal-lateral transcytosis. DEVELOPMENT than wild type in cells near the source (1.3±0.3-fold within the first four cell rows, n=3). This suggests that much of the laterally accumulating Wg on Dlp-overexpressing cells is inaccessible to internalization. 315 RESEARCH ARTICLE Wingless, Fz2 and Arrow are internalized mainly from the apical surface (despite their steady-state basal-lateral localization), suggesting that basal Wingless endocytosis must be independent of these proteins. One possibility is that membrane association of Wg via Palmitate is sufficient to allow its endocytosis – perhaps by mechanisms similar to those used by gpi-linked proteins (Sabharanjak et al., 2002). Alternatively, Wg bound to Lipoprotein particles (Panáková et al., 2005) might be internalized via Lipoprotein receptors. We were surprised to observe that the Wg accumulating on the basal side of disc epithelial cells after Rab5SN expression did not spread onto the lateral membrane, as no barrier to diffusion has been identified between these domains. Three possible explanations occur to us. (1) There may indeed be a ‘fence’ separating the lateral and basal sides of disc epithelial cells. Neurons have a fence that prevents diffusion of lipid and lipid-linked proteins between the axon and the cell body, although it does not resemble a classical intercellular junction (Kobayashi et al., 1992; Nakada et al., 2003). (2) It may be that the receptor(s) that normally internalize Wg basally are linked to cytoskeletal components, or interact with extracellular matrix (ECM), and are not free to diffuse. If these Wg receptors were of sufficiently high affinity, they might trap Wg before it could move laterally. (3) Perhaps Wg itself interacts efficiently with basal ECM components. While it seems that endocytosis restricts the spread of Wg on the apical and basal surfaces, it is not yet clear which receptors might be responsible. A simple model would predict that removing such a receptor should produce phenotypes similar to Rab5SN expression, i.e. increased and more extensive extracellular Wg, and less Wg in endosomes. Conversely, overexpression might be expected to compress the range of Wg distribution and decrease extracellular Wg. None of the known receptors behaves in this way. Previous studies showed that at least a fraction of Wg is still internalized in the absence of both Fz1 and Fz2 (Baeg et al., 2004). Furthermore, overexpression of Fz2 causes extracellular Wg accumulation over longer distances (Cadigan et al., 1998; Baeg et al., 2004). We have shown here that loss of Arrow actually increases the amount of Wg present in Rab5- and Rab7-positive endosomes – more consistent with a role in Wg degradation after endocytosis. The complexity of these observations may reflect different mechanisms of Wg endocytosis on the apical and basal sides of the cell – understanding both these pathways and their interplay will be necessary to understanding how the Wg gradient forms. While internalization limits the range of Wg accumulation, the glypican Dlp extends it. It has been proposed that Dlp allows Wg to interact with disc cells, increasing local Wg concentration and restricting its diffusion to the epithelium (Han et al., 2005). Our data, however, show that disc cells can accumulate high levels of Wg on their surface in the absence of Dlp as long as Rab5-dependent internalization is blocked. This observation is not consistent with a model in which Dlp traps Wg on the cell surface or helps it transfer from cell to cell. Instead, it suggests that Dlp normally stabilizes Wg at the cell surface by antagonizing the effects of Rab5-dependent internalization (see model in Fig. 8). While Wg is normally internalized from the apical- and basal-most surfaces of disc cells, Dlp overexpression recruits Wg to the lateral cell surface. This raises the possibility that Dlp stabilizes Wg and increases its range by changing its subcellular localization to protect it from endocytosis. Polarizing the distribution of morphogens within an epithelium may have a key regulatory role in the trafficking events leading to gradient formation. Development 133 (2) Note added in proof A recent paper (Piddini et al., 2005) from the Vincent Laboratory has demonstrated that Arrow promotes Wingless degradation after internalization by Frizzled 2. We are grateful to Phil Beachy, Hugo Bellen, Steve Cohen, Susan Cumberledge, Steve di Nardo, Marcos González-Gaitán and Pernille Rørth for providing antibodies and constructs. We acknowledge Sarah Bowman for developing the co-localization protocol and Christina Eugster for constructing wg-GFP. We thank Christian Dahmann, Giovanna Mottola, Daniela Panáková, Sophie Quintin, Annette Schenck, Kai Simons, Christoph Thiele and Marino Zerial for critical comments on the manuscript. E.M. was supported by an A. von Humboldt fellowship and a grant from the Deutsche Forschungsgemeinschaft (EA 4/2-1). Supplementary material Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/133/2/307/DC1 References Ang, A. L., Taguchi, T., Francis, S., Folsch, H., Murrells, L. J., Pypaert, M., Warren, G. and Mellman, I. (2004). Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J. Cell Biol. 167, 531-543. Baeg, G. H., Lin, X., Khare, N., Baumgartner, S. and Perrimon, N. (2001). Heparan sulfate proteoglycans are critical for the organization of the extracellular distribution of Wingless. Development 128, 87-94. Baeg, G. H., Selva, E. M., Goodman, R. M., Dasgupta, R. and Perrimon, N. (2004). The Wingless morphogen gradient is established by the cooperative action of Frizzled and Heparan Sulfate Proteoglycan receptors. Dev. Biol. 276, 89-100. Bejsovec, A. and Wieschaus, E. (1995). Signalling activities of the Drosophila wingless gene are separately mutable and appear to be transduced at the cell surface. Genetics 139, 309-320. Belenkaya, T. Y., Han, C., Yan, D., Opoka, R. J., Khodoun, M., Liu, H. and Lin, X. (2004). Drosophila Dpp morphogen movement is independent of dynaminmediated endocytosis but regulated by the glypican members of heparan sulfate proteoglycans. Cell 119, 231-244. Bivona, T. G. and Philips, M. R. (2003). Ras pathway signaling on endomembranes. Curr. Opin. Cell Biol. 15, 136-142. Brand, A. H. and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401415. Bucci, C., Thomsen, P., Nicoziani, P., McCarthy, J. and van Deurs, B. (2000). Rab7: a key to lysosome biogenesis. Mol. Biol. Cell 11, 467-480. Cadigan, K. M., Fish, M. P., Rulifson, E. J. and Nusse, R. (1998). Wingless repression of Drosophila frizzled 2 expression shapes the Wingless morphogen gradient in the wing. Cell 93, 767-777. Classen, A., Anderson, K., Marois, E. and Eaton, S. (2005). Hexagonal packing of drosophila wing epithelial cells by the planar cell polarity pathway. Dev. Cell 9, 805-817. de Renzis, S., Sonnichsen, B. and Zerial, M. (2002). Divalent Rab effectors regulate the sub-compartmental organization and sorting of early endosomes. Nat. Cell Biol. 4, 124-133. Dubois, L., Lecourtois, M., Alexandre, C., Hirst, E. and Vincent, J. P. (2001). Regulated endocytic routing modulates wingless signaling in Drosophila embryos. Cell 105, 613-624. Edinger, A. L., Cinalli, R. M. and Thompson, C. B. (2003). Rab7 prevents growth factor-independent survival by inhibiting cell-autonomous nutrient transporter expression. Dev. Cell 5, 571-582. Emery, G., Hutterer, A., Berdnik, D., Mayer, B., Wirtz-Peitz, F., Gaitan, M. G. and Knoblich, J. A. (2005). Asymmetric rab11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell 122, 763-773. Entchev, E. V., Schwabedissen, A. and González-Gaitán, M. (2000). Gradient formation of the TGF-beta homolog Dpp. Cell 103, 981-991. Franch-Marro, X., Marchand, O., Piddini, E., Ricardo, S., Alexandre, C. and Vincent, J. P. (2005). Glypicans shunt the Wingless signal between local signalling and further transport. Development 132, 659-666. Giraldez, A. J., Copley, R. R. and Cohen, S. M. (2002). HSPG modification by the secreted enzyme Notum shapes the Wingless morphogen gradient. Dev. Cell 2, 667-676. González-Gaitán, M. (2003). Signal dispersal and transduction through the endocytic pathway. Nat. Rev. Mol. Cell. Biol. 4, 213-224. González-Gaitán, M., Capdevila, M. P. and Garcia-Bellido, A. (1994). Cell proliferation patterns in the wing imaginal disc of Drosophila. Mech. Dev. 46, 183-200. Greco, V., Hannus, M. and Eaton, S. (2001). Argosomes: a potential vehicle for the spread of morphogens through epithelia. Cell 106, 633-645. DEVELOPMENT 316 Han, C., Yan, D., Belenkaya, T. Y. and Lin, X. (2005). Drosophila glypicans Dally and Dally-like shape the extracellular Wingless morphogen gradient in the wing disc. Development 132, 667-679. Jékely, G. and Rørth, P. (2003). Hrs mediates downregulation of multiple signalling receptors in Drosophila. EMBO Rep. 4, 1163-1168. Jones, S. M., Howell, K. E., Henley, J. R., Cao, H. and McNiven, M. A. (1998). Role of dynamin in the formation of transport vesicles from the trans-Golgi network. Science 279, 573-577. Kirkpatrick, C. A., Dimitroff, B. D., Rawson, J. M. and Selleck, S. B. (2004). Spatial regulation of Wingless morphogen distribution and signaling by Dally-like protein. Dev. Cell 7, 513-523. Kobayashi, T., Storrie, B., Simons, K. and Dotti, C. G. (1992). A functional barrier to movement of lipids in polarized neurons. Nature 359, 647-650. Kreuger, J., Perez, L., Giraldez, A. J. and Cohen, S. M. (2004). Opposing activities of Dally-like glypican at high and low levels of Wingless morphogen activity. Dev. Cell 7, 503-512. Lazzarino, D. A., Blier, P. and Mellman, I. (1998). The monomeric guanosine triphosphatase rab4 controls an essential step on the pathway of receptormediated antigen processing in B cells. J. Exp. Med. 188, 1769-1774. Lock, J. G. and Stow, J. L. (2005). Rab11 in Recycling Endosomes Regulates the Sorting and Basolateral Transport of E-Cadherin. Mol Biol. Cell 16, 1744-1755. Lum, L., Yao, S., Mozer, B., Rovescalli, A., Von Kessler, D., Nirenberg, M. and Beachy, P. A. (2003). Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science 299, 2039-2045. Maxfield, F. R. and McGraw, T. E. (2004). Endocytic recycling. Nat. Rev. Mol. Cell. Biol. 5, 121-132. McGuire, S. E., Le, P. T., Osborn, A. J., Matsumoto, K. and Davis, R. L. (2003). Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302, 17651768. Miaczynska, M., Pelkmans, L. and Zerial, M. (2004). Not just a sink: endosomes in control of signal transduction. Curr. Opin. Cell Biol. 16, 400-406. Nakada, C., Ritchie, K., Oba, Y., Nakamura, M., Hotta, Y., Iino, R., Kasai, R. S., Yamaguchi, K., Fujiwara, T. and Kusumi, A. (2003). Accumulation of anchored proteins forms membrane diffusion barriers during neuronal polarization. Nat. Cell Biol. 5, 626-632. Neumann, C. and Cohen, S. (1997). Morphogens and pattern formation. BioEssays 19, 721-729. Oda, H., Uemura, T., Harada, Y., Iwai, Y. and Takeichi, M. (1994). A Drosophila homolog of cadherin associated with armadillo and essential for embryonic cellcell adhesion. Dev. Biol. 165, 716-726. Panáková, D., Sprong, H., Marois, E., Thiele, C. and Eaton, S. (2005). Lipoprotein particles carry lipid-linked proteins and are required for long-range Hedgehog and Wingless signalling. Nature 435, 58-65. Pelissier, A., Chauvin, J. P. and Lecuit, T. (2003). Trafficking through Rab11 endosomes is required for cellularization during Drosophila embryogenesis. Curr. Biol. 13, 1848-1857. Pepinsky, R. B., Zeng, C., Wen, D., Rayhorn, P., Baker, D. P., Williams, K. P., Bixler, S. A., Ambrose, C. M., Garber, E. A., Miatkowski, K. et al. (1998). Identification of a palmitic acid-modified form of human Sonic hedgehog. J. Biol. Chem. 273, 14037-14045. Pfeiffer, S., Ricardo, S., Manneville, J. B., Alexandre, C. and Vincent, J. P. (2002). Producing cells retain and recycle Wingless in Drosophila embryos. Curr. Biol. 12, 957-962. RESEARCH ARTICLE 317 Piddini, E., Marshall, F., Dubois, L., Hirst, E. and Vincent, J. P. (2005). Arrow (LRP6) and Frizzled2 cooperate to degrade Wingless in Drosophila imaginal discs. Development 132, 5479-5489. Porter, J. A., Young, K. E. and Beachy, P. A. (1996). Cholesterol modification of Hedgehog signaling proteins in animal development. Science 274, 255-259. Rink, J., Ghigo, E., Kalaidzidis, Y. and Zerial, M. (2005). Rab conversion as a mechanism of progression from early to late endosomes. Cell 122, 735-749. Sabharanjak, S., Sharma, P., Parton, R. G. and Mayor, S. (2002). GPI-anchored proteins are delivered to recycling endosomes via a distinct cdc42-regulated, clathrin-independent pinocytic pathway. Dev. Cell 2, 411-423. Satoh, A. K., O’Tousa, J. E., Ozaki, K. and Ready, D. F. (2005). Rab11 mediates post-Golgi trafficking of rhodopsin to the photosensitive apical membrane of Drosophila photoreceptors. Development 132, 1487-1497. Scholpp, S. and Brand, M. (2004). Endocytosis controls spreading and effective signaling range of Fgf8 protein. Curr. Biol. 14, 1834-1841. Seachrist, J. L., Anborgh, P. H. and Ferguson, S. S. (2000). beta 2-adrenergic receptor internalization, endosomal sorting, and plasma membrane recycling are regulated by rab GTPases. J. Biol. Chem. 275, 27221-27228. Sever, S. (2002). Dynamin and endocytosis. Curr. Opin. Cell Biol. 14, 463-467. Simmonds, A. J., dosSantos, G., Livne-Bar, I. and Krause, H. M. (2001). Apical localization of wingless transcripts is required for wingless signaling. Cell 105, 197-207. Stenmark, H., Valencia, A., Martinez, O., Ullrich, O., Goud, B. and Zerial, M. (1994). Distinct structural elements of rab5 define its functional specificity. EMBO J. 13, 575-583. Strigini, M. and Cohen, S. M. (2000). Wingless gradient formation in the Drosophila wing. Curr. Biol. 10, 293-300. Torroja, C., Gorfinkiel, N. and Guerrero, I. (2004). Patched controls the Hedgehog gradient by endocytosis in a dynamin-dependent manner, but this internalization does not play a major role in signal transduction. Development 131, 2395-2408. van Dam, E. M. and Stoorvogel, W. (2002). Dynamin-dependent transferrin receptor recycling by endosome-derived clathrin-coated vesicles. Mol. Biol. Cell 13, 169-182. van der Sluijs, P., Hull, M., Webster, P., Male, P., Goud, B. and Mellman, I. (1992). The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell 70, 729-740. Willert, K., Brown, J. D., Danenberg, E., Duncan, A. W., Weissman, I. L., Reya, T., Yates, J. R., 3rd and Nusse, R. (2003). Wnt proteins are lipidmodified and can act as stem cell growth factors. Nature 423, 448-452. Wu, J., Klein, T. J. and Mlodzik, M. (2004). Subcellular localization of frizzled receptors, mediated by their cytoplasmic tails, regulates signaling pathway specificity. PLoS Biol. 2, E158. Yu, S. Y., Yoo, S. J., Yang, L., Zapata, C., Srinivasan, A., Hay, B. A. and Baker, N. E. (2002). A pathway of signals regulating effector and initiator caspases in the developing Drosophila eye. Development 129, 3269-3278. Zerial, M. and McBride, H. (2001). Rab proteins as membrane organizers. Nat. Rev. Mol. Cell. Biol. 2, 107-117. Zhai, L., Chaturvedi, D. and Cumberledge, S. (2004). Drosophila wnt-1 undergoes a hydrophobic modification and is targeted to lipid rafts, a process that requires porcupine. J. Biol. Chem. 279, 33220-33227. DEVELOPMENT Endocytosis and the Wingless gradient Curriculum vitae Article 5: 2010 PLoS Biology The Major Yolk Protein Vitellogenin Interferes with the Anti-­ Plasmodium Response in the Malaria Mosquito Anopheles gambiae. Rono, M., Whitten, M., Oulad-­‐Abdelghani, M., Levashina, E., Marois, E. The Major Yolk Protein Vitellogenin Interferes with the Anti-Plasmodium Response in the Malaria Mosquito Anopheles gambiae Martin K. Rono1,2,3¤a, Miranda M. A. Whitten1,2,3¤b, Mustapha Oulad-Abdelghani4, Elena A. Levashina1,2,3, Eric Marois1,2,3* 1 INSERM, U963, Strasbourg, France, 2 CNRS, IBMC, UPR9022, Strasbourg, France, 3 Université de Strasbourg, UMR 963, Strasbourg, France, 4 INSERM, U964/CNRS, UMR 7104/Université de Strasbourg, IGBMC, Illkirch, France Abstract When taking a blood meal on a person infected with malaria, female Anopheles gambiae mosquitoes, the major vector of human malaria, acquire nutrients that will activate egg development (oogenesis) in their ovaries. Simultaneously, they infect themselves with the malaria parasite. On traversing the mosquito midgut epithelium, invading Plasmodium ookinetes are met with a potent innate immune response predominantly controlled by mosquito blood cells. Whether the concomitant processes of mosquito reproduction and immunity affect each other remains controversial. Here, we show that proteins that deliver nutrients to maturing mosquito oocytes interfere with the antiparasitic response. Lipophorin (Lp) and vitellogenin (Vg), two nutrient transport proteins, reduce the parasite-killing efficiency of the antiparasitic factor TEP1. In the absence of either nutrient transport protein, TEP1 binding to the ookinete surface becomes more efficient. We also show that Lp is required for the normal expression of Vg, and for later Plasmodium development at the oocyst stage. Furthermore, our results uncover an inhibitory role of the Cactus/REL1/REL2 signaling cassette in the expression of Vg, but not of Lp. We reveal molecular links that connect reproduction and immunity at several levels and provide a molecular basis for a longsuspected trade-off between these two processes. Citation: Rono MK, Whitten MMA, Oulad-Abdelghani M, Levashina EA, Marois E (2010) The Major Yolk Protein Vitellogenin Interferes with the Anti-Plasmodium Response in the Malaria Mosquito Anopheles gambiae. PLoS Biol 8(7): e1000434. doi:10.1371/journal.pbio.1000434 Academic Editor: David S. Schneider, Stanford University, United States of America Received December 17, 2009; Accepted June 10, 2010; Published July 20, 2010 Copyright: ß 2010 Rono et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Funding: This work was supported by grants from the Centre National de la Recherche Scientifique (UPR 9022 du CNRS), the Institut National de la Santé et de la Recherche Médicale (U963 INSERM), the Fondation pour la Recherche Médicale (FRM) to EM, the Schlumberger Foundation for Education and Research (FSER) to EAL and MMAW, from the EMBO Young Investigator Program (to EAL), and from the EC FP6th Networks of Excellence ‘‘BioMalPar’’ (to EAL and MR), EC FP7 HEALTH Collaborative Project ‘‘MALVECBLOK’’, EC FP7 Capacities Specific Programme Research Infrastructures ‘‘INFRAVEC’’, and EC FP7 Networks of Excellence ‘‘EVIMalar’’. EAL is an international research scholar of the Howard Hughes Medical Institute. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Competing Interests: The authors have declared that no competing interests exist. Abbreviations: dsRNA, double-stranded RNA; ELISA, enzyme-linked immunosorbent assay; GFP, green fluorescent protein; GST, glutathione S-transferase; HDL, high-density lipoprotein; hpi, hours post infection; Imd, Immune deficiency; IP, immuno-precipitation; KBr, potassium bromide; kDa, kiloDalton; Lp, Lipophorin (AGAP001826); LDL, low-density lipoprotein; LRR, leucine-rich repeat; MALDI, matrix-assisted laser desorption/ionization; PCR, polymerase chain reaction; qRT, quantitative real-time; RNAi, RNA interference; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel; TEP1, thioester-containing protein 1; TOF, time-of-flight; Vg, Vitellogenin (AGAP004203); TOR, Target of Rapamycin * E-mail: E.Marois@unistra.fr ¤a Current address: KEMRI - Wellcome Trust Research Programme, Kilifi, Kenya ¤b Current address: Department of Pure and Applied Ecology, School of the Environment and Society, Swansea University, Swansea, United Kingdom between 16 and 48 h post infection (hpi). Once they reach the hemolymph-bathed basal side of the midgut, ookinetes round up and transform into oocysts, protected capsules within which asexual multiplication of the parasite takes place. Previous studies have established that the ookinete is the parasite stage most vulnerable to the mosquito immune response [1,2]. As a consequence of this response, most mosquito species efficiently eliminate all the invading ookinetes, thereby aborting the parasite cycle [3]. In a few parasite/mosquito combinations, up to 20% of ookinetes survive and the disease can be further transmitted. A number of mosquito humoral antiparasitic proteins have been characterized (reviewed in [4]). The molecularly best characterized and phenotypically most prominent defense pathway mediating the killing of Plasmodium berghei in A. gambiae involves a thioester-containing protein (TEP1) homologous to vertebrate Introduction Malaria is a mosquito-borne parasitic disease affecting annually an estimated 250 million people, of which close to 1 million (mostly children in sub-Saharan Africa) succumb to the disease (World Health Organization fact sheet #94, April 2010; http:// www.who.int/mediacentre/factsheets/fs094/en/index.html). Several Plasmodium species cause malaria, the most deadly being P. falciparum transmitted mainly by the Anopheles gambiae mosquito. As mosquito females require a blood meal to produce eggs, feeding on a malaria-infected host simultaneously activates oogenesis and triggers immune responses to malaria parasites. In the midgut, ingested Plasmodium gametocytes differentiate within minutes into gametes. After fertilization, zygotes rapidly transform into ookinetes, i.e. motile cells that traverse the midgut epithelium PLoS Biology | www.plosbiology.org 1 July 2010 | Volume 8 | Issue 7 | e1000434 Reproduction versus Immunity in Anopheles addition to lipids, Lp particles serve as a vehicle for morphogen proteins in the imaginal discs of Drosophila larvae [20]. Interestingly, human HDL has been shown to host a fraction of complement factor C3 [21] as well as trypanosome-killing protein complexes [22]. In mosquitoes, recent studies [23–25] have implicated Lp in both mosquito reproduction and Plasmodium survival. In particular, experimental depletion of Lp by RNAi inhibited oogenesis and also reduced the number of developing Plasmodium oocysts in the mosquito midgut [23]. This could point to a nutritional requirement for Lp in the early stages of parasite development. Indeed, Lp has recently been detected by in vitro approaches inside developing P. gallinaceum oocysts, suggesting that it provides parasites with a source of lipids [26]. An intriguing alternative explanation is that the increasing levels of Lp following a blood meal may negatively impact mosquito immunity against parasites. Artificially blocking the physiological rise in Lp levels would then allow the immune system to exert its full strength against the parasite. In the mosquito fat body, two distinct pathways are required for optimal expression of proteins involved in vitellogenesis: (i) the nutrient-sensing TOR pathway and (ii) a hormonal cascade that oversees production of 20-hydroxyecdysone [14,27,28]. Furthermore, in Ae. aegypti mosquitoes infected with microbes and Plasmodium, the NF-kB factor REL1 positively regulates expression of Lp and its receptor [24], suggesting that the NF-kB pathway may also contribute to the regulation of oogenesis in addition to its known role in mosquito immunity [29–31]. However, our understanding of how oogenesis and immunity impact each other remains incomplete: on one hand depletion of Lp strongly inhibits development of P. gallinaceum; on the other hand over-expression of Lp resulting from the depletion of the REL1 inhibitor Cactus in Ae. aegypti is insufficient to rescue the complete block in parasite development [24]. Here, we investigated the role of the two major nutrient transport proteins Lp and Vg in mosquito antiparasitic responses using a common laboratory model of malaria transmission: A. gambiae mosquitoes infected with the GFP-expressing rodent parasite P. berghei [32]. We show that similarly to Lp, Vg depletion reduces parasite survival in mosquito tissues. Strikingly however, Lp and Vg are no longer required for parasite survival if TEP1 is depleted, suggesting that the low parasite survival phenotype associated with the Lp/Vg knockdowns requires TEP1 function. We propose that Lp and Vg exert distinct non-redundant roles in reproduction and immunity: Lp is crucial for oogenesis and is required for normal Vg expression after an infectious blood meal, whereas Vg contributes to oogenesis and negatively impacts TEP1 binding to the ookinetes. We suggest that the reported negative impact of Lp depletion on ookinete survival is indirect and is mediated by reduced levels of Vg. We further demonstrate that the NF-kB factors REL1 and REL2 limit the expression of Vg after an infectious blood meal. These results reveal an unexpected network of interactions whereby Plasmodium killing in mosquitoes is potentiated by NF-kB pathways at two levels: (i) activation of anti-Plasmodium genes and (ii) inhibition of the expression of the nutrient transport protein Vg. Author Summary Malaria annually claims the lives of almost 1 million infants and imposes a major socio-economic burden on Africa and other tropical regions. Meanwhile, the detailed biological interactions between the malaria parasite and its Anopheles mosquito vector remain largely enigmatic. What we do know is that the majority of malaria parasites are normally eliminated by the mosquito’s immune response. Mosquitoes accidentally acquire an infection by sucking parasite-laden blood, but this belies the primary function of the blood in the provisioning of nutrients for egg development in the insect’s ovaries. We have found that the molecular processes involved in delivering bloodacquired nutrients to maturing eggs diminish the efficiency of parasite killing by the mosquito immune system. Conversely, molecular pathways that set the immune system on its maximal capacity for parasite killing preclude the efficient development of the mosquito’s eggs. Our results reveal some of the molecules that underpin this example of the trade-offs between reproduction and immunity, a concept that has long intrigued biologists. complement factor C3 [2,5,6]. Depletion of TEP1 by RNA interference (RNAi) renders mosquitoes hypersusceptible to Plasmodium infections, resulting in abnormally high infection levels. Two leucine-rich repeat (LRR) proteins, LRIM1 and APL1C, act as TEP1 control proteins to stabilize the mature form of TEP1 in the hemolymph [7,8] and show the same RNAi phenotype as TEP1 in P. berghei infections [9–12]. The depletion of either protein results in precocious deposition of TEP1 on self tissues and completely aborts its binding to the ookinetes [7]. Therefore, it appears that LRR proteins regulate maintenance of mature TEP1 in circulation; however, the factors that control TEP1 targeting to the parasite surface remain unknown. Simultaneously to the midgut crossing by ookinetes, the physiology of the mosquito is profoundly modified by a blood meal in preparation for the laying of a clutch of eggs. Within 2 to 3 d after a blood meal, the massive ovary growth allows maturation of 50–150 oocytes, a process called vitellogenesis (reviewed in [13]). The blood meal provides the mosquito with amino acids and lipids that are transferred through midgut cells to the hemolymph and signal via the Target of Rapamycin (TOR) pathway to initiate massive synthesis of nutrient transport proteins in the mosquito fat body [14]. These transport proteins include the lipid transporter lipophorin (Lp, AGAP001826) (also known as apolipoprotein II/I or retinoic and fatty acid binding protein, RFABG/P) and vitellogenin (Vg, AGAP004203), a precursor of the yolk storage protein vitellin. Both proteins are secreted into the hemolymph and transported to the ovaries. Vg is a large phospholipoglycoprotein encoded in A. gambiae by a small family of nearly-identical genes. Insect Vg harbors potential sites for lipidation, glycosylation, and phosphorylation and is internalized by developing oocytes where it is proteolytically cleaved to generate vitellin, a nutrient source for the developing embryo (reviewed in [15,16]). Lp, encoded by a single transcript and posttranslationally cleaved, is composed of two subunits of 250 and 80 kDa that together scaffold a lipidic particle. Similar to vertebrate low- and high-density lipoproteins (LDL and HDL, respectively), mosquito Lp particles contain a core of fatty acids and sterols, surrounded by an outer leaflet of phospholipids [17,18]. These particles function to deliver lipids and fatty acids to energy-consuming tissues, including rapidly growing imaginal discs in larvae, muscles, and the ovary in adult females [19]. In PLoS Biology | www.plosbiology.org Results Lp and Vg Depletion Reduce Parasite Survival in a TEP1Dependent Manner Lp knockdown causes a decrease in parasite loads and simultaneously arrests oogenesis [23]. We examined whether the Lp knockdown phenotype requires the antiparasitic factor TEP1. To this end, we compared the numbers of surviving parasites in 2 July 2010 | Volume 8 | Issue 7 | e1000434 Reproduction versus Immunity in Anopheles from a triplet running between 160 and 200 kDa was unequivocally identified as Vg, and the bands running at ,250 and 80 kDa were unequivocally identified as the large and small subunits of Lp, respectively. In addition, a protein running at ,70 kD and showing an expression pattern identical to that of the ,200 kD Vg band (including after RNAi silencing) was identified as the N-terminal fragment of the polypeptide encoded by Vg mRNA (visible in Figures 3C, 4C, and S1). This fragment was not recognized by our Vg antibody, raised against a C-terminal Vg fragment. Its existence is consistent with the cleavage of Ae. aegypti Vg prior to secretion [33–35]. No contaminating proteins were detected at these sizes in the mass spectrometry analysis. Therefore, Lp and Vg proteins can be readily visualized after hemolymph electrophoresis and Coomassie staining of SDSPAGE gels even without immunoblotting. The efficiency of TEP1 silencing was also confirmed by immunoblotting (Figure 1D). We next investigated whether Vg and Lp cooperate to sustain oogenesis and parasite development or are involved in independent processes. We performed double-knockdown experiments by simultaneously injecting dsVg-dsLp to compare to single injections of dsVg and dsLp as controls. As expected, dsLp completely blocked oogenesis and the same was observed in concomitant dsLp-dsVg knockdowns (Figure 1F). Moreover, single dsVg (p = 0.0001) and double dsLp-dsVg (p,0.0001) knockdowns caused comparable reductions in oocyst counts; these reductions in oocyst numbers were stronger than in the single dsLp knockdown (p = 0.024) (Figure 1E). These results suggest that the influences of Lp and Vg on reproduction and immunity are balanced differently. Lp may be more crucial for oogenesis than Vg, whereas Vg influences Plasmodium survival more strongly than does Lp. In most experiments, the effect of Vg and Lp knockdowns on parasite counts did not appear to be additive (Figures 1E, 2A, and unpublished data). Although this observation is not supported by strong statistical significance, it raises the possibility that the two proteins may be involved in a single process benefiting ookinete survival in the physiological situation. To determine whether similarly to Lp, the effect of Vg on parasite development required TEP1 function, we performed triple knockdown experiments by injecting combinations of dsTEP1, dsVg, dsLp, or control dsLacZ. Again, total inhibition of oogenesis was observed in all dsRNA combinations that included dsLp, suggesting that oogenesis is not influenced by TEP1 function but absolutely requires Lp (Figure 2B). In striking contrast, high parasite loads similar to that detected in the dsTEP1 single knockdown were obtained when TEP1 was depleted simultaneously to Vg (unpublished data) or to both Vg and Lp (Figure 2A, Figure S3C). These findings imply that blocking the transport of lipids and vitellogenin-derived nutrients does not limit parasite survival when the immune defense is suppressed; instead, the observed reduction in parasite numbers in dsLp and dsVg knockdowns is dependent on TEP1. We conclude that TEP1dependent parasite killing is more efficient when Lp and/or Vg levels are low and that the TEP1-mediated immune pressure exerted by the vector is a bigger impediment to the establishment of a Plasmodium infection than nutrient availability. If this constraint is removed via TEP1 depletion, Plasmodium parasites can effectively exploit even reduced vector resources and proceed with the formation of viable oocysts. We next examined at which level Vg and Lp genetically interact with TEP1. Binding of mature TEP1 to the parasite surface is one of the first steps leading to parasite killing; either increasing or reducing this event greatly influences the outcome of infection [31,36]. Therefore, we gauged the efficiency of TEP1 binding to single TEP1 or Lp knockdown mosquitoes and in double TEP1/Lp knockdowns by injecting double-stranded RNA (dsRNA) resulting in RNAi. Four days after dsRNA injection, mosquitoes were fed on a mouse infected with GFP-expressing parasites. Mosquitoes were dissected 8 to 10 d later to gauge prevalence of infection and mean oocyst numbers per midgut (Figure 1A, Figure S3A). As reported earlier, Lp silencing strongly reduced the number of developing oocysts. Strikingly, silencing TEP1 at the same time as Lp annihilated the effect of Lp silencing, i.e. yielded the high oocyst numbers typically observed upon silencing of TEP1 alone. Therefore, the low oocyst counts observed in Lp-depleted mosquitoes are not due to a nutritional dependence of ookinetes on Lp-derived lipids but are a consequence of TEP1 activity. This result also suggests that the increased parasite killing in Lpdepleted mosquitoes takes place at the ookinete stage, since TEP1 binding does not kill oocysts. Further, these results imply that the loss of Lp renders ookinetes more vulnerable to TEP1-dependent killing. To explain these data, we initially hypothesized that Lp particles might physically sequester components of the TEP1 machinery in an inactive state, but a search for Lp-associated immune factors was unsuccessful (with the notable exception of prophenoloxidase), suggesting that TEP1-containing complexes are not carried in the hemolymph by Lp particles (see Text S1 and Figure S1). To investigate whether the adverse effect on immunity is a specific property of Lp or may be manifested as well by other nutrient transport factors, we injected mosquitoes with dsVg and compared parasite development with dsLacZ and dsTEP1-injected mosquitoes. A 4-fold reduction in mean parasite numbers was observed in the dsVg group compared to dsLacZ controls (p,0.001, p,0.001, p,0.05, and p,0.05 depending on the replicate of this experiment; Figure 1B and Figure S3B). This effect was more profound than the effect of dsLp (Figure 1A and 1E). We then examined whether depletion of the major yolk protein would compromise oogenesis. In contrast to Lp silencing, which resulted in total abortion of ovary development, roughly 50% of mosquito females still developed eggs after silencing of Vg compared to 80% in dsLacZ control mosquitoes (Figure 1C), though ovaries that did mature usually contained only a few eggs bearing melanotic spots (unpublished data). When given a chance to lay, Vg-silenced females did lay a few eggs, the majority of which never hatched (unpublished data). The difference in strength between the Lp and Vg silencing phenotypes regarding egg development suggests either that Lp is more crucial than Vg for egg development or that the efficiency of Lp silencing is greater than the efficiency of Vg silencing. Residual Vg protein may allow the development of a few eggs in dsVg-treated mosquitoes. It is interesting to note that the strengths of the silencing phenotypes are reversed when considering parasite survival. To verify the efficiency of RNAi-mediated depletion of Lp and Vg, we used specific antibodies directed against the large and small subunits of Lp, and against Vg. RNAi silencing caused Lp and Vg protein amounts to drop below 10% of control levels (Figure S2). Subsequently, we systematically controlled for Lp and Vg silencing efficiency and noted that Vg depletion was somewhat more variable than Lp depletion, residual Vg protein sometimes approaching 20% of control levels (unpublished data). Strikingly, this analysis revealed that the major protein bands detected in hemolymph samples by Coomassie staining of SDS-PAGE gels (or of PVDF membranes after protein transfer) correspond to the Vg and Lp signals detected by specific antibodies (Figure S2). We excised these easily visualized bands from Coomassie-stained protein gels and submitted them to MALDI mass spectrometry. The peptide mass spectra were searched against the NCBInr database. Each band PLoS Biology | www.plosbiology.org 3 July 2010 | Volume 8 | Issue 7 | e1000434 Reproduction versus Immunity in Anopheles Figure 1. Effects of Lp and Vg silencing on parasite counts and oogenesis. Mosquitoes were injected with the indicated double-stranded RNA and infected with P. berghei. Parasite development was gauged 7–9 d post infection by counting GFP-expressing oocysts. Each dot represents the number of oocysts counted in one midgut. Ovaries containing mature eggs were counted 7 d post infection. Pie charts show the percentage of mosquitoes containing mature eggs (grey) versus percentage of mosquitoes containing only undeveloped oocytes (black). (A) Effect of concomitant silencing of TEP1 and Lp on parasite survival. (B) Effect of Vg silencing on parasite survival. In (A) and (B), one representative experiment out of 4 independent replicates is shown. The additional replicates are shown in Figure S3. (C) Effect of Vg silencing on oogenesis. (D) Coomassie staining (top panel) of a PVDF membrane allows visualization of Vg and Lp in control, Vg-, and Lp-depleted mosquitoes. Hemolymph proteins were separated on a denaturing SDS-polyacrylamide gel and transferred to a PVDF membrane. Western blotting analysis of hemolymph of Vg- and Lp-depleted mosquitoes 0, 24, or 48 h after infection using anti-TEP1 antibody (bottom panel). (E, F) Parasite counts (E) and oogenesis (F) in Lp- and Vg-depleted mosquitoes. doi:10.1371/journal.pbio.1000434.g001 PLoS Biology | www.plosbiology.org 4 July 2010 | Volume 8 | Issue 7 | e1000434 Reproduction versus Immunity in Anopheles Figure 2. Vg and Lp are involved in TEP1-dependent parasite killing. Mosquitoes were injected with the indicated combinations of dsLacZ, dsVg, dsLp, dsTEP1. (A) Parasite counts. TEP1 silencing rescues the effect of Vg depletion on parasite loss. (B) Oogenesis. TEP1 knockdown doesn’t rescue the effect of Lp/Vg depletion on oogenesis. (C) Mosquitoes infected with P. berghei were dissected 24 and 48 h post blood meal. Midguts were fixed and immunostained with anti-TEP1 antibodies. The percentages of live GFP-expressing parasites (green), dying parasites (GFP-positive but partly covered by TEP1), and dead parasites (GFP negative, TEP1-covered) were determined on microscope images. 48 hpi, the percentage of dead, TEP1-labelled ookinetes is markedly higher in dsLp or dsLp-Vg mosquitoes than in the dsLacZ controls. In the LacZ control, the percentage of live parasites increases at 48 h because of the progressive clearance of already dead parasites. See Table S1 for parasite numbers scored in each of three independent repeats of this experiment. (D) Lp is required for oocyst maturation. Parasite development was gauged 8 dpi by estimating the size of oocysts in mosquitoes after the depletion of Lp, Vg, or double KD Lp-Vg compared to TEP1 and LacZ knockdown controls. Pictures of dissected midguts were analyzed using Axiovision. Parasite sizes were estimated by the surface area of each individual oocyst and averaged as mean oocyst size per dsRNA treatment, yielding the graph to the right. Lp depletion alone or with Vg significantly reduced oocyst sizes compared to controls. doi:10.1371/journal.pbio.1000434.g002 PLoS Biology | www.plosbiology.org 5 July 2010 | Volume 8 | Issue 7 | e1000434 Reproduction versus Immunity in Anopheles ookinetes in dsLp- and dsLp-Vg-injected mosquitoes. At early time points (24 hpi) TEP1 binding to ookinetes did not differ in the Lp or Lp-Vg -depleted versus control mosquitoes; but at 48 hpi 70% to 86% of ookinetes were TEP1 positive (i.e., either dead or moribund) in dsLp- or dsVg-Lp-injected mosquitoes versus only 41% to 68% in dsLacZ controls (Figure 2C and Table S1, p = 0.005 or less by chi-square analysis). Thus, TEP1 binding to parasites is more efficient in the absence of Lp/Vg. This strongly suggests that physiological levels of Vg and Lp interfere with the efficient binding of TEP1 to ookinetes once the invasion phase is completed. To see if we could also detect an effect of Lp and Vg depletion at a later stage of parasite development, we examined oocyst growth. Strikingly, oocyst size 9 d after infection was markedly reduced when Lp, but not Vg, was depleted (Figure 2D). In contrast to oocyst numbers, silencing TEP1 at the same time as Lp did not rescue oocyst growth (unpublished data), indicating that the small oocyst size does not result from TEP1 activity in Lpdeficient mosquitoes. This supports the hypothesis that Lp contributes nutrients to oocyst development [26]. Therefore, Lp benefits Plasmodium development at two independent levels: an early effect favoring ookinete survival by protecting against TEP1dependent killing, and a later effect favoring normal oocyst growth. The latter effect does not require Vg or TEP1 function. Vg and Lp Do Not Affect TEP1 Expression or Cleavage, but Lp Is Necessary for Proper Vg Expression Previous work [7,31] has demonstrated that boosting mosquito basal immunity via depletion of the inhibitory IkB protein Cactus up-regulates components of the TEP1 pathway (including TEP1, LRIM1, and APL1C) and completely blocks parasite development. Therefore, we asked whether the knockdown of Vg and Lp could mimic the effect of Cactus depletion and elevate TEP1 expression levels, providing an explanation to the above observations. We silenced Lp and/or Vg and examined the transcript levels of TEP1 before and after blood feeding using quantitative real-time polymerase chain reaction (qRT-PCR). Silencing of the two nutrient transport genes did not alter TEP1 expression (Figure 3A). We then evaluated the effect of Lp and Vg silencing on TEP1 protein amounts and TEP1 cleavage in the hemolymph by immunoblotting using polyclonal anti-TEP1 antibodies. This analysis did not reveal any marked increase in the amounts of full-length or mature TEP1 protein (Figures 3C and 1D). Surprisingly, silencing of Lp reproducibly lowered the expression of Vg mRNA (Figure 3B and unpublished data). At the protein level, Lp depletion strongly reduced Vg levels at 47 h (but not 24 h) post-infectious feeding compared with the controls (Figure 3C), confirming that Lp is indeed required for full Vg Figure 3. Lp is required for normal Vg expression. Mosquitoes were injected with dsLp, dsVg, or dsLp+dsVg. (A, B) TEP1, Vg, and Lp expression, respectively, was measured at several time points after P. berghei infection using quantitative RT-PCR. (C) Lp and Vg protein levels in mosquito hemolymph were gauged by Coomassie staining; TEP1 (full length and processed) by immunoblotting. PPO2 served as a loading control. Note that levels of Vg protein are strongly reduced at 47, but not 24, h after infection specifically in dsLp-treated mosquitoes. doi:10.1371/journal.pbio.1000434.g003 PLoS Biology | www.plosbiology.org 6 July 2010 | Volume 8 | Issue 7 | e1000434 Reproduction versus Immunity in Anopheles expression above the levels in the dsLacZ control (Figure 4A). Interestingly, concomitant silencing of Cactus/Rel1 and Cactus/Rel2 restored Vg expression to physiological levels (Figure 4B), indicating that REL1 and REL2 contribute to the regulation of Vg expression. At the protein level, Vg amounts were unchanged at 24 h but strongly reduced 43 h after infectious blood feeding specifically in dsCactus-injected-mosquitoes (Figure 4C), confirming the qPCR data and revealing a clear delay between mRNA and protein fluctuations. Thus, in the dsCactus background, while TEP1 expression is upregulated, Vg expression is directly or indirectly repressed by REL1/2. Therefore, the Cactus protein affects TEP1 and Vg levels in opposite directions. We extended our analysis to Lp, but in contrast to the situation reported for Ae. aegypti [24], its expression was unaffected by the knockdown of the NF-kB-like factors (Figure 4A). Since Vg silencing alone, unlike Cactus silencing, is not sufficient to completely block oogenesis, other molecules required by developing mosquito oocytes may be regulated by Cactus in the same manner as Vg. Taken together, our findings uncover the complex phenotype of Cactus depletion. It leads to a lower level of Vg expression after a blood meal, thereby contributing to the arrest in oogenesis seen in expression between day 1 and day 2 post-infectious blood-feeding. In contrast, the depletion of Vg had no effect on Lp expression (Figure 3B) or protein levels (Figure 3C). Depletion of Cactus Represses Vg Expression The unexpected observation that Lp and Vg knockdown simultaneously arrests oogenesis and facilitates TEP1 binding to ookinetes led us to re-examine the previously observed striking phenotype of dsCactus, which boosts basal immunity while arresting oogenesis ([31] and unpublished data). Depleting the IkB-like repressor protein Cactus increases the activity of NF-kB factors REL1 and REL2, leading to elevated expression of TEP1 and other immune factors. Therefore, we investigated whether REL1, REL2, and Cactus influence the expression of Vg and/or Lp. To this end, mosquitoes were injected with either dsRel1, dsRel2, dsCactus, or co-injected with dsRel1-dsRel2, dsRel1-dsCactus, dsRel2dsCactus, and dsLacZ control. Mosquitoes were fed on an infected mouse, and subsequently, the expression of Vg and Lp was monitored by qRT-PCR. Strikingly, Vg expression was almost abolished in dsCactus mosquitoes at 24 hpi; conversely, the depletion of REL1 or REL2 at this time point elevated Vg Figure 4. Vg expression is repressed by NF-kB factors REL1/REL2. Mosquitoes were injected with the indicated combinations of dsCactus, dsRel1, dsRel2, and the expression levels of Vg and Lp examined by qRT-PCR at the indicated time points after infection and compared to dsLacZ control. Gene expression is expressed relative to the LacZ control at time 0. (A) Vg expression was inhibited in dsCactus but increased in dsRel1/Rel2 mosquitoes. (B) Concomitant depletion of Cactus and Rel1 restored Vg expression. (C) Coomassie staining of hemolymph proteins after electrophoresis on a 7% SDS-PAGE gel and transfer to a PVDF membrane. No change in Vg protein is seen 24 h after infection, but at 43 h dsCactus completely blocks Vg expression. Protein identities are indicated to the left. The identity of the subunits of Lp and Vg, and the identity of APL1C and LRIM1 proteins were established by mass spectrometry of the Coomassie-stained bands and by immunoblotting. APL1C and LRIM1 over-expression confirms the efficiency of Cactus silencing. doi:10.1371/journal.pbio.1000434.g004 PLoS Biology | www.plosbiology.org 7 July 2010 | Volume 8 | Issue 7 | e1000434 Reproduction versus Immunity in Anopheles Cactus knockdown mosquitoes. On the other hand, it stimulates the mosquito antiparasitic defense at least at two different levels: (i) by lowering the level of Vg, rendering TEP1-mediated killing more efficient, and (ii) by elevating the levels of TEP1 pathway proteins. depletion of Lp and, more strikingly, of Vg resulted in more efficient TEP1 binding to the surface of ookinetes at 48 hpi, promoting their killing. One explanation could be that Vg (and perhaps Lp, to a lesser extent) are recruited to the parasite surface, where they might mask TEP1 binding sites. Consistent with this idea, fish vitellogenin has recently been found to bind microorganisms and to opsonize them for phagocytosis [40]. Mosquito Vg may behave non-productively in a similar manner and outcompete TEP1 from the ookinete surface. Alternatively, a physical interaction between TEP1 and Vg could inhibit TEP1 activity, a hypothesis that should be further investigated. Yet another possible explanation is that transient interactions of ookinetes with Vg might alter the lipid composition in the ookinetes’ membrane, rendering them less visible to the TEP1 machinery. The parasite molecules to which TEP1 covalently attaches are currently unknown, but hydroxyl residues on surface lipids could be good targets for thioester-dependent TEP1 covalent binding. We further observed a retarded oocyst growth in Lp-deficient mosquitoes 9 d post infection. This phenotype was specific to Lp, as parasites developed normally in Vg-deficient mosquitoes. Therefore, Lp is a probable lipid source for developing oocysts. Indeed, Lp was detected inside P. gallinaceum oocysts in vitro, suggesting that oocysts tap some of the host’s Lp for their development [26]. Taken together, Lp appears to regulate parasite development at two distinct stages by two independent mechanisms: (i) providing an indirect protection to ookinetes via regulation of Vg levels after a blood meal and thereby dampening TEP1 binding to ookinetes, and (ii) exerting a direct nutritional role by supplying lipids to growing oocysts. The quantitative RT-PCR and protein expression results reported here added the IkB/NF-kB-like factors Cactus/REL1 and REL2, previously known to control immunity [29–31], to the list of factors that influence Vg expression. We propose that Cactus depletion boosts TEP1 parasite killing by simultaneously increasing TEP1 expression [31] and decreasing the expression of Vg, in the absence of which TEP1-mediated killing is more efficient. Previously, the reason why Cactus depletion blocked oogenesis while boosting anti-Plasmodium immunity was unknown. Our results shed new light on this phenomenon by suggesting that Cactus activity is necessary for the expression of Vg, and probably of additional factors involved in vitellogenesis. Although many mosquito genes showing antiparasitic activity are induced by the NF-kB-like factors REL1 and REL2 [12,29– 31,41], it is currently unclear whether parasite invasion of mosquito tissues actually activates the NF-kB pathways. However, the expression of nutrient transport molecules is affected by signals arising from the parasite’s invasion, in addition to being influenced by hormone signaling, the TOR pathway, and NF-kB factors. Indeed, ookinete invasion of the midgut induces Lp mRNA expression further than does an uninfected blood meal in A. gambiae and Ae. aegypti [23,24]. At the protein level, we did not observe a corresponding increase in Lp amounts using specific antibodies (unpublished data), which may reflect consumption of the additionally produced Lp by parasites and/or by the midgut wound healing response to parasite invasion. This implies that Lp protein homeostasis is under tight physiological regulation. Conversely, Ahmed et al. [42] reported that parasite invasion reduces the abundance of the Vg transcript in A. gambiae, while Vg protein levels were only transiently reduced before accumulating in the hemolymph. Therefore, the production of both proteins is subjected to multiple physiological switches. The reported changes in Vg levels correlated with apoptosis of patches of ovarian follicular cells, which was prominent following infections and immune stimulation. Dying ovarian follicles stop secreting Discussion The first indication that nutrient transport after a blood meal influences mosquito susceptibility to P. berghei was provided by Vlachou et al. [23], who demonstrated that experimental depletion of the lipid carrier protein Lp by RNAi reduces the number of developing oocysts in the mosquito midgut. Recently, these results were extended to P. falciparum [25]. However, how and at which stage of development the parasites were eliminated in Lp-deficient mosquitoes remained to be determined. We show here that the major yolk protein Vg shows a similar but more drastic knockdown phenotype than Lp on Plasmodium survival and that the Lp and Vg depletion phenotypes require the function of the immune factor TEP1, which targets ookinetes for killing. Further, high numbers of parasites actually survive and turn into oocysts even in the context of Lp and/or Vg depletion, as long as TEP1 is also experimentally depleted. From these observations, we infer that physiological levels of both nutrient transport proteins following a blood meal somehow dampen the strength of the immune defense and protect ookinetes against destruction by the TEP1 pathway. The effects of Lp and Vg depletion on TEP1mediated parasite killing are similar, and we find that Lp is required for the full induction of Vg expression on day 2 following an infectious blood meal. We therefore propose that Lp may indirectly affect ookinete survival by influencing Vg expression, while Vg impinges either directly or more closely than Lp on the TEP1-killing mechanism. The induction of Vg expression after a blood meal requires both the TOR pathway and ecdysone signaling [14]. It is unclear why Lp depletion reduces the expression of Vg after an infectious blood meal. One possible explanation is that an Lp shortage precludes ovarian follicle development, preventing the normal secretion of ecdysone by follicle cells; thus leading to the reduction in Vg expression. However, attempts to rescue the Lp silencing effect on Vg expression with exogenously provided 20-hydroxyecdysone were unsuccessful. As the lower level of Vg expression in Lpdeficient A. gambiae is reminiscent of the situation observed in adult Ae. aegypti mosquitoes malnourished during larval life [37], it would be interesting to determine if Plasmodium survival is compromised in such malnourished mosquitoes in laboratory and field settings. The GFP-tagged P. berghei strain used in this study provides a good model and enables analyses of vectorial capacity that are much more demanding with wild malaria parasites. However, recent studies indicate that the mosquito response to P. berghei and to P. falciparum differ in important ways [10,38]. In addition, the P. berghei–A. gambiae model is an unnatural host-parasite association. Therefore, it will be important to see whether our observations hold true in the A. gambiae–P. falciparum relationship. Importantly though, the TEP1 pathway does limit P. falciparum survival in A. gambiae natural infections ([39] and Levashina et al., unpublished results) and the Lp knockdown was shown to have similar effects in both systems [25]. What is the molecular basis of the negative effect of the two nutrient transport proteins on the TEP1 pathway? We initially hypothesized that Lp-scaffolded lipidic particles could sequester components of the TEP1 pathway in an inactive state. However, TEP1 and its interacting partners LRIM1 and APL1C were not detectable in Lp extracts, suggesting that the Plasmodium-killing machinery is not carried by Lp particles. Instead, RNAi-mediated PLoS Biology | www.plosbiology.org 8 July 2010 | Volume 8 | Issue 7 | e1000434 Reproduction versus Immunity in Anopheles IgG2ak) recognizing the 80 kDa Lp subunit was selected and ascites fluid was prepared by injection of 26106 hybridoma cells into pristane-primed BALB/c mice. The resulting antibody efficiently immuno-precipitated the 80 kDa Lp subunit and coimmunoprecipated the 250 kDa subunit. The identity of both immuno-precipitated subunits, excised from Coomassie-stained protein gels, was confirmed by mass spectrometry. Similarly, we prepared a monoclonal antibody (2C6) recognizing the large Lp subunit. Rabbit polyclonal antibodies specific to Vg were obtained by immunizing rabbits with a purified recombinant Vg fragment fused to GST. The Vg gene fragment used for protein production was amplified from mosquito cDNA using attB-site (capital letters)containing primers GGGGACAAGTTTGTACAAAAAAGCAGGCTtcaagtttgtgctgcagcacaagcag and GGGGACCACTTTGTACAAGAAAGCTGGGTCCTAagcgcaagatggatggtagtttc. The PCR product was cloned into pDEST15 (Invitrogen) using the Gateway technology. Protein was produced in E. coli BL21-AI. ecdysteroids and taking up Vg protein, which may explain both the drop in Vg transcription and the accumulation of Vg protein in the hemolymph [43,44]. It would be interesting to identify infection-dependent signals arising at the midgut and triggering ovarian follicle apoptosis. In Drosophila, pathogenesis is also reported to trigger cell death in ovaries [45]. In the presence or absence of an infection, activation of the Immune deficiency (Imd) pathway (e.g., by injection of dead bacteria) negatively impacted oogenesis. This effect depended on the immune status, as oogenesis remained normal in Imd pathway mutants injected with dead bacteria [46]. The mosquito Cactus/REL1/REL2 NFkB pathway is related to the Drosophila Toll and Imd immune pathways; its targets would therefore represent attractive candidates as modulators of mosquito reproduction. A full understanding of the interactions between reproductive and immune functions in mosquitoes will require a thorough study of the molecular pathways influencing the transcription of immune and vitellogenic factors, and how these pathways are affected by blood meals, immune defense, and parasite invasion. To our knowledge, Vg and Cactus are the first molecules reported to occupy a central position at the interface between reproduction and immunity, providing a molecular handle to further explore the long-suspected trade-off between these two processes. Immunoprecipitation 120 adult mosquitoes were severed by opening the thorax and abdomen cuticles with fine forceps and bled on ice in 1 ml IP buffer (TRIS pH 7.9 50mM, NaCl 100 mM, EDTA 2 mM, BSA 0.1 mg/ml) + Complete protease inhibitors (Roche). Carcasses and cellular debris were removed by two successive 2,500 g centrifugation steps (for 2 min at 4uC); the extract was further cleared by three 16,500 g centrifugations (2 min each). The sample was precleared for 1 h at 4uC under gentle rocking with 2 mg of an irrelevant mouse IgG2ak antibody that was removed by incubation at 4uC with 35 ml protein A-sepharose slurry (Pharmacia) for 1 h followed by centrifugation. Supernatant was split in two aliquots, one subjected to a 1 h incubation with specific antibody and the other with a non-specific antibody of the same immunoglobulin class. 35 ml of protein A-Sepharose were added to each sample, further rocked at 4uC for 1 h, centrifuged. The supernatant was saved (post-IP supernatant sample). Sepharose beads were washed 5610 min in TE buffer with or without 500 mM KCl, successively. Lipophorin and associated proteins were eluted from the beads using SDS-PAGE sample buffer and submitted to Western blotting. Material and Methods Potassium Bromide Gradient Purification of Lipophorin Particles Approximately 0.5 g of mosquito adults (ca. 330 mosquitoes) were roughly ground with a Polytron electric homogenizer in 2 ml ice-cold TNE buffer (100 mM Tris-HCl pH 7.5, 0.2 mM EGTA, 150 mM NaCl) + Complete protease inhibitors (Roche). Debris were centrifuged at 4uC in a tabletop centrifuge. The supernatant was transferred to 2.2 ml ultracentrifuge tubes and spun for 3 h at 120,000 g at 4uC in a Sorvall ultracentrifuge equipped with an S55-S rotor. The cleared supernatant was recovered, completed with solid potassium bromide to a final concentration of 0.34 g/ ml, overlayed with 0.5 ml TNE buffer+0.33 g/ml KBr, and centrifuged in 2.2 ml PET ultracentrifuge tubes (Hitachi Koki) at 250,000 g, 10uC, for at least 36 h. The top layer of fat was discarded and 5 or 6 fractions of 0.5 ml were carefully collected starting from the top. Lipophorin particles were present in the top fraction, while the majority of other proteins fractionated into the fourth. Hemolymph Protein Samples for SDS-PAGE At least 8 anesthetized mosquitoes were aligned on ice under the binocular microscope. Their proboscis was clipped with dissection scissors. Each mosquito was gently pressed on the thorax with forceps and the hemolymph droplet forming at the tip of the cut proboscis was collected into 16 sample (Laemmli) buffer. An hemolymph amount equivalent to that collected from 4 mosquitoes was loaded in each lane of SDS-PAGE gels. Lipophorin and Vitellogenin Antibodies The top fraction of a potassium bromide gradient prepared using a scale-up of the above method was desalted on a Pharmacia PD-10 column according to the manufacturer’s instructions. The two subunits of Lp were the predominant proteins in the extract according to Coomassie staining of an SDS-PAGE gel. Protein amount was quantified with a Bradford assay. Six-week-old female BALB/c mice were injected intraperitoneally with 40 mg of these lipophorin particles and 100 mg of poly I/C as adjuvant. Three injections were performed at 2-wk intervals. Four days prior to hybridoma fusion, mice with positively reacting sera were reinjected. Spleen cells were fused with Sp2/0.Agl4 myeloma cells as described [47]. Hybridoma culture supernatants were tested at day 10 by ELISA for cross-reaction with purified Lp particles. Positive supernatants were then tested by Western blot on mosquito extracts. All ELISA-positive supernatants recognized peptides corresponding in size to either the large (250 kDa) or the small (80 kDa) Lp subunit. Specific cultures were cloned twice on soft agar. A hybridoma clone (2H5, immunoglobulin subclass PLoS Biology | www.plosbiology.org RNAi and Infections The 741 bp long HincII fragment of Vg1 (AGAP004203) and the 431 bp long BspHI/BsgI fragment of Lp (AGAP001826) were cloned from cDNA library clones into the pLL10 vector. RNAi constructs for TEP1 and NF-kB factors have been described (Frolet et al. 2006) [31]. Potential cross-silencing effects of the chosen sequences were analyzed using the Deqor software ([48]; http://deqor.mpi-cbg.de/) with the predicted A. gambiae transcriptome ENSEMBL database. DsRNA was synthesized as previously described [36]. A. gambiae susceptible G3 strain were maintained at 28uC, 75%–80% humidity, and a 12/12 h light/ dark cycle. Two-day-emerged adult female mosquitoes from the same cohort were injected with 0.2 mg of dsRNA using a Nanoject II injector (Drummond, http://www.drummondsci.com). Coinjection experiments were performed by injecting a double 9 July 2010 | Volume 8 | Issue 7 | e1000434 Reproduction versus Immunity in Anopheles volume of 1:1 mixtures of 3 mg/ml solutions of dsRNAs. Four days after dsRNA injection mosquitoes were fed on a mouse carrying P. berghei GFP-con 259cl2 as previously described [36,37]. Statistical significance was determined with a Kruskall-Wallis test for nonparametric data followed by Dunn’s post-test. The indicated p values are those obtained with Dunn’s test. Autoflex III Smartbeam (Bruker-Daltonik GmbH, Bremen, Germany) matrix-assisted laser desorption/ionization time-offlight mass spectrometer (MALDI-TOF TOF) used in reflector positive mode. The resulting peptide mass fingerprinting data and peptide fragment fingerprinting data were combined by Biotools 3 software (Bruker Daltonik) and transferred to the search engine MASCOT (Matrix Science, London, UK). Peptide mass error was limited to 50 ppm. Proteins were identified by searching data against NCBI non-redundant protein sequence database. Assessment of Ovary Development The ovaries of dissected females were observed under the binocular microscope. Ovaries containing 3 fully grown eggs or more were scored as positive. Ovaries with only undeveloped oocytes or less than 3 fully grown eggs were scored negative. Supporting Information Figure S1 Prophenoloxidase but not TEP1 or LRIM1 associates with Lp particles. (A, top panel) Coomassiestained polyacrylamide gel resolving mosquito proteins fractionated on a potassium bromide gradient. Molecular weight standards are indicated on the left. Lp subunits (ApoI and ApoII, circled red in lane 1) are the main proteins detectable in top gradient fractions. Fractions 1, 2, 3 are 10-fold concentrated compared to fractions 4, 5, 6. (A, middle and bottom panel) Western blotting with anti-TEP1 and LRIM1 antibodies reveal TEP1 and LRIM1 proteins only in higher density fractions. TEP1F, full-length TEP1; TEP1-C, C-terminal TEP1 fragment. (B) Western blotting analysis of KBr fractions using anti-PPO2 antibody. A fraction of PPO fractionates with Lp particles. (C) Immunoblotting analysis of Lp particles purified by immunoprecipitation 0, 4, or 14 d after a P. berghei infection (dpi) with mouse anti-Lp (ApoLpII) monoclonal antibody. Non-specific mouse antibody (NS) is used as an immunoprecipitation control. TEP1 does not associate with purified Lp and is found only in post-IP (unbound) supernatants. Found at: doi:10.1371/journal.pbio.1000434.s001 (0.92 MB TIF) qRT-PCR Total RNA from 10 mosquitoes was extracted with Trizol reagent (Invitrogen) before and after dsRNA injection or after blood feeding. 2–8 mg of RNA was reverse transcribed using MMLV enzyme and random primers (Invitrogen). Specific primers (Table 1) were used at 300 nM for qRT-PCR reactions. Ribosomal protein L19 (RPL19) served as an internal control to normalize gene expression. The reactions were run on an Applied Biosystems 7500 Fast Real-Time PCR System using Power SYBR Green Mastermix (http://www.appliedbiosystems.com). Fluorescence Microscopy In order to count the surviving GFP-expressing parasites, mosquito midguts were dissected between 7 and 10 dpi and prepared as previously described [36,37] and observed under a fluorescence microscope. To assess TEP1 binding to ookinetes, mosquito midguts were dissected at 18, 24, and 48 hpi, fixed in 4% formaldehyde at room temperature for 45 min, then washed with phosphate buffered saline, and stained with anti-TEP1 antibodies as previously described [31,36]. Parasite numbers and TEP1 labeling were scored using a Zeiss fluorescence microscope (Axiovert 200M) equipped with a Zeiss Apotome module (http:// www.zeiss.com). GFP-expressing parasites were considered live while dead parasites were GFP negative. Differential TEP1 staining on ookinete were gauged at 18, 24, and 48 hpi. At least three independent experiments were conducted per treatment group with a minimum of five mosquito midguts per treatment. For each midgut, all ookinetes visible in 4 fields covering most of the midgut were scored. Table S1 summarizes the ookinete counts from three independent experiments. Figure S2 Lp and Vg proteins are readily visualized by Coomassie staining. Mosquitoes were injected with dsLacZ, dsLp, or dsVg as indicated and offered a blood meal 4 d later to induce Vg expression. Hemolymph was collected 24 h after a blood meal from clipped mosquito proboscises. Hemolymph from the equivalent of 4 mosquitoes as well as 5- and 10-fold dilutions of the control dsLacZ hemolymph (2 lanes at the right of the gel) was resolved by electrophoresis on a 7% SDS-PAGE gel and transferred to a PVDF membrane. The membrane was subjected to staining with Coomassie brilliant blue (top panel). Proteins were subsequently revealed with the indicated antibodies (lower panels). Molecular weight markers are indicated on the right. Protein bands revealed by the antibodies superpose perfectly with the protein bands revealed by Coomassie staining. The protein identities were confirmed by a mass spectrometric analysis. The intensities of antibody signals in the 5- and 10-fold diluted sample indicate that residual Vg and Lp protein levels are less than 10% of the control level in the corresponding RNAi samples. Found at: doi:10.1371/journal.pbio.1000434.s002 (2.34 MB TIF) MALDI Mass Spectrometry Coomassie-stained protein bands excised from SDS-PAGE gels were digested with trypsin. Tryptic peptides eluted from the gel slices were subjected to MALDI mass measurement on an Table 1. Primers used for qRT-PCR. Gene Primers for qRT-PCR TEP1 AAAGCTACGAATTTGTTGCGTCA TTCTCCCACACACCAAACGAA Vg CCGACTACGACCAGGACTTC CTTCCGGCGTAGTAGACGAA Lp CAGCCAGGATGGTGAGCTTAA CACCAGCACCTTGGCGTT RPL19 CCAACTCGCGACAAAACATTC ACCGGCTTCTTGATGATCAGA Figure S3 (A) Three additional repeats of the experiment shown in Figure 1A. (See Figure 1A for legend.) (B) Three additional repeats of the experiment shown in Figure 1B. (See Figure 1B for legend.) (C) Two additional repeats of the experiment shown in Figure 2A. (See Figure 2A for legend.) Found at: doi:10.1371/journal.pbio.1000434.s003 (0.35 MB TIF) The table summarizes the parasite scores for three independent repeats of the experiment shown in Figure 2C. Shown are parasite percentages in each of the three possible classes (live, GFP positive; dying, GFP + TEP1 positive; dead, TEP1 positive). The total number of ookinetes scored for each treatment group is given in parentheses next to the injected Table S1 doi:10.1371/journal.pbio.1000434.t001 PLoS Biology | www.plosbiology.org 10 July 2010 | Volume 8 | Issue 7 | e1000434 Reproduction versus Immunity in Anopheles dsRNA. p values were obtained by chi-square analysis comparing parasite scores in dsLp and dsLacZ-injected mosquitoes or comparing parasite scores in dsLp-Vg and dsLacZ-injected mosquitoes. For this analysis, we summed all TEP1-positive ookinetes (dead + dying). Figure 2C was generated with Experiment 3. Found at: doi:10.1371/journal.pbio.1000434.s004 (0.04 MB XLS) Acknowledgments The authors thank J. Soichot, M.E. Moritz, and C. Kappler for help with the mosquito colony and parasite cultures; C. Guillet and P. Hammann (IBMC mass spectrometry platform) for protein identification; V. Andres and N. Jung for hybridoma cultures and subcloning; and F. Catteruccia and members of her and our group for fruitful discussions and critical reading of the manuscript. Text S1 The supplemental text describes lipophorin particle purification from adult mosquitoes by potassium bromide gradient fractionation or immuno-precipitation and a search for immune factors that co-purify with lipophorin. Found at: doi:10.1371/journal.pbio.1000434.s005 (0.10 MB DOC) Author Contributions The author(s) have made the following declarations about their contributions: Conceived and designed the experiments: MKR MMW EAL EM. Performed the experiments: MKR MMW EM. Analyzed the data: MKR MMW EAL EM. Contributed reagents/materials/analysis tools: MOA. Wrote the paper: MKR EAL EM. References 23. Vlachou D, Schlegelmilch T, Christophides GK, Kafatos FC (2005) Functional genomic analysis of midgut epithelial responses in Anopheles during Plasmodium invasion. Curr Biol 15: 1185–1195. 24. Cheon HM, Shin SW, Bian G, Park JH, Raikhel AS (2006) Regulation of lipid metabolism genes, lipid carrier protein lipophorin, and its receptor during immune challenge in the mosquito Aedes aegypti. J Biol Chem 281: 8426–8435. 25. Mendes AM, Schlegelmilch T, Cohuet A, Awono-Ambene P, De Iorio M, et al. (2008) Conserved mosquito/parasite interactions affect development of Plasmodium falciparum in Africa. PLoS Pathog 4: doi:10.1371/journal.ppat.1000069. 26. Atella GC, Bittencourt-Cunha PR, Nunes RD, Shahabuddin M, Silva-Neto MA (2009) The major insect lipoprotein is a lipid source to mosquito stages of malaria parasite. Acta Trop. 27. Zhu J, Chen L, Raikhel AS (2003) Posttranscriptional control of the competence factor betaFTZ-F1 by juvenile hormone in the mosquito Aedes aegypti. Proc Natl Acad Sci U S A 100: 13338–13343. 28. Zhu J, Chen L, Sun G, Raikhel AS (2006) The competence factor beta Ftz-F1 potentiates ecdysone receptor activity via recruiting a p160/SRC coactivator. Mol Cell Biol 26: 9402–9412. 29. Bian G, Shin SW, Cheon HM, Kokoza V, Raikhel AS (2005) Transgenic alteration of Toll immune pathway in the female mosquito Aedes aegypti. Proc Natl Acad Sci U S A 102: 13568–13573. 30. Meister S, Kanzok SM, Zheng XL, Luna C, Li TR, et al. (2005) Immune signaling pathways regulating bacterial and malaria parasite infection of the mosquito Anopheles gambiae. Proc Natl Acad Sci U S A 102: 11420–11425. 31. Frolet C, Thoma M, Blandin S, Hoffmann JA, Levashina EA (2006) Boosting NF-kappaB-dependent basal immunity of Anopheles gambiae aborts development of Plasmodium berghei. Immunity 25: 677–685. 32. Franke-Fayard B, Trueman H, Ramesar J, Mendoza J, van der Keur M, et al. (2004) A Plasmodium berghei reference line that constitutively expresses GFP at a high level throughout the complete life cycle. Mol Biochem Parasitol 137: 23–33. 33. Bose SG, Raikhel AS (1988) Mosquito vitellogenin subunits originate from a common precursor. Biochem Biophys Res Commun 155: 436–442. 34. Sappington TW, Raikhel AS (1998) Molecular characteristics of insect vitellogenins and vitellogenin receptors. Insect Biochem Mol Biol 28: 277–300. 35. Dhadialla TS, Raikhel AS (1990) Biosynthesis of mosquito vitellogenin. J Biol Chem 265: 9924–9933. 36. Blandin S, Levashina EA (2004) Mosquito immune responses against malaria parasites. Curr Opin Immunol 16: 16–20. 37. Shiao SH, Hansen IA, Zhu J, Sieglaff DH, Raikhel AS (2008) Juvenile hormone connects larval nutrition with target of rapamycin signaling in the mosquito Aedes aegypti. J Insect Physiol 54: 231–239. 38. Jaramillo-Gutierrez G, Rodrigues J, Ndikuyeze G, Povelones M, Molina-Cruz A, et al. (2009) Mosquito immune responses and compatibility between Plasmodium parasites and anopheline mosquitoes. BMC Microbiol 9: 154. 39. Dong Y, Aguilar R, Xi Z, Warr E, Mongin E, et al. (2006) Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog 2: e52. doi:10.1371/journal.ppat.0020052. 40. Li Z, Zhang S, Liu Q (2008) Vitellogenin functions as a multivalent pattern recognition receptor with an opsonic activity. PLoS ONE 3: doi:10.1371/ journal.pone.0001940. 41. Garver LS, Dong Y, Dimopoulos G (2009) Caspar controls resistance to Plasmodium falciparum in diverse anopheline species. PLoS Pathog 5: doi:10.1371/journal.ppat.1000335. 42. Ahmed AM, Maingon R, Romans P, Hurd H (2001) Effects of malaria infection on vitellogenesis in Anopheles gambiae during two gonotrophic cycles. Insect Mol Biol 10: 347–356. 43. Hopwood JA, Ahmed AM, Polwart A, Williams GT, Hurd H (2001) Malariainduced apoptosis in mosquito ovaries: a mechanism to control vector egg production. J Exp Biol 204: 2773–2780. 44. Ahmed AM, Hurd H (2006) Immune stimulation and malaria infection impose reproductive costs in Anopheles gambiae via follicular apoptosis. Microbes Infect 8: 308–315. 1. Sinden RE (1999) Plasmodium differentiation in the mosquito. Parassitologia 41: 139–148. 2. Blandin S, Shiao SH, Moita LF, Janse CJ, Waters AP, et al. (2004) Complementlike protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 116: 661–670. 3. Habtewold T, Povelones M, Blagborough AM, Christophides GK (2008) Transmission blocking immunity in the malaria non-vector mosquito Anopheles quadriannulatus species A. PLoS Pathog 4: doi:10.1371/journal.ppat.1000070. 4. Blandin SA, Marois E, Levashina EA (2008) Antimalarial responses in Anopheles gambiae: from a complement-like protein to a complement-like pathway. Cell Host Microbe 3: 364–374. 5. Levashina EA, Moita LF, Blandin S, Vriend G, Lagueux M, et al. (2001) Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell 104: 709–718. 6. Baxter RH, Chang CI, Chelliah Y, Blandin S, Levashina EA, et al. (2007) Structural basis for conserved complement factor-like function in the antimalarial protein TEP1. Proc Natl Acad Sci U S A 104: 11615–11620. 7. Fraiture M, Baxter RH, Steinert S, Chelliah Y, Frolet C, et al. (2009) Two mosquito LRR proteins function as complement control factors in the TEP1mediated killing of Plasmodium. Cell Host Microbe 5: 273–284. 8. Povelones M, Waterhouse RM, Kafatos FC, Christophides GK (2009) Leucinerich repeat protein complex activates mosquito complement in defense against Plasmodium parasites. Science 324: 258–261. 9. Osta MA, Christophides GK, Kafatos FC (2004) Effects of mosquito genes on Plasmodium development. Science 303: 2030–2032. 10. Cohuet A, Osta MA, Morlais I, Awono-Ambene PH, Michel K, et al. (2006) Anopheles and Plasmodium: from laboratory models to natural systems in the field. EMBO Rep 7: 1285–1289. 11. Riehle MM, Markianos K, Niare O, Xu J, Li J, et al. (2006) Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science 312: 577–579. 12. Riehle MM, Xu J, Lazzaro BP, Rottschaefer SM, Coulibaly B, et al. (2008) Anopheles gambiae APL1 is a family of variable LRR proteins required for Rel1-mediated protection from the malaria parasite, Plasmodium berghei. PLoS ONE 3: doi:10.1371/journal.pone.0003672. 13. Attardo GM, Hansen IA, Raikhel AS (2005) Nutritional regulation of vitellogenesis in mosquitoes: implications for anautogeny. Insect Biochem Mol Biol 35: 661–675. 14. Hansen IA, Attardo GM, Park JH, Peng Q, Raikhel AS (2004) Target of rapamycin-mediated amino acid signaling in mosquito anautogeny. Proc Natl Acad Sci U S A 101: 10626–10631. 15. Raikhel AS, Dhadialla TS (1992) Accumulation of yolk proteins in insect oocytes. Annu Rev Entomol 37: 217–251. 16. Tufail M, Takeda M (2008) Molecular characteristics of insect vitellogenins. J Insect Physiol 54: 1447–1458. 17. Canavoso LE, Jouni ZE, Karnas KJ, Pennington JE, Wells MA (2001) Fat metabolism in insects. Annu Rev Nutr 21: 23–46. 18. Arrese EL, Canavoso LE, Jouni ZE, Pennington JE, Tsuchida K, et al. (2001) Lipid storage and mobilization in insects: current status and future directions. Insect Biochem Mol Biol 1: 7–17. 19. Atella GC, Silva-Neto MA, Golodne DM, Arefin S, Shahabuddin M (2006) Anopheles gambiae lipophorin: characterization and role in lipid transport to developing oocyte. Insect Biochem Mol Biol 36: 375–386. 20. Panáková D, Sprong H, Marois E, Thiele C, Eaton S (2005) Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature 435: 58–65. 21. Vaisar T, Pennathur S, Green PS, Gharib SA, Hoofnagle AN, et al. (2007) Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest 117: 746–756. 22. Pays E, Vanhollebeke B (2009) Human innate immunity against African trypanosomes. Curr Opin Immunol 21: 493–498. PLoS Biology | www.plosbiology.org 11 July 2010 | Volume 8 | Issue 7 | e1000434 Reproduction versus Immunity in Anopheles 45. Brandt SM, Schneider DS (2007) Bacterial infection of fly ovaries reduces egg production and induces local hemocyte activation. Dev Comp Immunol 31: 1121–1130. 46. Zerofsky M, Harel E, Silverman N, Tatar M (2005) Aging of the innate immune response in Drosophila melanogaster. Aging Cell 4: 103–108. PLoS Biology | www.plosbiology.org 47. St Groth FS, Scheidegger D (1980) Production of monoclonal antibodies: strategy and tactics. J Immunol Methods 35: 1–21. 48. Henschel A, Buchholz F, Habermann B (2004) DEQOR: a web-based tool for the design and quality control of siRNAs. Nucleic Acids Res 32: W113–W120. 12 July 2010 | Volume 8 | Issue 7 | e1000434 Article 6 (review): 2011 Current Opinion in Molecular Biology The multifaceted mosquito anti-­Plasmodium response. Marois, E. Author's personal copy Available online at www.sciencedirect.com The multifaceted mosquito anti-Plasmodium response Eric Marois Plasmodium development within its mosquito vector is an essential step in malaria transmission, as illustrated in world regions where malaria was successfully eradicated via vector control. The innate immune system of most mosquitoes is able to completely clear a Plasmodium infection, preventing parasite transmission to humans. Understanding the biological basis of this phenomenon is expected to inspire new strategies to curb malaria incidence in countries where vector control via insecticides is unpractical, or inefficient because insecticide resistance genes have spread across mosquito populations. Several aspects of mosquito biology that condition the success of the parasite in colonizing its vector begin to be understood at the molecular level, and a wealth of recently published data highlights the multifaceted nature of the mosquito response against parasite invasion. In this brief review, we attempt to provide an integrated view of the challenges faced by the parasite to successfully invade its mosquito host, and discuss the possible intervention strategies that could exploit this knowledge for the fight against human malaria. Address INSERM U963, CNRS UPR9022, 15 rue René Descartes, 67084 Strasbourg, Cedex, France Corresponding author: Marois, Eric (E.Marois@unistra.fr) Current Opinion in Microbiology 2011, 14:429–435 This review comes from a themed issue on Host-microbe interactions: Parasites Edited by Isabelle Tardieux Available online 27th July 2011 1369-5274/$ – see front matter # 2011 Elsevier Ltd. All rights reserved. DOI 10.1016/j.mib.2011.07.016 Human malaria is caused by 5 different species of Plasmodium parasites (P. falciparum, P. vivax, P. malariae, P. ovale and P. knowlesi) vectored by about 30 mosquito species within the Anopheles genus. 225 million people were infected and 780,000 (mainly African children) died in 2009 (WHO malaria report, 2010), mostly owing to P. falciparum. Humans in biological terms are the ‘secondary’ or ‘intermediary’ host for Plasmodium: Anopheles mosquitoes constitute the ‘primary’ or ‘definitive’ host, in which the parasite undergoes sexual reproduction. When a mosquito female takes a blood meal on an infected host, Plasmodium gametes fertilize within the blood bolus. The zygote differentiates into an ookinete, a motile cell able to traverse the intestinal epithelium, on the basal side of which it rests and, if surviving mosquito defenses that www.sciencedirect.com most efficiently target this stage, transforms into an oocyst. Within each oocyst, asexual multiplications produce thousands of sporozoites, single cells that are released into the hemolymph (mosquito blood) and journey from the midgut to the salivary glands. There, sporozoites become competent for injection into another host during a subsequent blood meal. Plasmodium infection has a subtle but documented impact on mosquito reproduction, fitness and survival [1–5], and the insect’s innate immune system reacts strongly to the parasite. Out of some 400 mosquito species in the Anopheles genus, only about 40 allow the development of Plasmodium parasites [6]. Neither mosquitoes belonging to other genera, nor other blood-sucking insects can vector human Plasmodium parasites (some avian Plasmodium species are vectored by Aedes and Culex mosquitoes); a fact that underlines the existence of molecular determinants of parasite–vector specificity and the co-evolution between vectors and parasites. Part of this specificity is based on stringent molecular matches between Plasmodium ookinete proteins required for invasion and their unknown mosquito targets [7–9]. If the invasion step is bypassed by experimental injection of ookinetes into insect hemolymph, even Drosophila can support oocyst development [10] and was used to identify molecules involved in the insect/parasite interaction [11]. Another basis of the specificity is the crucial role played by the mosquito immune system. Illustrating this point, knocking down immune factors in the TEP1 pathway (described below) in the non-vector mosquito Anopheles quadriannulatus is sufficient to convert it into a competent P. berghei vector [12]. Interestingly, even in laboratorycultured strains of the main African malaria vector A. gambiae, individual mosquitoes that took similar amounts of infected blood will display hugely variable numbers of parasites successfully colonizing their intestine. For P. berghei, a rodent model of malaria, the numbers of oocysts counted within the midguts of sibling, untreated mosquitoes can range from 0 to more than 1000, away from a normal (Gaussian) distribution. Interference with immune defense pathways in a batch of mosquito adults shifts the mean and median oocyst numbers, but usually preserves a great variability. What differences between individual mosquitoes may underlie this extreme variability? We can group the diverse factors that influence parasite success in individual mosquitoes into the following categories: genetic diversity in mosquito genes encoding immune factors, variation in the expression level of immune genes due to each mosquito’s life history, molecular interferences between immune factors and other physiological processes, and the influCurrent Opinion in Microbiology 2011, 14:429–435 Author's personal copy 430 Host-microbe interactions: Parasites Figure 1 Gut flora: ROS, NO Immune elicitors BLOOD MEAL ook. di-Tyr. ntwk O S m. e. c. R Epithelial cells dual oxidase /peroxidase ROS, NO antimicrobial compounds N O p. mx NO b. lam. spz. ooc. hem. Hemocytes soluble immune proteins (TEP1, LRIM1, APL1...) Fat Body Vitellogenic factors HEMOLYMPH Current Opinion in Microbiology A synthetic view of mosquito factors influencing Plasmodium survival. Schematic respresentation of a small section of the mosquito midgut epithelium with the ingested blood meal at the top, mosquito hemolymph at the bottom. Plasmodium and the factors favoring its survival are depicted in green, factors promoting its killing in red. Ook.: ookinete; ooc.: oocyst, p. mx.: peritrophic matrix, di-Tyr. Ntwk: di-tyrosine network; m. e. c.: midgut epithelial cell; b. lam.: basal lamina, hem.: hemocyte; NO: nitric oxide; ROS: reactive oxygen species. ence on Plasmodium infection of microbes residing within the mosquito intestine (Figure 1). an infection depends partly on the genetic makeup of both the vector and the parasite. Immunity factors The development of the gene silencing technology by RNA interference (RNAi) was instrumental for the identification of Anopheles antiparasitic genes and their regulatory pathways [[14], reviewed in [15]]. It allows silencing a chosen gene in mosquito adults by injecting gene-specific double-stranded RNA into the insect’s thorax. Many antiparasitic factors have been identified using this technique and their regulatory pathways outlined [reviewed in detail in [16]]. A prominent antiparasitic factor is Thioester-Containing Protein 1 (TEP1). This homologue of vertebrate complement factor C3 labels the parasite surface and specifies its killing by a still unknown mechanism [17,18]. It is also an important component of the mosquito antibacterial defense, and may have evolved primarily to fight bacterial infections. The protein is secreted as a precursor that matures into two disulfide-bonded fragments, chaperoned in mosquito hemolymph by the two leucine-rich repeat containing The first hint that genetic diversity among mosquito immune factors largely controls the outcome of parasite infection came from the observation that Plasmodiumsensitive and Plasmodium-refractory lines could be derived from a single, genetically heterogeneous mosquito population. A laboratory strain of A. gambiae initially isolated from the Gambia in 1975 was thus submitted to selection for mosquitoes permissive or refractory to the simian parasite P. cynomolgi, yielding the commonly used G3 and L3-5 laboratory strains, respectively [13]. P. berghei infection in the mouse is now widely used as a safe laboratory model of malaria. G3 permits the development of large numbers of P. berghei oocysts, whereas L3-5 usually kills and melanizes 100% of invading ookinetes. However, L3-5 is susceptible to some P. falciparum strains, indicating that some parasites have the ability to escape the immune defense, and that the outcome of Current Opinion in Microbiology 2011, 14:429–435 www.sciencedirect.com Author's personal copy Mosquito anti-Plasmodium response Marois 431 proteins LRIM1 and APL1C [19,20]. The APL1C antiparasitic function was not identified in RNAi screens but by genetic mapping of a refractoriness locus [21,22]. The three proteins are all strictly required for P. berghei killing. Interestingly, Plasmodium-susceptible G3 and refractory L3-5 mosquitoes differ in the TEP1 allele they carry. The allelic form of TEP1 accounts largely for the difference in susceptibility between the two strains [17,23]. In these strains, the presence of TEP1 ‘refractory alleles’ correlates with complete abortion of the development of P. berghei, while ‘susceptible alleles’ still determine the killing of about 80% of invading ookinetes, as shown by the 5-fold average increase in parasite survival when TEP1 is silenced by RNAi. The TEP1 pathway is also active against P. falciparum [24,25] though its impact in the field appears to be more subtle than in the laboratory (Levashina and colleagues, personal communication). Polymorphism at the TEP1 locus influences the level of P. falciparum infection in laboratory mosquitoes [26]. Interestingly, the heterodimer formed by LRIM1 and APL1C is dispensable for TEP1-mediated P. falciparum killing [27,28], contrasting the requirement for these two proteins in fighting P. berghei infections. Importantly, APL1A, a gene closely related to APL1C, plays a role in killing P. falciparum [28] and polymorphism at the APL1A-C locus also correlates with differences in susceptibility to P. falciparum [21]. It is unclear at present if the antiparasitic functions of APL1A and TEP1 depend on each other; further work is needed to determine whether TEP1 requires APL1A and an unknown LRIM1 substitute in killing P. falciparum or completely distinct chaperoning complexes. A new twist to the current model of TEP1 function is the observation by Baxter et al. [29] that APL1C and LRIM1 interact only with a form of TEP1 carrying an inactivated thioester site, at least in vitro. The thioester site is thought to serve in covalent binding to the parasite surface, so why would a heterotrimeric complex comprising a thioester-less TEP1 be important for parasite labeling and killing? In analogy with the complement system, the complex could actually be a convertase that must activate additional, intact, free TEP1 molecules and direct them to the parasite surface. Further studies are required to test this idea, but distinct convertases might direct TEP1 against the surface of different microorganisms, explaining the absence of requirement for LRIM1 and APL1C in fighting P. falciparum infection. LRIM1 and APL1C appear to also chaperon other proteins including other TEP1 family members [30]. This, as well as the existence of a large family of LRIM1-like proteins [31], is consistent with the potential existence of multiple convertase complexes of various specificities. Many other mosquito genes were shown to contribute to the mosquito response to Plasmodium [reviewed in [16,32]]. Notably, depleting lipophorin I/II (Lp), a lipid www.sciencedirect.com transport protein that provides energy to tissues and nutrients to developing eggs, was shown to decrease parasite survival [33]. It came as a surprise to see that this effect is mediated by the TEP1 pathway; and that another nutrient transporter, vitellogenin (Vg), behaves in the same manner [34]. As Lp silencing resulted in a decrease in Vg expression, the Lp knockdown phenotype could be mediated by the drop in Vg levels. Why do nutrient transporters activated by the blood meal appear to dampen the parasite-killing activity of the TEP1 pathway? Hypotheses include a physical interaction of the Vg protein with TEP1 or TEP1 target sites on the parasite surface, implying that the parasite may use blood-induced vector molecules as a decoy. Increasing levels of nutrient transport proteins in the hemolymph could also reduce the expression of unknown immune factors required for TEP1 activity. Alternatively, Vg levels in the hemolymph reach such an enormous level after a blood meal that the function of other hemolymph proteins, including TEP1, could simply suffer as their relative concentration plummets. Nevertheless, we showed that regulatory cascades controlling the expression of immune and reproductive genes are interconnected: increasing NF-kB transcription factor activity boosts basal immunity by elevating the levels of TEP1, LRIM1, APL1C and other genes. Simultaneously, it decreases the expression of the Vg gene, impairing egg development [34]. This example of a trade-off between immunity and reproduction shows that the vector’s physiological state can profoundly affect its ability to eliminate the parasite: allocating a large fraction of mosquito resource to reproduction rather than to immunity will allow the parasite to establish more successful infections, while an elevated mosquito immune response will decrease reproductive success. Reactive oxygen species and nitric oxide In parallel to the TEP1 pathway, an apparently independent important mosquito antiparasitic response is the production of reactive oxygen species (ROS) that are toxic to the parasite. An uninfected blood meal alone induces ROS production, the level of which is higher in refractory than in susceptible mosquitoes [35]. This is thought to contribute to the refractory phenotype, as experimentally elevating ROS in susceptible mosquitoes makes them more refractory [36], while administrating antioxidants to refractory mosquitoes reduces parasite encapsulation [35]. Plasmodium traversal of midgut epithelial cells exacerbates ROS production, and is accompanied by the induction in the fat body of several ROSdetoxifying enzymes such as catalase and superoxide dismutase, that keep global levels of ROS in check [37]. On the contrary, at the parasite invasion site in the midgut, a specific reduction in catalase expression is observed. The resulting local elevation of ROS is thought to reduce the parasite population [37]. Silencing the ROS-detoxifying enzymes or their positive regulators [11,37,38] decreases parasite survival, whereas providCurrent Opinion in Microbiology 2011, 14:429–435 Author's personal copy 432 Host-microbe interactions: Parasites ing antioxidants to the mosquito increases it [37]. This defense mechanism is a two-edged sword from the point of view of the mosquito, as ROS are damaging to longevity and reproduction [39]. This constitutes another example of trade-off between mosquito immunity and reproduction. It is intriguing that refractory mosquitoes are characterized both by greater basal levels of ROS and by refractory alleles of the TEP1 gene. Are ROS production and the TEP1 pathway really two completely independent parasite-killing mechanisms? Would the stimulation of ROS (and nitric oxide, see below) productions still have adverse effects on parasite success in the context of a TEP1 knockdown? These exciting questions await to be tackled. Plasmodium presence in the blood meal, possibly via parasite molecules such as glycosylphosphatidylinositol [40] or hemozoin [41] also induces the expression of nitric oxide synthase (NOS) in the mosquito midgut. In Anopheles stephensi, midgut epithelial cells that are traversed by ookinetes undergo apoptosis and extrude from the epithelium after expressing elevated levels of NOS [[42], reviewed in [43]]. In A. gambiae, the process of midgut cell traversal has not been observed to involve extrusion of apoptotic cells in electron microscopy studies [18]. Mosquito-produced nitric oxide (NO), a cytotoxic molecule, plays a complex role in controlling Plasmodium invasion success. Within the blood meal, it induces apoptosis of a large fraction of ookinetes [44]; but then appears to be necessary for the success of the surviving ookinetes in traversing the midgut epithelium [45]. Subsequently, NO is again detrimental to the early oocyst stage, as silencing NOS or feeding the mosquitoes a NOS inhibitor increases the survival of young oocysts [45]. The latter data document a previously uncharacterized, NOmediated mosquito response directed against the early oocyst stage. Fine-tuned biochemical mechanisms ensure that midgut cells are generally protected against microorganism-induced NO and oxidative stress. In particular, a blood meal and bacterial elicitor-induced oxidase-peroxidase enzyme couple synthesizes a dityrosine protein polymer network that insulates epithelial cells and ensures that NO and ROS synthesis only fire if this barrier is breached by ookinetes [46]. The ookinete-to-oocyst transition represents the main bottleneck in parasite population, and as such represents an attractive target for interventions against malaria. Once parasite numbers have again amplified several thousandfold during oocyst development, a second bottleneck occurs at the transition between oocyst sporozoites and infectious, salivary-gland sporozoites [reviewed in [47]]. This bottleneck is probably due, in a large part, to the scattering and loss of migrating sporozoites throughout the mosquito body. However, a mosquito immune response directed against the sporozoite stage is also likely to take place, as silencing certain immune-related Current Opinion in Microbiology 2011, 14:429–435 genes such as Serpin6 [48] elevates salivary gland sporozoite counts. Much work is needed to better understand this anti-sporozoite mosquito response, which may offer another target for anti-malarial interventions. Gut microbiota The crucial importance of ROS in Plasmodium resistance was recently underlined in an unexpected manner with the demonstration that resident gut bacteria produce ROS that directly suppress P. falciparum survival [49]. A fraction of field-caught A. gambiae individuals harbored an Enterococcus species in their intestine, and reactive oxygen species produced by these bacteria efficiently reduced Plasmodium invasion without the intervention of mosquito immune factors. It is amusing that the effect of the mosquito gut microbial communities on pathogenic infection is recognized just as similar phenomena in humans receive increasing attention [50]. A less direct effect, that is, bacterial stimulation of the mosquito immune system, is also recognized as an important factor in the anti-Plasmodium mosquito response. Mosquitoes rendered unable to activate the PGRPLC-dependent anti-bacterial defense were found to be more permissive to Plasmodium infections, and mosquito treatment with the antibiotic gentamycin had the same effect. Conversely, feeding the mosquitoes extra bacteria inhibited parasite development, unless PGRPLC was inactivated [51]. These results are in agreement with the fact that the same immune mechanisms are shared between the antibacterial and antiparasitic responses. Generally elevating the expression of immune defense genes by boosting the activity of NF-kB factors completely aborts parasite development in mosquitoes [52]. The authors suggested that basal immunity, the presence of immune factors at a baseline level in preparation for fighting a microbial infection as soon as it occurs, could actually be primed by former encounters with bacteria even during larval life. A mechanism for immune priming during adult mosquito life, involving immune memory mediated by bacteria-induced hemocyte differentiation, has been proposed [53]. Microsporidia, which are unicellular fungi, also have a negative impact on parasite development [54]. Somatic infection of mosquito tissue with intracellular bacteria of the Wolbachia genus also affect P. falciparum survival by stimulating the mosquito immune response, but vertical transmission of this potentially useful infection to the offspring seems difficult to achieve [55,56] though a similar approach was successful in the dengue fever mosquito Aedes aegypti [57]. Since Plasmodium is phylogenetically related to algae, it would be tempting to investigate whether mosquitoes respond to algal symbionts (often observed to colonize laboratory-grown larvae) with even greater adverse consequences on Plasmodium invasion. In summary, bacteria and other microorganisms that colonize mosquitoes influence Plasmodium success both by directly interfering with Plasmodium development and indirectly by activating the mosquito immune system. www.sciencedirect.com Author's personal copy Mosquito anti-Plasmodium response Marois 433 Potential applications As illustrated above, many aspects of mosquito biology are now understood to participate in reducing Plasmodium infection. How could these processes be exploited in intervention strategies to curb malaria incidence in endemic countries? Targeting the Plasmodium-transmitting mosquito populations on a continental scale, such as in Africa, is an immense task. First of all, mapping Plasmodium genetic susceptibility (which can vary geographically and seasonally) in field mosquito populations should orient interventions, such as insecticide treatment, against the most infectious mosquito subpopulations. Such mapping work is currently underway in several countries. If microorganisms identified to render mosquitoes resistant to Plasmodium can be harnessed to colonize wild mosquitoes, they could be introduced in breeding sites or via artificial adult feeders to target mosquito subpopulations that insecticides cannot reach. Much research is still required to define if any naturally-occurring microbe with a Plasmodium-killing potential can be co-opted in intervention strategies: how easy would it be to drive massive colonization of wild mosquitoes with the chosen organism? Would this organism be transmitted from larvae to adults, and from adults to offspring? Following this line, some laboratories are investigating the potential of entomophilic fungi [58], or of natural Anopheles gut bacteria [59] that can be genetically modified to express anti-Plasmodium compounds and might have a capacity to spread across mosquito populations, a technique known as paratransgenesis. The sterile insect technique (SIT), which consists in flooding a target population with laboratory-raised males rendered sterile by irradiation, could be used to reduce vector populations, and thus Plasmodium transmission to humans, in areas with an easy access from mosquito production units. Of note, the release of sterile females is not desirable since any female may participate in the transmission of parasites. Male mosquitoes genetically engineered to be sterile or to have sterile female progeny constitute an interesting alternative approach to irradiation for SIT, as illustrated in current field trials against dengue fever with transgenic Aedes aegypti mosquitoes [60,61]. Transgenic fluorescent sexing markers [62], once combined with genetic sterility, could facilitate the automated selection of very large numbers of sterile males at the larval stage. Unfortunately, SIT interventions are unrealistic in vast regions of high malaria incidence where human populations are scattered and remotely accessible. There, along with classical insecticide treatment and the continuous use of insecticide-impregnated bednets, strategies involving the release of transgenic mosquitoes engineered to be malaria-refractory are an interesting alternative. Constant progress in the understanding of Plasmodium resistance in the mosquito will certainly lead to the design of genetically fully refractory mosquitoes in the near future, with encouraging steps in this direction www.sciencedirect.com [63,64]. Successful transgenes may exploit immune molecules such as components of the TEP1 pathway, boost ROS or NO production [44], or encode synthetic antiPlasmodium compounds derived from exogenous biomolecules [65,66]. Each proposed refractoriness-promoting synthetic construct should be subjected to careful risk assessment: is the increase in mosquito immunity specific to Plasmodium or might GM mosquitoes undesirably proliferate due to increased general resistance to their natural pathogens? Is resistance to viral pathogens altered? Could novel Plasmodium strains arise under the selective pressure of the transgene? The next challenge will be to drive those successful modifications into wild mosquito populations. ‘Gene drive systems’, based on selfish genetic elements such as homing endonuclease genes [67] or on dual expression of toxin/antidote [68,69] are under development to force the spread of desired transgenes into entire target populations, and have already yielded promising results in cage trials [67]. Computer modeling shows that theoretical drive systems may be fine-tuned to control the dynamics of transgene spread, potentially offering the possibility to choose between generalized transgene spread or programmed transgene extinction [68,69]. Bringing together laboratories that develop these powerful genetic tools and laboratories that identify efficient refractorinesspromoting genetic constructs will be key to implementing these strategies, which have the potential to bring a major contribution in the multifactorial fight against the millennial scourge. Acknowledgements The author thanks Elena Levashina for constant support and Stéphanie Blandin for critical reading of the manuscript. Work in our laboratory is funded by INSERM, CNRS, The University of Strasbourg and the following European FP7 programmes: EviMalar (Networks of Excellence), MALVECBLOK (Collaborative projects), INFRAVEC (Infrastructures). References and recommended reading Papers of particular interest, published within the period of review, have been highlighted as: of special interest of outstanding interest 1. Hogg JC, Hurd H: The effects of natural Plasmodium falciparum infection on the fecundity and mortality of Anopheles gambiae s. l. in north east Tanzania. Parasitology 1997, 114(Pt 4):325-331. 2. Anderson RA, Knols BG, Koella JC: Plasmodium falciparum sporozoites increase feeding-associated mortality of their mosquito hosts Anopheles gambiae s.l. Parasitology 2000, 120(Pt 4):329-333. 3. Ahmed AM, Maingon R, Romans P, Hurd H: Effects of malaria infection on vitellogenesis in Anopheles gambiae during two gonotrophic cycles. Insect Mol Biol 2001, 10:347-356. 4. Ahmed AM, Hurd H: Immune stimulation and malaria infection impose reproductive costs in Anopheles gambiae via follicular apoptosis. Microbes Infect 2006, 8:308-315. 5. Ferguson HM, Read AF: Why is the effect of malaria parasites on mosquito survival still unresolved? Trends Parasitol 2002, 18:256-261. Current Opinion in Microbiology 2011, 14:429–435 Author's personal copy 434 Host-microbe interactions: Parasites 6. Service MW: Mosquitoes (Culicidae). In Medical Insects and Arachnids. Edited by Lane RP, Crosskey RW. Chapman & Hall; 1993:120-140. 7. Dessens JT, Beetsma AL, Dimopoulos G, Wengelnik K, Crisanti A, Kafatos FC, Sinden RE: CTRP is essential for mosquito infection by malaria ookinetes. EMBO J 1999, 18:6221-6227. 8. 9. Dessens JT, Siden-Kiamos I, Mendoza J, Mahairaki V, Khater E, Vlachou D, Xu XJ, Kafatos FC, Louis C, Dimopoulos G et al.: SOAP, a novel malaria ookinete protein involved in mosquito midgut invasion and oocyst development. Mol Microbiol 2003, 49:319-329. Kadota K, Ishino T, Matsuyama T, Chinzei Y, Yuda M: Essential role of membrane-attack protein in malarial transmission to mosquito host. Proc Natl Acad Sci USA 2004, 101:16310-16315. 10. Schneider D, Shahabuddin M: Malaria parasite development in a Drosophila model. Science 2000, 288:2376-2379. 11. Brandt SM, Jaramillo-Gutierrez G, Kumar S, Barillas-Mury C, Schneider DS: Use of a Drosophila model to identify genes regulating Plasmodium growth in the mosquito. Genetics 2008, 180:1671-1678. 12. Habtewold T, Povelones M, Blagborough AM, Christophides GK: Transmission blocking immunity in the malaria non-vector mosquito Anopheles quadriannulatus species A. PLoS Pathog 2008, 4:. 13. Collins FH, Sakai RK, Vernick KD, Paskewitz S, Seeley DC, Miller LH, Collins WE, Campbell CC, Gwadz RW: Genetic selection of a Plasmodium-refractory strain of the malaria vector Anopheles gambiae. Science 1986, 234:607-610. 14. Blandin S, Moita LF, Köcher T, Wilm M, Kafatos FC, Levashina EA: Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO Rep 2002, 3:852-856. 15. Catteruccia F, Levashina EA: RNAi in the malaria vector, Anopheles gambiae. Methods Mol Biol 2009, 555:63-75. 16. Cirimotich CM, Dong Y, Garver LS, Sim S, Dimopoulos G: Mosquito immune defenses against Plasmodium infection. Dev Comp Immunol 2010, 34:387-395. 17. Blandin S, Shiao SH, Moita LF, Janse CJ, Waters AP, Kafatos FC, Levashina EA: Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 2004, 116:661-670. basis of resistance to malaria parasites in Anopheles gambiae. Science 2009, 326:147-150. The strength of various TEP1 alleles in mediating Plasmodium resistance is finely measured by performing allele-specific RNAi in the progeny of crosses that generate different allele combinations. 24. Dong Y, Aguilar R, Xi Z, Warr E, Mongin E, Dimopoulos G: Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog 2006, 2:e52. 25. Garver LS, Dong Y, Dimopoulos G: Caspar controls resistance to Plasmodium falciparum in diverse anopheline species. PLoS Pathog 2009, 5:. 26. White BJ, Lawniczak MK, Cheng C, Coulibaly MB, Wilson MD, Sagnon N, Costantini C, Simard F, Christophides GK, Besansky NJ: Adaptive divergence between incipient species of Anopheles gambiae increases resistance to Plasmodium. Proc Natl Acad Sci USA 2011, 108:244-249. 27. Cohuet A, Osta MA, Morlais I, Awono-Ambene PH, Michel K, Simard F, Christophides GK, Fontenille D, Kafatos FC: Anopheles and Plasmodium: from laboratory models to natural systems in the field. EMBO Rep 2006, 7:1285-1289. 28. Mitri C, Jacques JC, Thiery I, Riehle MM, Xu J, Bischoff E, Morlais I, Nsango SE, Vernick KD, Bourgouin C: Fine pathogen discrimination within the APL1 gene family protects Anopheles gambiae against human and rodent malaria species. PLoS Pathog 2009, 5:e1000576. 29. Baxter RH, Steinert S, Chelliah Y, Volohonsky G, Levashina EA, Deisenhofer J: A heterodimeric complex of the LRR proteins LRIM1 and APL1C regulates complement-like immunity in Anopheles gambiae. Proc Natl Acad Sci USA 2010, 107: 16817-16822. Crystal structure of the LRIM1/APL1C protein complex and molecular details of their interaction. The in vitro interaction of this complex with a series of TEP1 forms is examined using gel filtration. The TEP1 convertase hypothesis is described. 30. Povelones M, Upton LM, Sala KA, Christophides GK: Structurefunction analysis of the Anopheles gambiae LRIM1/APL1C complex and its interaction with complement C3-like protein TEP1. PLoS Pathog 2011, 7:e1002023. 31. Waterhouse RM, Povelones M, Christophides GK: Sequencestructure-function relations of the mosquito leucine-rich repeat immune proteins. BMC Genomics 2010, 11:531. 32. Blandin SA, Marois E, Levashina EA: Antimalarial responses in Anopheles gambiae: from a complement-like protein to a complement-like pathway. Cell Host Microbe 2008, 3:364-374. 18. Shiao SH, Whitten MM, Zachary D, Hoffmann JA, Levashina EA: Fz2 and cdc42 mediate melanization and actin polymerization but are dispensable for Plasmodium killing in the mosquito midgut. PLoS Pathog 2006, 2:e133. 33. Vlachou D, Schlegelmilch T, Christophides GK, Kafatos FC: Functional genomic analysis of midgut epithelial responses in Anopheles during Plasmodium invasion. Curr Biol 2005, 15:1185-1195. 19. Fraiture M, Baxter RH, Steinert S, Chelliah Y, Frolet C, Quispe Tintaya W, Hoffmann JA, Blandin SA, Levashina EA: Two mosquito LRR proteins function as complement control factors in the TEP1-mediated killing of Plasmodium. Cell Host Microbe 2009, 5:273-284. RNAi knockdowns reveal the coordinated action and TEP1-stabilizing function of the LRIM1 and APL1C proteins. Precocious TEP1 deposition on mosquito tissue is observed in their absence. 34. Rono MK, Whitten MM, Oulad-Abdelghani M, Levashina EA, Marois E: The major yolk protein vitellogenin interferes with the anti-plasmodium response in the malaria mosquito Anopheles gambiae. PLoS Biol 2010, 8:e1000434. Yolk precursor proteins are shown to dampen the efficiency of the TEP1mediated mosquito anti-Plasmodium response and the NF-kB pathway is found to regulate reproduction and immunity in opposite manners. 20. Povelones M, Waterhouse RM, Kafatos FC, Christophides GK: Leucine-rich repeat protein complex activates mosquito complement in defense against Plasmodium parasites. Science 2009, 324:258-261. 35. Kumar S, Christophides GK, Cantera R, Charles B, Han YS, Meister S, Dimopoulos G, Kafatos FC, Barillas-Mury C: The role of reactive oxygen species on Plasmodium melanotic encapsulation in Anopheles gambiae. Proc Natl Acad Sci USA 2003, 100:14139-14144. 21. Riehle MM, Markianos K, Niare O, Xu J, Li J, Toure AM, Podiougou B, Oduol F, Diawara S, Diallo M et al.: Natural malaria infection in Anopheles gambiae is regulated by a single genomic control region. Science 2006, 312:577-579. 36. Oliveira JH, Goncalves RL, Oliveira GA, Oliveira PL, Oliveira MF, Barillas-Mury C: Energy metabolism affects susceptibility of Anopheles gambiae mosquitoes to Plasmodium infection. Insect Biochem Mol Biol 2011, 41:349-355. 22. Riehle MM, Xu J, Lazzaro BP, Rottschaefer SM, Coulibaly B, Sacko M, Niare O, Morlais I, Traore SF, Vernick KD: Anopheles gambiae APL1 is a family of variable LRR proteins required for Rel1-mediated protection from the malaria parasite, Plasmodium berghei. PLoS ONE 2008, 3:. 37. Molina-Cruz A, DeJong RJ, Charles B, Gupta L, Kumar S, Jaramillo-Gutierrez G, Barillas-Mury C: Reactive oxygen species modulate Anopheles gambiae immunity against bacteria and Plasmodium. J Biol Chem 2008, 283:3217-3223. The expression of ROS detoxifying enzymes in the midgut and fat body is studied during Plasmodium infection compared to the uninfected blood meal. Knockdown of these enzymes and dietary supplementation with antioxidants reveal their role in mosquito immunity. 23. Blandin SA, Wang-Sattler R, Lamacchia M, Gagneur J, Lycett G, Ning Y, Levashina EA, Steinmetz LM: Dissecting the genetic Current Opinion in Microbiology 2011, 14:429–435 www.sciencedirect.com Author's personal copy Mosquito anti-Plasmodium response Marois 435 38. Jaramillo-Gutierrez G, Molina-Cruz A, Kumar S, Barillas-Mury C: The Anopheles gambiae oxidation resistance 1 (OXR1) gene regulates expression of enzymes that detoxify reactive oxygen species. PLoS ONE 2010, 5:e11168. 39. DeJong RJ, Miller LM, Molina-Cruz A, Gupta L, Kumar S, BarillasMury C: Reactive oxygen species detoxification by catalase is a major determinant of fecundity in the mosquito Anopheles gambiae. Proc Natl Acad Sci USA 2007, 104:2121-2126. 40. Lim J, Gowda DC, Krishnegowda G, Luckhart S: Induction of nitric oxide synthase in Anopheles stephensi by Plasmodium falciparum: mechanism of signaling and the role of parasite glycosylphosphatidylinositols. Infect Immun 2005, 73:2778-2789. 41. Akman-Anderson L, Olivier M, Luckhart S: Induction of nitric oxide synthase and activation of signaling proteins in Anopheles mosquitoes by the malaria pigment, hemozoin. Infect Immun 2007, 75:4012-4019. 42. Luckhart S, Vodovotz Y, Cui L, Rosenberg R: The mosquito Anopheles stephensi limits malaria parasite development with inducible synthesis of nitric oxide. Proc Natl Acad Sci USA 1998, 95:5700-5705. 43. Han YS, Barillas-Mury C: Implications of Time Bomb model of ookinete invasion of midgut cells. Insect Biochem Mol Biol 2002, 32:1311-1316. 44. Ali M, Al-Olayan EM, Lewis S, Matthews H, Hurd H: Naturally occurring triggers that induce apoptosis-like programmed cell death in Plasmodium berghei ookinetes. PLoS ONE 2010, 5:. 45. Gupta L, Molina-Cruz A, Kumar S, Rodrigues J, Dixit R, Zamora RE, Barillas-Mury C: The STAT pathway mediates latephase immunity against Plasmodium in the mosquito Anopheles gambiae. Cell Host Microbe 2009, 5:498-507. Careful examination of the involvement of the STAT pathway in Plasmodium resistance, found to be mediated by NOS expression. An unexpected dual role of NOS is reported: one favoring ookinete midgut invasion, one mediated the killing of young oocysts. 46. Kumar S, Molina-Cruz A, Gupta L, Rodrigues J, Barillas-Mury C: A peroxidase/dual oxidase system modulates midgut epithelial immunity in Anopheles gambiae. Science 2010, 327:1644-1648. The formation of a dityrosine-crosslinked protein network in addition to the peritrophic matrix, controlled by a dual oxidase and a peroxidase, is described along with its importance in limiting immune signaling in the midgut – conferring protection for the gut flora, the gut epithelial cells and Plasmodium. 47. Sinden RE: Plasmodium differentiation in the mosquito. Parassitologia 1999, 41:139-148. 48. Pinto SB, Kafatos FC, Michel K: The parasite invasion marker SRPN6 reduces sporozoite numbers in salivary glands of Anopheles gambiae. Cell Microbiol 2008, 10:891-898. 49. Cirimotich CM, Dong Y, Clayton AM, Sandiford SL, Souza Neto JA, Mulenga M, Dimopoulos G: Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 2011, 332:855-858. Evidence is presented for the direct anti-Plasmodium activity of ROS secreted by certain midgut bacteria. 50. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM et al.: Enterotypes of the human gut microbiome. Nature 2011, 473:174-180. 51. Meister S, Agianian B, Turlure F, Relogio A, Morlais I, Kafatos FC, Christophides GK: Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathog 2009, 5:e1000542. Detailed characterization of the role of PGRPLC in mosquito resistance against bacterial infections. The PGRPLC-mediated antibacterial immunity is shown to affect the development of both Plasmodium berghei and falciparum. 52. Frolet C, Thoma M, Blandin S, Hoffmann JA, Levashina EA: Boosting NF-kappaB-dependent basal immunity of Anopheles www.sciencedirect.com gambiae aborts development of Plasmodium berghei. Immunity 2006, 25:677-685. 53. Rodrigues J, Brayner FA, Alves LC, Dixit R, Barillas-Mury C: Hemocyte differentiation mediates innate immune memory in Anopheles gambiae mosquitoes. Science 2010, 329:1353-1355. 54. Bargielowski I, Koella JC: A possible mechanism for the suppression of Plasmodium berghei development in the mosquito Anopheles gambiae by the microsporidian Vavraia culicis. PLoS ONE 2009, 4:e4676. 55. Kambris Z, Blagborough AM, Pinto SB, Blagrove MS, Godfray HC, Sinden RE, Sinkins SP: Wolbachia stimulates immune gene expression and inhibits plasmodium development in Anopheles gambiae. PLoS Pathog 2010, 6:. 56. Hughes GL, Koga R, Xue P, Fukatsu T, Rasgon JL: Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in anopheles gambiae. PLoS Pathog 2011, 7:e1002043. 57. McMeniman CJ, O’Neill SL: A virulent Wolbachia infection decreases the viability of the dengue vector Aedes aegypti during periods of embryonic quiescence. PLoS Negl Trop Dis 2010, 4:e748. 58. Fang W, Vega-Rodriguez J, Ghosh AK, Jacobs-Lorena M, Kang A, St Leger RJ: Development of transgenic fungi that kill human malaria parasites in mosquitoes. Science 2011, 331:1074-1077. 59. Bisi DC, Lampe DJ: Secretion of anti-plasmodium effector proteins from a natural isolate pantoea agglomerans using pelb and hlya secretion signals. Appl Environ Microbiol 2011, 77:4669-4675. 60. Wise de Valdez MR, Nimmo D, Betz J, Gong HF, James AA, Alphey L, Black WCt: Genetic elimination of dengue vector mosquitoes. Proc Natl Acad Sci USA 2011, 108:4772-4775. 61. Subbaraman N: Science snipes at Oxitec transgenic-mosquito trial. Nat Biotechnol 2011, 29:9-11. 62. Catteruccia F, Benton JP, Crisanti A: An Anopheles transgenic sexing strain for vector control. Nat Biotechnol 2005, 23:1414-1417. 63. Moreira LA, Ito J, Ghosh A, Devenport M, Zieler H, Abraham EG, Crisanti A, Nolan T, Catteruccia F, Jacobs-Lorena M: Bee venom phospholipase inhibits malaria parasite development in transgenic mosquitoes. J Biol Chem 2002, 277:40839-40843. 64. Ito J, Ghosh A, Moreira LA, Wimmer EA, Jacobs-Lorena M: Transgenic anopheline mosquitoes impaired in transmission of a malaria parasite. Nature 2002, 417:452-455. 65. Isaacs AT, Li F, Jasinskiene N, Chen X, Nirmala X, Marinotti O, Vinetz JM, James AA: Engineered resistance to Plasmodium falciparum development in transgenic Anopheles stephensi. PLoS Pathog 2011, 7:e1002017. 66. Li C, Marrelli MT, Yan G, Jacobs-Lorena M: Fitness of transgenic Anopheles stephensi mosquitoes expressing the SM1 peptide under the control of a vitellogenin promoter. J Hered 2008, 99:275-282. 67. Windbichler N, Menichelli M, Papathanos PA, Thyme SB, Li H, Ulge UY, Hovde BT, Baker D, Monnat RJ Jr, Burt A et al.: A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature 2011, 473:212-215. Proof-of-principle that homing endonucleases can be employed in gene drive systems in transgenic Anopheles. 68. Marshall JM, Hay BA: Inverse Medea as a novel gene drive system for local population replacement: a theoretical analysis. J Hered 2011, 102:336-341. 69. Marshall JM, Pittman GW, Buchman AB, Hay BA: Semele: a killer-male, rescue-female system for suppression and replacement of insect disease vector populations. Genetics 2011, 187:535-551. Modeling of theoretical gene drive systems. Current Opinion in Microbiology 2011, 14:429–435 Article 7: 2012 Malaria Journal High-­throughput sorting of mosquito larvae for laboratory studies and for future vector control interventions. Marois E, Scali C, Soichot J, Kappler C, Levashina EA, Catteruccia F. Marois et al. Malaria Journal 2012, 11:302 http://www.malariajournal.com/content/11/1/302 METHODOLOGY Open Access High-throughput sorting of mosquito larvae for laboratory studies and for future vector control interventions Eric Marois1, Christina Scali2, Julien Soichot1, Christine Kappler1, Elena A Levashina1,3*† and Flaminia Catteruccia4,5*† Abstract Background: Mosquito transgenesis offers new promises for the genetic control of vector-borne infectious diseases such as malaria and dengue fever. Genetic control strategies require the release of large number of male mosquitoes into field populations, whether they are based on the use of sterile males (sterile insect technique, SIT) or on introducing genetic traits conferring refractoriness to disease transmission (population replacement). However, the current absence of high-throughput techniques for sorting different mosquito populations impairs the application of these control measures. Methods: A method was developed to generate large mosquito populations of the desired sex and genotype. This method combines flow cytometry and the use of Anopheles gambiae transgenic lines that differentially express fluorescent markers in males and females. Results: Fluorescence-assisted sorting allowed single-step isolation of homozygous transgenic mosquitoes from a mixed population. This method was also used to select wild-type males only with high efficiency and accuracy, a highly desirable tool for genetic control strategies where the release of transgenic individuals may be problematic. Importantly, sorted males showed normal mating ability compared to their unsorted brothers. Conclusions: The developed method will greatly facilitate both laboratory studies of mosquito vectorial capacity requiring high-throughput approaches and future field interventions in the fight against infectious disease vectors. Keywords: Malaria, Mosquito, Transgenesis, Fluorescence-assisted sorting, Sexing, Anopheles gambiae, piggyBac Background Vector-borne infectious diseases are a major scourge for humanity. Malaria alone, caused by Plasmodium parasites transmitted by the bite of infected female Anopheles mosquitoes, annually infects 250 million people worldwide and kills close to one million, mostly children in sub-Saharan Africa [1]. Past attempts at curbing the disease by the massive use of insecticides and of insecticideimpregnated bed nets have promoted the spread of genetic resistance to a wide range of insecticides across mosquito populations. This is reducing the impact of * Correspondence: Levashina@mpiib-berlin.mpg.de; flacat@unipg.it † Equal contributors 1 Institut de Biologie Moléculaire et Cellulaire, INSERM U963, CNRS UPR9022, 15 rue René Descartes, 67084, Strasbourg, France 3 Department of Vector Biology, Max-Planck Institute for Infection Biology, Chariteplatz 1, 10117, Berlin, Germany Full list of author information is available at the end of the article insecticide-based control methods, and novel approaches to control vector populations are urgently needed to roll back the disease. A similar challenge is posed by the control of Aedes mosquito species transmitting viral diseases including yellow fever, dengue and chikungunya. Vector control methods that specifically target the desired species represent a valid and environmentally friendly alternative to insecticides. Dating from the 1950s, the sterile insect technique (SIT) is a species-specific control strategy that has been successfully used to reduce the population size of insect pests such as the screw worm and the Mediterranean fruit fly [2]. For mosquito SIT ([3]; reviewed in [4,5]), large numbers of male insects must be produced in breeding facilities, sterilized by γ-ray irradiation, and released in the field to compete with wild males for mating with wild females. The SIT approach is well suited for Anopheles mosquitoes, as the majority of © 2012 Marois et al.; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Marois et al. Malaria Journal 2012, 11:302 http://www.malariajournal.com/content/11/1/302 females mate once in their lifetime and use the sperm stored in their spermatheca to fertilize egg batches produced every time they take a blood meal [6]. This implies that if a female copulates with a sterile male, she will not mate again and will lay only infertile eggs. Repeated releases of sterile males over a given area would, therefore, achieve a local drop in vector populations and a consequent decrease in malaria transmission. However, labourintensive procedures for selection of male-only populations and a large variability in the efficiency of sterilization have to date posed vast blocks for a massive application of this approach. Since the turn of the century, vast progress has been made in the generation of molecular and genetic tools for studies on Anopheles mosquitoes. The sequencing of the Anopheles gambiae genome [7] has allowed the identification of thousands of genes that shape the biology and behaviour of this main malaria vector. These genomic advances, combined with the development of transgenic technologies [8] to modify the mosquito genome and the possibility of silencing gene expression through RNAi [9], are facilitating studies on the biological processes that are crucial to the ability of Anopheles mosquitoes to transmit malaria. Importantly, they also offer a basis for novel vector control strategies to complement insecticide treatments. Among the advocated novel approaches are genetic control strategies related to SIT, such as the release of insects carrying a dominant lethal (RIDL), that are currently being developed for the dengue vector, Aedes aegypti [10,11]. In RIDL, released male mosquitoes carry a transgene that makes their female progeny unviable or infertile. Further alternative strategies propose to alter the genetic make-up of mosquito populations to reduce their vectorial capacity traits, for instance by rendering mosquitoes resistant to human pathogens [12-14]. The introgression of natural genetic traits or of synthetic transgenic constructs interfering with the mosquito vectorial capacity would lead to the replacement of natural diseasetransmitting populations with mosquitoes that do not transmit infectious agents, a process that could be enhanced by artificial gene drive systems [15,16]. Regardless of the final goal of the control programme (population eradication through SIT/RIDL or population replacement), when targeting mosquito vectors male-only populations must be released, since females may (i) contribute to an unwanted increase of the mosquito populations if not sterile, (ii) mate with the released males thereby reducing the efficacy of the trial, and (iii) participate in disease transmission. This underlines the need for highthroughput sexing tools for mosquitoes: males carrying the desired sterility or disease-resistance trait need to be produced on an industrial scale to reach the vast numbers (millions) necessary for a release programme. It is essential that the released males, whether sterile or not, are fully Page 2 of 9 competitive for mating with field females. Therefore, the mechanism utilized for inducing sterility/refractoriness, the mass rearing conditions and the sex sorting procedures must have no impact on overall male fitness. Proof-of-principle evidence that automated separation of the sexes is achievable in A. gambiae mosquitoes was previously provided using a flow cytometry machine (COPASW, Union Biometrica) to separate males from females based on the expression of a sperm-specific fluorescence marker in the testis [17]. The usefulness of this system was limited by the fact that male individuals could be identified only during late larval stages, at the onset of sperm development. Here, the full potential of this high-throughput system is achieved by the use of early sex-specific transgenic markers. This system allows the efficient and fully accurate separation of the desired phenotypes at early stages of development. Large larval populations can be sorted into populations of males/females; transgenics/non-transgenics, heterozygous/ homozygous, transgenic females/non-transgenic males. Further, the property of the system to quantify transgene copy number offers a new approach for mosquito sexing based on X-linked insertions at any stage of larval development. The sorting procedure has no impact on the mating ability of the resulting adult males. The system, here tested on A. gambiae mosquitoes, could be easily adapted to all mosquito species that are amenable to transgenesis. Methods Mosquito strains and rearing Anopheles gambiae mosquitoes (G3 and Ngousso strains and their transgenic derivatives) were maintained at 28°C and 70-80% humidity in a 12/12 h day/night cycle. Anesthetized CD1 mice were used for blood feeding and larvae were fed finely ground Tetra Goldfish food (Tetra, Germany). The DSX transgenic strain, obtained in the G3 background, has been previously described [18]. The FK transgenesis plasmid was assembled by recombining three entry plasmids and one piggyBac destination plasmid using three-fragment Multisite Gateway cloning (Invitrogen) according to the manufacturer’s instructions. The resulting construct contained the attB1, attB2, attB3 and attB4 Gateway seam sites that delimit each of the three cassettes contributed by the entry plasmids (for the full annotated construct sequence refer to Additional file 1). The Vitellogenin (AGAP004203) promoter was amplified from A. gambiae genomic DNA using the following primers: 5’-TGACCTCGAGTTCAACTCGACC-3’ and 5’-GATATCGATGGTTCGGTTGTTCGCAGTTG-3’. The amplified fragment was cloned into Xho I and Cla I restriction sites of the YFP-containing entry vector. The AGAP002620 promoter region was amplified using primers: 5-CCGTCTAGACCGGGCTCTACAAAGTC-3’ and 5’-CAGCTCTCGAGCAGGAGGATCGTT-3’ and cloned Marois et al. Malaria Journal 2012, 11:302 http://www.malariajournal.com/content/11/1/302 as an Xba I - Xho I fragment into the tdTomato-containing entry vector. Embryos (n = 120) of the Ngousso strain were injected with a 200 ng/μl solution of the transgenesis plasmid and 20 surviving adults were back-crossed to Ngousso mosquitoes. A single transgenic mosquito male was recovered from the back-cross progeny. Further genetic crosses revealed that the transgene insertion was X-linked. The piggyBac insertion was mapped by inverse PCR as follows: 500 ng of genomic DNA were digested with Sau3AI or cocktails of blunt end restriction enzymes (ScaI HincII, DraI, SmaI PvuII, Fermentas), and re-ligated with T4 DNA ligase (Fermentas) in a final volume of 500 μl. The sample was ethanol-precipitated, resuspended in 20 μl water, of which 2 μl were subjected to PCR. The piggyBac 5’ border of the insertion site was mapped by sequencing a product amplified with primers 5’-TGCACAGCGACGGATTCGCGCTATT-3’ and 5’-AGGACATCTCAGTCGCCGCTTGGA-3’, followed by nested PCR with 5’-CGCGCTATTTAGAAAGAGA GAG-3’ and 5’-GAACTATAACGACCGCGTGAGTC-3’; or with 5’-GAACTATAACGACCGCGTGAGTC-3’ and 5’CAGTGACACTTACCGCATTGACA-3’. The piggyBac 3’ border of the insertion site was mapped by sequencing the product amplified with primers 5’-CGAGGTTTATTT ATTAATTTGAATAGATATTAAG-3’ and 5’-CGATATA CAGACCGATAAAACACATGCGT-3’, followed by nested PCR with 5’-GCGTCAATTTTACGCATGATTATCTTT3’ and 5’-ATTTACACTTACATACTAATAATAAATT CAAC-3’. Amplified fragments were compared by BLAST to the Anopheles gambiae genome (VectorBase). The transposon insertion was mapped within a 232-base pair repeated element on the X chromosome. This short repeated element is broadly distributed in the genome, but at the position X: 22463464 is present in the Ngousso strain and absent from the G3 and PEST strains. The transgenic construct carried the EGFP gene under the control of a 3xP3 promoter [19] as a transgenesis selection marker, and two additional reporter genes: (i) YFPvenus [20] under the control of the A. gambiae Vitellogenin (AGAP004203) promoter and (ii) tdTomato [21] under the control of the AGAP002620 gene promoter [22]. The detailed characterization of these additional reporter constructs will be described elsewhere. Mosquitoes were reared and blood-fed on anesthetized mice in compliance with French and European laws on animal house procedures (agreement #E67-482-2 of the Direction of Veterinary services of the French Ministry of Agriculture). COPAS-assisted larval sorting The blood-fed mosquitoes (two to four days after a blood meal) were offered an egg dish made of a conical, 90 mm diameter, hardened filter paper (#50 Whatman, Page 3 of 9 GE Healthcare, UK) dipped in a glass bowl filled with water. Larvae were allowed to hatch directly in the egg dish and were recovered from water leaving most of the empty eggshells on the filter paper. Larvae were transferred to the reservoir of a Complex Object Parametric Analyzer and Sorter (COPAS) large particles flow cytometry SELECT instrument (Union Biometrica, Holliston, MA, USA), and analysed with the Biosort5281 software. For laser-assisted sorting, 488 or 514 nm emission filters were used indifferently with the following acquisition parameters: Green PMT 500, Yellow PMT 500, Red PMT 600, Delay 8; Width 6, pure mode selection with superdrops. Detection thresholds were set to 100 (signal) and 150 (time of flight). “Sheath” pressure (demineralized water was used instead of Sheath medium) was kept at a value of around 3, sorter pressure 3.3, while sample pressure varied between 3 and 4.5 depending on the density of the larvae. The flow rate was kept below 18 detected objects per second. Selected larvae were collected into a Petri dish placed underneath the flow cell outlet. Though result diagrams can be directly viewed in the COPASW software, the LMD files generated by the COPAS analysis were imported in the WinMDI freeware for data analyses and diagram generation. Fluorescence microscopy Small larvae were spotted in a drop of water in the wells of a CEL-LINE teflon-coated, 24-well diagnostic microscope slide (Erie Scientific, Menzel GmbH, Braunschweig; Germany) and observed with the 5x objective of an Axiovert 200 M Zeiss fluorescence microscope. To immobilize larvae for photography, an anaesthetics solution of final concentration 5% tricaine and 0.5% tetramizole was added to the water 15 min before observation. Results Rapid and precise establishment of homozygous transgenic Anopheles gambiae lines by COPAS sorting The goal of this work was to evaluate the possibility of performing accurate, fast and high-throughput larval screening and sorting using a flow cytometry machine, the Complex Object Parametric Analyzer and Sorter (COPASW, Union Biometrica). To this end, the DSX:: EGFP transgenic line (thereafter, DSX) [18] was used, as it comprises a combination of fluorescent markers that allow the differentiation between male and female larvae as early as the first instar stage (based on higher expression levels of an EGFP reporter gene in the male midgut), and between heterozygous and homozygous individuals (based on higher expression levels of the selectable DsRed marker in the central nervous system of homozygotes) (Figure 1). To test the efficacy and sensitivity of the COPAS machine to sort different classes of larvae, Marois et al. Malaria Journal 2012, 11:302 http://www.malariajournal.com/content/11/1/302 Page 4 of 9 Figure 1 Sex-specific expression of the Dsx-GFP transgene. A mix of wild-type and heterozygous DSX first instar larvae observed under a fluorescence microscope with a 5x objective. Red fluorescence (upper panel, left) denotes larvae carrying the DsRed transgene (the upper larva shows its dorsal side, the four additional red larvae show their ventral side). Green fluorescence intensity (upper panel, right) allows distinguishing between females (less bright, arrowheads) and males (showing stronger fluorescence). Lower panels: left, transmission image; right; overlay of images on upper panels. non-transgenic mosquitoes were crossed to DSX mosquitoes and the resulting F2 larvae were analysed. Such progeny was expected to segregate into five fluorescence classes according to single gene Mendelian inheritance and sex-specific expression of the GFP marker: 12.5% of homozygous transgenic females, 12.5% of homozygous transgenic males, 25% of heterozygous females, 25% of heterozygous males, and 25% of homozygous nontransgenic males and females (for which no fluorescencebased sex separation was possible). First instar larvae were COPAS-analysed and the number and ratio of larval classes were quantified by COPAS software. First, the area corresponding to live larvae was determined using light extinction and time of flight parameters. Signals beyond this area were identified by microscopy as eggshells and debris from the larval breeding water. The selected area was next analysed by fluorescence and the five expected larval classes were identified (Figure 2). Out of 4,036 larvae screened, 493 (12.2%) showed high green (EGFP-positive) and high red (DsRed-positive) fluorescence (homozygous transgenic males), 520 (12.9%) were low green and high red (homozygous transgenic females), 1,003 (24.9%) were high green and low red (heterozygous transgenic males), 958 (23.7%) were low green and low red (heterozygous transgenic females) and 1,062 (26.3%) were negative for any fluorescence (Figure 2). These values are consistent with the expected percentages (goodness of fit χ2 =6.072, p = 0.19). About 500 larvae from each of the four transgenic populations were sorted separately (in less than 30 min) and the accuracy of sex sorting was verified by visual examination of the resulting adults. All individuals were of the predicted sex. Next, male and female homozygous transgenic adults obtained from the sorting process were crossed and their progeny was screened (again using the COPAS instrument) to search for potential heterozygous individuals arising from inefficient sorting of homozygotes. Heterozygotes were absent, suggesting that the initial sorting of homozygous individuals by COPAS was 100% efficient, thereby permitting the establishment of a homozygous transgenic mosquito line in one generation. These results demonstrate the potential of the COPAS machine for compensating the lack of balancer chromosomes in mosquitoes for selection and maintenance of a desired transgenic genotype, thereby permitting a major gain in time and precision. COPAS-assisted high-throughput sexing of early mosquito larvae To test the efficiency and reliability of sorting single-sex mosquito populations, first instar homozygous DSX larvae were analysed with the COPAS machine, which detected two distinct populations of larvae displaying high and low levels of green fluorescence that were expected to correspond to male and female populations, respectively (Figure 3A). After sorting, 2,000 larvae of each population were raised separately to the pupal stage and resulting pupae Marois et al. Malaria Journal 2012, 11:302 http://www.malariajournal.com/content/11/1/302 Page 5 of 9 Figure 2 COPAS-assisted analysis of larval populations. The progeny of DSX/+ mosquitoes, containing non-transgenic as well as heterozygous and homozygous transgenic mosquitoes, was analysed by COPAS. The left diagram (Extinction vs Time of Flight) shows all detected objects. Mosquito larvae were empirically determined to be located inside the gated area (R1). The right diagram (red vs green fluorescence) decomposes the larval population into five categories: non-transgenic (+/+, R6), heterozygous females (XX; DSX/+, R5), heterozygous males (XY; DSX/+, R4), homozygous females (XX; DSX, R3), homozygous males (XY; DSX, R2), as indicated. were placed in two different cages. Careful and repeated visual examination of the adults emerged in each cage confirmed that all adults from the larvae displaying high GFP fluorescence were males and all adults from the larvae showing low GFP fluorescence were females (Figure 3B). In an independent experiment, groups of 150 larvae were recovered in four successive passages of the same individuals through the COPAS machine, sorting for male and for female larvae successively. These larvae were raised to adulthood and the sex of each mosquito verified. Again, visual examination confirmed the 100% accuracy of sorting at each of the four passages; none of the sorted groups contained an adult of the wrong sex. Importantly, the number of larvae from the fourth sorting that developed to adulthood (>100 individuals) was similar to the number of larvae that underwent 0, 1, 2 or 3 COPAS treatments (Table 1). These results suggest that the survival rate of COPAS-sorted larvae was not affected by the sorting procedure itself repeated up to three times. COPAS sorting does not impair male mating competitiveness The next step was to examine whether COPAS treatment negatively affected fitness and reproductive capability of sorted male insects. To this end, the reproductive success of males raised in standard conditions was compared to that of males that had passed three times through the COPAS machine. Two crosses were assembled in separate cages. Each cage contained 100 non-fluorescent, wild-type virgin females, 100 non-fluorescent competitor males, and 100 DSX transgenic males that were either untreated or had been sorted three times with COPAS at the early larval stage. Freshly emerged mosquitoes of each kind were simultaneously placed in the two cages and kept together for five days. Females received a blood meal on day 5, and on day 8 they were isolated into single plastic tubes to oviposit on shallow water. Freshly hatched larvae from individual females were examined under the fluorescence microscope to score the identity of their father. No significant difference was detected in the number of progeny fathered by the COPAS-treated vs control DSX males (Table 2), suggesting that COPAS sorting does not impair male competitiveness. Note that these experiments revealed a significant proportion of females fertilized by more than one male (15 progenies, out of 80 analysed, arose from females inseminated by at least two males of different genotypes, suggesting an overall rate of multiple matings in these experiments of at least 37.5%). This result is in agreement with previous laboratory-based reports showing that multiple inseminations occur in crowded cages [23], possibly due to repeated female exposure to males in the first 24 h after mating before the mechanisms of refractoriness to further matings are fully activated. Three additional independent repeats of this experiment were performed with smaller mosquito numbers (45–60 mosquitoes for each group). These confirmed the absence of significant loss of fitness and mating competitiveness for males that passed through the machine compared to control males of the same genotype (data not shown). These results suggest that repeated COPAS sorting does not confer any obvious mating disadvantage to the males, at least in laboratory conditions. Marois et al. Malaria Journal 2012, 11:302 http://www.malariajournal.com/content/11/1/302 Page 6 of 9 Figure 3 COPAS profile of the homozygous DSX strain and sexual selection. A. About 5,000 freshly hatched larvae of the homozygous DSX line were subjected to COPAS analysis and sorting. The data generated by COPAS was treated with WinMDI software to analyse the results and to express results as artificially colored regions corresponding to males (red) and to females (green) on the fluorescence diagram (on the right). Dot colours are the same as in the time-of-flight vs extinction diagram (left). B. A total of 2,000 larvae of each sex were selected with COPAS and grown to adulthood. Careful visual examination of the cages did not reveal the presence of any adult of the wrong sex. Left panels: male mosquitoes, as identified by the hairy antennae; right panels: female mosquitoes. Bottom panels are close-ups of top panels. Isolation of non-transgenic male-only populations from transgenic colonies For certain types of vector control interventions (release of sterile males for non-transgenic SIT or release of selected natural disease-resistance traits), it will be desirable to obtain large populations of non-transgenic mosquito males. This would be essential in all instances where the release of transgenic insects may not be possible for regulatory reasons. A simple COPAS-based strategy was designed to obtain a large, non-transgenic, male-only population based on the inheritance of an Xlinked gene in the F1 generation. This strategy made use of a newly established transgenic mosquito line, the FKX line, carrying a GFP-expressing transgene on the X chromosome (see Methods). A total of 120 FKX males were crossed to 200 non-transgenic virgin females. In the F1 progeny, all males inherit their fathers' Y chromosome and are non-transgenic, while all females inherit a copy of the GFP-expressing X chromosome. Taking advantage of this property, the COPAS sorter was set to select only Marois et al. Malaria Journal 2012, 11:302 http://www.malariajournal.com/content/11/1/302 Page 7 of 9 Table 1 Efficiency of COPAS sorting and survival of sorted larvae Number of COPAS sortings Sorted sex 0 1 2 3 4 - males females males females Sorted larvae 150 154 150 153 153 Survivors 110 118 117 112 129 Survival rate 73.3% 76.6% 78.0% 73.2% 84.3% Sorting efficiency 100% 100% 100% 100% 100% A population of newly hatched DSX larvae was passed through the COPAS machine the indicated number of times and larvae of the indicated sex were requested successively. The number of larvae surviving to adulthood was scored and the sex of the resulting adults was verified. No error was found and survival was not affected by the number of passes through the machine. the non-fluorescent F1 male larvae (Figure 4A). For quality control, the sorted individuals were immediately re-analysed by the COPAS. All objects fell in the nonfluorescent region of the diagram, confirming the absence of any contaminating transgenic female larvae (Figure 4B). Visual examination of the resulting cage of adult males confirmed their purity. Therefore, the use of COPAS allows the selection of non-transgenic, male-only larvae from a cross between non-transgenic females and males from an X-linked transgenic line. The limiting step of this procedure for high-throughput applications is the separation of female-only wild type individuals for the crosses: this limitation would be overcome by the generation of Y-linked transgenic lines allowing to sort (again by the use of fluorescence-assisted sorting) large numbers of non-transgenic virgin females from their transgenic siblings. Discussion The possibility to employ genetic control strategies to roll back malaria and other mosquito-borne diseases calls for new technological approaches that overcome the current blocks in mass-rearing of mosquitoes with the desired phenotypes. The work reported here demonstrates that the technology of large particle flow cytometry provides an extremely efficient, fast and reliable method for the selection of male-only (or female-only) mosquito populations early in their developmental cycle, and for the rapid and effective establishment of homozygous transgenic lines. This work builds on an earlier report that provided a proof-of-principle demonstration of the potential of COPAS for genetic sexing of mosquito larvae [17]. This initial study was restricted to the separation of the sexes and achieved incomplete accuracy and low speed of sorting due to the advanced stage of larvae development. Here, these limitations are overcome by exploiting a sex-specific fluorescent marker expressed at the very early stages of larval development. Moreover, it is shown here that X-linked transgenes efficiently substitute for early sex-specific markers for mosquito sexing. Currently, sexing in Culicidae is achieved manually by microscopic sorting of pupae based on the dimorphism of the male and female terminal abdominal segments. Another method for separation of the sexes exploits differences in size of male and female pupae characteristic of Aedes mosquitoes (http://www.oxitec.com/news-andviews/topic-pages-safety-and-sustainability/what-happens-if-female-mosquitoes-are-released/). This approach cannot be used for A. gambiae, a mosquito species in which the size difference between male and female pupae is too subtle to warrant accurate separation of females from males. Therefore, to date COPAS sorting is the only reliable method that allows high-throughput sexing in Anopheles. The performance of the current COPAS sorters (40,000 male larvae per hour) would allow the separation of almost 2 million male mosquitoes per week (considering a 20% larval mortality rate and an 8-hour daily use of a single machine). Based on past SIT attempts [24], this number would be sufficient for most release strategies. For interventions requiring larger number of released males, the sorting method described here would need to be scaled up. The further improvement of sorters specifically designed for mosquitoes (upgraded COPAS instruments or similar sorters that may be developed by other manufacturers) may render this task achievable in the near future. Table 2 Effect of COPAS on male reproductive competitiveness Non-sorted DSX males Sorted DSX males Progenies of DSX male 18 19 Progenies of a competitor male 15 13 Mixed Progenies 8 7 Total number of progenies 41 39 Two crosses were assembled in separate cages. Each cage contained 100 wild-type virgin females, 100 wild-type competitor males, and 100 DSX males that were either raised normally (non-sorted DSX males) or sorted three times with the COPAS (sorted DSX males). Freshly emerged mosquitoes of each kind were simultaneously placed in the cages and kept together through the blood meal on day 5 until day 8, when females were isolated into single plastic tubes to oviposit on shallow water. Freshly hatched larvae were examined under the fluorescence microscope to score the identity of their father (100% fluorescent progeny indicated a DSX male, 100% non-fluorescent a competitor male, mixed progenies arose from females fertilized by at least two males of different genotypes). Results from the two crosses are not statistically different (χ2 = 1.053, p = 0.5906). Several repeats of this experiment confirmed that COPAS-sorted males are not noticeably inferior in their reproductive competitiveness. Marois et al. Malaria Journal 2012, 11:302 http://www.malariajournal.com/content/11/1/302 Page 8 of 9 Figure 4 COPAS-assisted sorting of a pure population of non-transgenic mosquito males. A. About 1,000 freshly hatched larvae of the progeny of a cross between males of the FKX line (having a GFP transgene inserted on the X chromosome) and wild type females were subjected to COPAS sorting. The lower area (gated) corresponds to wild-type male larvae, the upper area to heterozygous transgenic females. B. The 437 male larvae sorted in A were analysed again by the COPAS to assess the occurrence of female contamination. Ordinarily when a new transgenic mosquito line is obtained, inaccurate and lengthy visual screening of larvae from a heterogeneous F2 population must be performed by fluorescence microscopy to select homozygous individuals based on their stronger fluorescence phenotype. This process is subjective and prone to human error; in addition, given the limited speed of the manual process, only a small number of homozygous larvae can be obtained by one operator in a few hours. These limitations make obtaining stable homozygous transgenic lines a lengthy, tedious and inefficient process. As illustrated here for the DSX line, COPAS usage revolutionizes this step and yields a large population of homozygous individuals in one sorting session lasting less than 30 min, based on the differential expression level of the selectable marker in heterozygous and homozygous individuals. This method, now routinely used in the laboratory, facilitated the establishment of more than 20 distinct transgenic lines in two distinct A. gambiae genetic backgrounds, the G3 and Ngousso strains (data not shown), illustrating its general value for the development of transgenic mosquito lines. The availability of an effective and high-throughput technology to rapidly select homozygous individuals from mixed populations offers the possibility to increase the fitness of released insects. It is particularly important when considering the problems posed by mass-rearing transgenic insects, where inbreeding of the population causes large fitness costs. As a way to increase the genetic pool of inbred populations with a consequent positive effect on fitness, field mosquitoes can be introduced into the mass-reared population every few generations. Moreover, the sorting technology allows purifying a transgenic line in case of contamination with wild-type individuals, a frequently occurring problem even in the most experienced laboratories. The selection procedure described here and the use of multiple fluorescent markers such as GFP, YFP, CFP and DsRed proteins in transgenic larvae enable novel experimental schemes now routinely used by the authors. For example, multiple transgenes can be rapidly combined into a single mosquito line, or the frequency dynamics of multiple transgenes present in a single population can be monitored over generations and corrected at will. When using transgenic lines that allow separation of the sexes based on fluorescent phenotypes, COPASassisted sexing can produce large single-sex mosquito populations. In addition to enabling particular laboratory studies that require considerable numbers of virgin females or males, such a tool will be invaluable for the selection of male-only mosquito populations for genetic control strategies against vector-borne diseases such as malaria. In the current experiments, a limitation on the number of screened individuals (up to 20,000 in 30 min) is only imposed by the size of the mosquito colony and not by the throughput characteristics of the COPAS instrument used. Importantly, COPAS-selected males are as competitive for mating as their unsorted peers in cage conditions. In addition, the authors’ empirical experience in managing more than 20 independent transgenic A. gambiae strains using routine COPAS sorting indicates that sorted populations are often healthier than their unsorted peers, possibly because COPAS sorting eliminates microbe-rich debris from progenitor mosquitoes. Conclusions The powerful capacity of the COPAS system to distinguish gene copy number by intensity of fluorescence is Marois et al. Malaria Journal 2012, 11:302 http://www.malariajournal.com/content/11/1/302 not limited to the use of DSX-like sex-specific constructs, and is applicable to a large number of insect species that are amenable to transgenesis. Novel COPASbased strategies will certainly further assist the development of transgenic applications, such as large-scale larval seeding in multi-well plates for high-throughput drug or small molecule screening. We are now entering a new era in which much anticipated genetics-based disease control strategies are finally taking shape. Considerable laboratory work is urgently needed to evaluate their promises and risks, and to determine the practical aspects of their implementation. The tools presented here remove many of the hurdles related to inaccurate and low-throughput sorting of mosquito populations and thereby pave the way for both laboratory studies and future field releases of sterile or disease-resistant vectors. Page 9 of 9 4. 5. 6. 7. 8. 9. 10. 11. 12. Additional file Additional file 1: Description of the FKX transgenic construct. 13. Competing interests The authors declare that they have no competing interests. Authors’ contributions CS and FC designed and constructed the DSX plasmid and the transgenic mosquito strain. EM designed methods, performed experiments and drafted the manuscript. JS and CK generated and mapped the FKX line, respectively. EAL and FC conceived the study, participated to its design and co-ordination and drafted the manuscript. All authors read and approved the final manuscript. Acknowledgements This work was supported by the Institut National de la Santé et de la Recherche Médicale (INSERM U963) and by the Centre National de la Recherche Scientifique (CNRS UPR9022), the European Commission FP7 projects INFRAVEC (grant agreement no. 228421) and EVIMalar (grant agreement no. 242095), by a Medical Research Council Career Development Award (Agreement ID 78415, File G0600062), by the European Research Council FP7 ERC Starting Grant project ‘Anorep’ (Grant ID: 260897). Author details Institut de Biologie Moléculaire et Cellulaire, INSERM U963, CNRS UPR9022, 15 rue René Descartes, 67084, Strasbourg, France. 2Division of Cell and Molecular Biology, Imperial College London, Imperial College Road, London SW7 2AZ, UK. 3Department of Vector Biology, Max-Planck Institute for Infection Biology, Chariteplatz 1, 10117, Berlin, Germany. 4Dipartimento di Medicina Sperimentale e Scienze Biochimiche, Università degli Studi di Perugia, Terni 05100, Italy. 5Department of Immunology and Infectious Diseases, Harvard School of Public Health, 665 Huntington Avenue, Boston, MA 02115, USA. 14. 15. 16. 17. 18. 19. 20. 1 Received: 25 May 2012 Accepted: 7 August 2012 Published: 28 August 2012 References 1. World Health Organization: Malaria Report. 2011. http://www.who.int/ malaria/publications/atoz/9789241564403/en/index.html. 2. Hendrichs J, Robinson A: Sterile Insect Technique. In Encyclopedia of Insects. Edited by Resh VH, Carde RT. London, Oxford, Boston: New York and San Diego: Elsevier Science; 2009:953–957. 3. Benedict M, Robinson AS, Knol BGJ: Development of the sterile insect technique for African malaria vectors. Malar J 2009, Suppl 2:8. 21. 22. 23. 24. Alphey L, Benedict M, Bellini R, Clark GG, Dame DA, Service MW, Dobson SL: Sterile-insect methods for control of mosquito-borne diseases: an analysis. Vector Borne Zoonotic Dis 2010, 10:295–311. Nolan T, Papathanos P, Windbichler N, Magnusson K, Benton J, Catteruccia F, Crisanti A: Developing transgenic Anopheles mosquitoes for the sterile insect technique. Genetica 2011, 139:33–39. Tripet F, Toure YT, Dolo G, Lanzaro GC: Frequency of multiple inseminations in field-collected Anopheles gambiae females revealed by DNA analysis of transferred sperm. Am J Trop Med Hyg 2003, 68:1–5. Holt RA, et al: The genome sequence of the malaria mosquito Anopheles gambiae. Science 2002, 298:129–149. Catteruccia F, Nolan T, Loukeris TG, Blass C, Savakis C, Kafatos FC, Crisanti A: Stable germline transformation of the malaria mosquito Anopheles stephensi. Nature 2000, 405:959–962. Blandin S, Moita LF, Köcher T, Wilm M, Kafatos FC, Levashina EA: Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO Rep 2002, 3:852–856. Fu G, Lees RS, Nimmo D, Aw D, Jin L, Gray P, Berendonk TU, White-Cooper H, Scaife S, Kim Phuc H, Marinotti O, Jasinskiene N, James AA, Alphey L: Female-specific flightless phenotype for mosquito control. Proc Natl Acad Sci U S A 2010, 107:4550–4554. Black WC, Alphey L, James AA: Why RIDL is not SIT. Trends Parasitol 2011, 27:362–370. Yoshida S, Shimada Y, Kondoh D, Kouzuma Y, Ghosh AK, Jacobs-Lorena M, Sinden RE: Hemolytic C-type lectin CEL-III from sea cucumber expressed in transgenic mosquitoes impairs malaria parasite development. PLoS Pathog 2007, 3:e192. Meredith JM, Basu S, Nimmo DD, Larget-Thiery I, Warr EL, Underhill A, McArthur CC, Carter V, Hurd H, Bourgouin C, Eggleston P: Site-specific integration and expression of an anti-malarial gene in transgenic Anopheles gambiae significantly reduces Plasmodium infections. PLoS One 2011, 6:e14587. Dong Y, Das S, Cirimotich C, Souza-Neto JA, McLean KJ, Dimopoulos G: Engineered anopheles immunity to Plasmodium infection. PLoS Pathog 2011, 7:e1002458. Marshall JM, Hay BA: Inverse Medea as a novel gene drive system for local population replacement: a theoretical analysis. J Hered 2011, 102:336–341. Marshall JM, Pittman GW, Buchman AB, Hay BA: Semele: a killer-male, rescue-female system for suppression and replacement of insect disease vector populations. Genetics 2011, 187:535–551. Catteruccia F, Benton JP, Crisanti A: An Anopheles transgenic sexing strain for vector control. Nat Biotechnol 2005, 23:1414–1417. Magnusson K, Mendes AM, Windbichler N, Papathanos PA, Nolan T, Dottorini T, Rizzi E, Christophides GK, Crisanti A: Transcription regulation of sex-biased genes during ontogeny in the malaria vector Anopheles gambiae. PLoS One 2011, 6:e21572. Sheng G, Thouvenot E, Schmucker D, Wilson DS, Desplan C: Direct regulation of rhodopsin 1 by Pax-6/eyeless in Drosophila: evidence for a conserved function in photoreceptors. Genes Dev 1997, 11:1122–1131. Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A: A variant of yellow fluorescent protein with fast and efficient maturation for cellbiological applications. Nat Biotechnol 2002, 20:87–90. Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY: Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 2004, 22:1567–1572. Rogers DW, Whitten MM, Thailayil J, Soichot J, Levashina EA, Catteruccia F: Molecular and cellular components of the mating machinery in Anopheles gambiae females. Proc Natl Acad Sci U S A 2008, 105:19390–19395. Thailayil J, Magnusson K, Godfray HC, Crisanti A, Catteruccia F: Spermless males elicit large-scale female responses to mating in the malaria mosquito Anopheles gambiae. Proc Natl Acad Sci U S A 2011, 108:13677–13681. Benedict MQ, Robinson AS: The first releases of transgenic mosquitoes: an argument for the sterile insect technique. Trends Parasitol 2003, 19:349–355. doi:10.1186/1475-2875-11-302 Cite this article as: Marois et al.: High-throughput sorting of mosquito larvae for laboratory studies and for future vector control interventions. Malaria Journal 2012 11:302. Article 8: 2013 PLoS One Targeted Mutagenesis in the Malaria Mosquito Using TALE Nucleases. Smidler AL, Terenzi O, Soichot J, Levashina EA, Marois E. Targeted Mutagenesis in the Malaria Mosquito Using TALE Nucleases Andrea L. Smidler1,2¤a, Olivier Terenzi1,2, Julien Soichot1,2, Elena A. Levashina1,2¤b, Eric Marois1,2* 1 Institut National de la Santé et de la Recherche Médicale U963, Strasbourg, France, 2 Centre National de la Recherche Scientifique UPR9022, Strasbourg, France Abstract Anopheles gambiae, the main mosquito vector of human malaria, is a challenging organism to manipulate genetically. As a consequence, reverse genetics studies in this disease vector have been largely limited to RNA interference experiments. Here, we report the targeted disruption of the immunity gene TEP1 using transgenic expression of Transcription-Activator Like Effector Nucleases (TALENs), and the isolation of several TEP1 mutant A. gambiae lines. These mutations inhibited protein production and rendered TEP1 mutants hypersusceptible to Plasmodium berghei. The TALEN technology opens up new avenues for genetic analysis in this disease vector and may offer novel biotechnology-based approaches for malaria control. Citation: Smidler ONE 8(8): e74511. doi:10.1371/journal.pone.0074511 Editor: George Dimopoulos, Johns Hopkins University, Bloomberg School of Public Health, United States of America Received June 24, 2013; Accepted August 7, 2013; Published August 15, 2013 Copyright: © 2013 Smid unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Funding: T Commission FP7 proj la Recherche collection and analysis, decision to publish, or preparation of the manuscript. Competing interests: The authors have declared that no competing interests exist. * E-mail: e.marois@unistra.fr ¤a Current add America ¤b Current address: Department of Vector Biology, Max-Planck Institute for Infection Biology, Berlin, Germany Introduction vulnerable points in the parasite cycle. On the other hand, the prospect of releasing engineered male mosquitoes to propagate malaria resistance genes through wild susceptible vector populations has been receiving increasing attention [3,4]. Novel methods to disrupt or alter target genes of interest in the malaria mosquito would promote rapid progress towards these goals. Furthermore, targeted genetic modifications that do not require the permanent introduction of transposons in the genome are particularly desirable, as they eliminate the potential risk of subsequent unplanned transposon mobilization by natural sources of transposition factors. Recently, a novel class of DNA-binding protein domain derived from the Xanthomonas Transcription Activator Like Effector (TALE) proteins [5,6] has been successfully harnessed to custom-design sequence-specific endonucleases [7,8]. These TALE nucleases (TALENs), in which the TAL DNAbinding domain is fused to the FokI endonuclease domain, are easy to engineer (e.g., [9,10]), highly predictable in their sequence specificity, and highly mutagenic [11] making them an attractive alternative to Zinc Finger Nucleases that have less predictable binding specificities and require in vitro optimization [12]. Mutations arise by imprecise repair of the Malaria is caused by Plasmodium parasites transmitted to their human hosts by the bite of anopheline mosquitoes. Malaria has been charged with causing more human deaths than any other disease in human history and continues to kill about 660,000 annually [1]. A reduction in the malaria death toll has been achieved thanks to vector control using insecticides and insecticide-impregnated bednets, better health care and progress in medical treatment, but this success is currently mitigated by the spread of resistance both of mosquitoes to insecticides and of Plasmodium to antimalarial drugs. Sequencing the genome of the main malaria vector, Anopheles gambiae [2], enabled the identification of hundreds of genes involved in the vector’s capacity to transmit Plasmodium. Altering the mosquito genome in a way that abates Plasmodium transmission, through transgenesis or other sophisticated genetic engineering tools, can offer new perspectives in the fight against malaria. On one hand, experimentally altering mosquito genes of interest will advance our fundamental understanding of the biological interactions between mosquito and parasite, and may help target PLOS ONE | www.plosone.org 1 August 2013 | Volume 8 | Issue 8 | e74511 TALEN-induced TEP1 mutations in Anopheles gambiae TALEN-generated double-stranded breaks by the nonhomologous end joining (NHEJ) repair pathway. In a number of animal species, injection of mRNA encoding TALENs have readily allowed researchers to generate mutants in their genes of interest [13–19]. Very recently, mutagenesis of an eye pigmentation gene was achieved in Aedes mosquitoes using this method [20]. In rice, disease resistant mutants have been produced by transgenic expression of the TALENs, whose respective transgenes were eliminated by subsequent genetic crosses once the desired mutation had been fixed [21]. We hypothesized that a similar approach would be applicable to insect vectors and set out to use TALENs to target the TEP1 gene, a key component of the mosquito immune system. and subjected the amplification product to a restriction digest with NcoI (Figure 1B). Of 310 screened F2 larvae, 16 (5.16%) carried a heterozygous mutation at the target locus, as evidenced by the appearance of a PCR product that NcoI was unable to cleave. Thus, at least 2.58% of TEP1 copies were mutated after exposure to one TALEN dose (i.e., one generation). This figure is a conservative estimate of mutation frequency, as we subsequently observed some mutations that left the NcoI recognition sequence intact. In order to obtain a mosquito population containing a higher frequency of mutations, we self-crossed successive generations of mosquitoes expressing both TALENs of the pair. This was facilitated by automated COPAS selection of larvae [28] that had inherited one copy each of the left and right TALEN genes, which are respectively associated with a red and yellow fluorescent marker expressed in the nervous system [29]. At least 1000 double-TALEN larvae were COPAS-selected and cultured for each generation. In the 7th generation (i.e., exposure to 6 TALEN mutagenic doses), 51% (49 out of 96) of the examined individual mosquitoes carried a heterozygous mutation in TEP1. This indicated that the frequency of mutations increased faster than predicted if mutations accumulated linearly from one generation to the next. This observation is consistent with a model where mosquitoes already carrying a heterozygous mutation employ homologous recombination to repair new TALEN-induced breaks in the wildtype chromosome, thereby effectively copying the existing mutation onto the newly damaged chromosome. This suggests that in addition to NHEJ, TALEN-caused breaks can also be repaired by homologous recombination. Therefore, the observed number of NHEJ mutations is an underestimation of the true rate of TALEN activity. We wondered if a single TALEN of the pair is capable of causing mutations in TEP1. To investigate this, we sampled mosquito larvae from lines carrying a single TALEN maintained at high population levels for 8 (right TALEN) or 10 (left TALEN) generations, and again sampled larvae after about 16 generations. We expected that such a high number of generations would have allowed rare “monotalenic” mutational events to accumulate in the population. Out of 96 individual larvae tested by PCR for each sample, none carried a mutation in the TEP1 NcoI site. The observed absence of mutations among 576 haploid genomes exposed to single TALEN activity for 7, 9 or 15 generations suggests that single TALENs never or rarely induce mutations at their target site. To strengthen this point, we purified DNA from 2600 pooled larvae whose genomes had been exposed to one TALEN of the pair for 7 or 9 generations, and PCR-amplified the target region. PCR products appeared to be fully cleaved by NcoI, pointing to the absence of TALEN-induced mutations. To increase the chance of detection of a minor fraction of mutated products, we purified the region of the gel in which uncleaved PCR products may exist, cloned them into a plasmid, and examined E. coli transformants containing single copies of the PCR products. Again, all cloned fragments were cleaved by NcoI. Although we note that deep sequencing of amplicons would provide a more sensitive assay to detect rare mutations, this result further suggests that single TALENs rarely or never generate mutants. Results Since the embryo microinjection procedure is technically challenging in Anopheles gambiae (as judged by poor survival and relatively low success rate of transgenesis in this species), we expected that mutant recovery after direct injection of TALEN-encoding plasmids or mRNA might be difficult. For this reason and to enable controlled mutagenesis experiments, we preferred transgenic expression of the TALENs in the mosquito germ cells. To obtain the proof-of-principle for gene targeting in A. gambiae via transgenic TALENs, we selected the wellcharacterized immune gene TEP1 as a target. TEP1, a protein similar to vertebrate complement factor C3, binds Plasmodium parasites as they invade the mosquito intestine and kills them in a manner probably dependent on its thioester site located in the C-terminus [22–25]. TEP1 mutants will be instrumental in further dissecting the antiparasitic complement-like system in mosquitoes. TALENs function in pairs, each member of which binds a chosen 12 to 24-nucleotide sequence. The two selected target sequences are separated by 14-16 nucleotides, the optimal distance for the two FokI domains of the TALENs to properly dimerize and create a double-stranded break. Cleavage of the bound DNA molecule occurs near the center of the sequence separating the two target sites. We designed a single pair of TALENs to target a site within the TEP1 gene centered on an NcoI restriction site (5’-CCATGG-3’), offering the possibility to easily screen individual mosquitoes for mutations that destroyed the NcoI site (Figure 1A). Mutations in this region are expected to strongly affect TEP1 protein function: frame-shifts resulting in premature stop codons would remove the Cterminal third of the protein including its thioester domain, while amino-acid deletions or insertions would alter the length of the alpha-helix connecting the CUB domain to the thioester domain [26], presumably resulting in destabilization of the protein’s structure. A distinct transgenic mosquito line was generated for each TALEN of the pair, the expression of which was driven by the A. gambiae Vasa promoter, active in the mosquito germline [27]. Therefore, F1 mosquitoes arising from a cross between the two lines will express both TALENs simultaneously and are expected to produce F2 gametes carrying mutations in the TEP1 gene. To screen individual larvae within the F2 progeny, we PCR-amplified a TEP1 fragment spanning the target site PLOS ONE | www.plosone.org 2 August 2013 | Volume 8 | Issue 8 | e74511 TALEN-induced TEP1 mutations in Anopheles gambiae Figure 1. TALEN mutagenesis of the TEP1 gene. A: Fragment from the TEP1 gene showing the target site of the TALEN pair. Nucleotides bound by each TALEN are underlined, TALEN repeats are color-coded to show repeat/nucleotide specificity. The NcoI restriction site centrally located at the TALEN cleavage site is highlighted. Inset: scheme of the entire TEP1 protein showing the location of TALEN-induced mutations (SP: signal peptide; CUB: CUB domain interrupted by the TED: thioester domain; the star indicates the position of the thioester site). B: PCR assay to identify TEP1 mutant mosquitoes. A PCR product spanning the TALEN target site is generated from individual mosquitoes (small larva or a leg from a living adult) and incubated with NcoI. Full cleavage of the PCR product (w) denotes a wild-type individual. Partial cleavage (h) denotes a heterozygous TEP1 mutant. Absence of cleavage (H) corresponds to a homozygous TEP1 mutant. C: TALEN-induced mutations in the TEP1 gene. Left and right TALEN target nucleotide sequences are shown in green and blue respectively, with the 15bp spacer sequence between the TALENs in black. The NcoI restriction site is highlighted in orange. Deletions are designated by a red dash or by Δ+number of missing bases. Insertions are shown in lowercase red letters. Uppercase red letters correspond to natural polymorphisms between multiple TEP1 alleles. doi: 10.1371/journal.pone.0074511.g001 PLOS ONE | www.plosone.org 3 August 2013 | Volume 8 | Issue 8 | e74511 TALEN-induced TEP1 mutations in Anopheles gambiae To examine the effect of TEP1 dosage in the process of parasite killing, we compared oocyst infection levels between TEP1 heterozygous mutant, homozygous mutant and control mosquitoes (Figure 3d). The heterozygous mutant had an intermediate susceptibility phenotype, which was closer to, but significantly different from, the control. This suggests that the efficiency of parasite killing depends on TEP1 protein levels. The antiparasitic role of TEP1 was discovered and characterized using RNA interference assays, in which synthetically produced double-stranded RNA homologous to a fragment of native TEP1 is injected at a high concentration in the body of adult mosquitoes. This technique was generalized for the functional characterization of hundreds of mosquito genes [34]. However, injection per se was reported to impinge on Plasmodium development by the potential induction of the wounding response [35]. Therefore the mutant TEP1 lines developed here will be useful in studies that must exclude the confounding effect of the injection procedure itself. To examine how the classical TEP1 RNAi phenotype compares with the mutant phenotype, we compared parasite loads in M∆T to dsTEP1-injected mosquitoes of the parental line (Figure 3e). The mutant and RNAi phenotypes were very similar, validating a posteriori the high efficiency of RNAi knockdown. The apparent absence of single TALEN background activity was likely facilitated by our design scheme in which we used obligate heterodimeric FokI domains in TALEN construction [30,31]. The TALEN-generated TEP1 mutations were very similar in nature to mutations obtained in other organisms (Figure 1C) and consisted mainly of small deletions and insertions (indels) that are the hallmark of imprecise NHEJ-mediated DNA repair. Some mutations deleted a number of nucleotides in multiples of three, resulting in the deletion of one to a few amino acids from the TEP1 protein. Other mutations introduced a frameshift in the TEP1 coding sequence, resulting in the loss of the entire C-terminal half of the protein, which contains features crucial for TEP1 function including the thioester domain. Using a PCR selection procedure from single legs taken from live mosquitoes, we recovered five homozygous mutant mosquito lines. These mutant lines were representative of the main classes of mutations: line M∆T and M∆MV are deletions of 1 (Threonine) and 2 (Methionine and Valine) amino-acids, respectively; line M∆3+6 lacks 3 endogenous amino-acids but gained an insertion of 6 exogenous ones, line M∆ct1 and M∆ct2 are frame-shift mutations causing the loss of the entire protein C-terminus (Figure 2A). Immunoblotting analysis failed to detect TEP1 in the Mut∆ct1 and M∆ct2 lines, while it revealed strongly reduced protein levels in M∆T, M∆MV and M∆3+6 lines (Figure 2B, top). Although small amounts of TEP1 protein could be detected in whole mosquito extracts of the M∆T, M∆MV and M∆3+6 lines, these proteins did not undergo cleavage and were impaired in their secretion to the hemolymph, as no TEP1 signal was observed in immunoblotting of hemolymph samples (Figure 2B, bottom). Therefore, the alpha helix connecting the CUB and thioester domains of TEP1 [26,32] seems to be very sensitive to insertion or deletion of single amino acids that destabilize the structure and prevent proper protein synthesis and secretion. We next assessed the phenotype of these mutant lines in comparison to the parental lines by infection assays using Plasmodium berghei-infected mice (Figure 3). Similar to the RNA interference knockdown phenotype of TEP1 [22], all mutations in the homozygous state resulted in a dramatic increase in the number of developing parasites within the mosquito gut. These results confirm the pivotal role of TEP1 in antiparasitic responses. While it cannot be fully excluded that RNAi-mediated TEP1 knockdown may also affect genes sharing some nucleotide identity with the TEP1 sequence, such as other closely-related genes in the TEP family [33], mutations in TEP1 should leave the expression of other genes unaffected. Interestingly, while midguts from TEP1 mutant mosquitoes often displayed impressive oocyst numbers that could exceed 1500, each experiment also yielded mutant midguts bearing no or only a few oocysts. Upon blood feeding on a single infected mouse, only well-gorged mosquito females were selected for subsequent dissection. It is therefore unlikely that some females ingested dramatically reduced numbers of P. berghei gametocytes. Thus, it is apparent that TEP1-independent mechanisms are at work to limit Plasmodium infection in a subset of mosquitoes. PLOS ONE | www.plosone.org Discussion Here, we obtained proof of principle that TALENs can be used for targeted mutagenesis in the genome of the malaria mosquito, which is notably difficult to manipulate genetically. Recently, Aryan et al. [20] reported disruption of the eye pigmentation kmo gene in the dengue vector mosquito Aedes aegypti by injection of TALEN-encoding plasmid DNA. In the same species, successful mutagenesis of GFP and of the odorant receptor co-receptor (orco) was achieved by injecting mRNA encoding Zinc Finger Nucleases [36]. In both cases and in other animal systems for which TALEN mutagenesis has been reported, mutants were obtained directly upon injection of DNA or RNA into embryos. In contrast, we employed transgenically-expressed TALENs for this purpose. Besides the assurance of expressing the pair of TALENs in all germ cells, this offered the possibility to increase the frequency of mutant alleles in successive TALEN-expressing generations of mosquitoes. We disrupted the antiparasitic gene TEP1 as a first target; future work will make use of the obtained hypersusceptible mutant lines to further dissect the role of this important anti-malarial factor in parasite killing. Beyond the research field of mosquito immunity, this study paves the way for numerous other applications such as obtaining mutations in Anopheles genes considered to be essential for Plasmodium parasite development [37–39]. Disrupting these genes, or altering specific domains on the encoded proteins, may render homozygous mutant mosquitoes unable to support parasite development. Such parasite-refractory mutant mosquitoes could be used in anti-malaria intervention schemes, including vector population replacement in endemic regions. Unless a gene-drive strategy is specifically designed to spread desired mutations, this could be achieved by the repeated release of mass-produced male mosquitoes. Where knockout mutants in 4 August 2013 | Volume 8 | Issue 8 | e74511 TALEN-induced TEP1 mutations in Anopheles gambiae Figure 2. TEP1 mutant proteins. A: Fragment of the TEP1 protein encompassing the TALEN-induced mutations is shown for the wild-type (WT) and for those mutants that we maintain as homozygous mosquito lines (M∆T, M∆MV, M∆3+6, M∆ct1 and M∆ct2). Gaps in the protein sequence denote amino acid deletions. Inserted exogenous amino acids are shown in red. Nonsense amino acids followed by a stop codon (*), resulting from frame-shift mutations, are shown in blue. B: Immunoblots to evaluate the presence of mutant TEP1 protein in whole mosquito extract (top panels) or hemolymph. Hemolymph prophenoloxidase (PPO) serves as a loading control. In the control samples, both TEP1 full-length (full) and C-terminal fragment (cleaved) are visible. Cross-reacting background bands, some running in close proximity to TEP1 fragments, are marked on the left with ‘>’ signs. doi: 10.1371/journal.pone.0074511.g002 genes for use in the Sterile Insect Technique to reduce vector populations [40]. Of note, the TALEN transgenes used to obtain a desired mutation can subsequently be discarded by selection, thereby rendering the obtained homozygous mutant mosquitoes transgene-free. This could facilitate mosquito a given target gene might compromise the fitness of the mosquitoes, TALEN mutagenesis offers the possibility of a gene therapy to cure mosquitoes of malaria by selecting mutations that prevent a protein’s interactions with parasite factors while preserving its other vital functions. TALEN mutagenesis could also be employed to knock out male fertility PLOS ONE | www.plosone.org 5 August 2013 | Volume 8 | Issue 8 | e74511 TALEN-induced TEP1 mutations in Anopheles gambiae Figure 3. TEP1 mutant mosquitoes are hypersusceptible to P. berghei. Mosquito females from five different homozygous mutant mosquito lines and from control parental lines were offered a blood meal on a P. berghei-infected mouse. Seven days after infection, the midgut was dissected and the number of oocysts developing in each midgut was evaluated. The statistical significance of differences in mean parasite numbers was measured with a Mann-Whitney test (mutant versus control) and with a Kruskall-Wallis test followed by Dunn’s post-test (to compare all groups in [d]). (a) M∆T mosquitoes are compared to the two parental, non-mutant TALEN lines. (b) 4 different mutant lines are compared to the parental mosquito line that initially served to produce TALEN transgenic lines. This control line was verified to show the same level of susceptibility to P. berghei as the two TALEN daughter lines (not M∆ct2 line did not, presumably due to a different physiological condition of this mosquito culture. In two independent experiments (c, d), we used mosquitoes of different genotypes marked with distinct fluorescence markers and cultured the larvae together in the same water to eliminate potential confounding factors due to rearing conditions. On the day of dissection, genotypes were separated on the basis of fluorescence. The same M∆ct2 line shows significantly elevated parasite numbers. (d) Heterozygous M∆ct2 mosquitoes are compared to control and homozygous M∆ct2 mosquitoes: the susceptibility phenotype of the heterozygote is intermediate. (e) Control mosquitoes of the parental line, mosquitoes of the parental line injected with TEP1 double-stranded RNA, and homozygous TEP1 mutant mosquitoes of the M∆T line are compared. TEP1 mutant and dsRNA-injected mosquitoes show comparable susceptibility to P. berghei. doi: 10.1371/journal.pone.0074511.g003 PLOS ONE | www.plosone.org 6 August 2013 | Volume 8 | Issue 8 | e74511 TALEN-induced TEP1 mutations in Anopheles gambiae release interventions that adhere to local regulations regarding genetically modified organisms. directly in the case of homozygous individuals, or cloned (CloneJET PCR Cloning Kit, Fermentas) and subsequently sequenced. Materials and Methods Mutagenesis and mutant line recovery Ethics statement For mutagenesis experiments, the two TALEN lines were crossed. From the F2 generation onwards, 1000 mosquito larvae that inherited a single copy of each TALEN were COPAS-purified to initiate the next generation. To isolate TEP1 mutants, mosquitoes from this population were out-crossed to the parental line. The progeny carried a single TALEN and was therefore no longer subjected to TALEN mutagenesis. Among this progeny, we screened single adult mosquitoes by PCR on one leg using the Phire direct animal tissue PCR kit (Thermo, Fisher). The identified heterozygous mutants were individually crossed to the parental line. In the F2 progeny, we identified homozygous mutants by leg PCR and pooled them to start distinct homozygous mutant families. Our experimental protocols were approved by Comité de Qualification Institutionnel (CQI), the ethics evaluation committee of INSERM (IRB00003888, FWA00005831). Mosquitoes were reared and blood-fed on anesthetized mice in compliance with French and European laws on animal house procedures (agreement E67-482-2 of the Direction of Veterinary services of the French Ministry of Agriculture). Assembling the TALENs TALENs targeting the sites shown in Figure 1 were constructed by Golden Gate Cloning according to [9]. For TALEN assembly, we prepared two transgenesis-compatible destination vectors (annotated sequences provided in Text S1) encoding a Venus yellow fluorescent and a DsRed fluorescent transgenesis reporter gene, respectively. These vectors also contain a phage ϕC31 attB site for genomic integration into transgenic lines harboring attP sites. The first module used in Golden Gate assembly provided the Vasa promoter characterized in [27]. The last module closed the TALEN assembly with either a FokI-DD or a FokI-RR domain [30], designed for obligate heterodimerization of the two TALENs and codon-optimized for A. gambiae. The annotated sequence of the three plasmids is provided in Text S1. The TALEN C and N-terminal domains flanking the repeat region were identical to those used in [7]. RNAi and infection assays RNAi silencing of TEP1 by double-stranded RNA injection into the thorax of adult mosquitoes and mosquito infections on mice carrying Plasmodium berghei GFP-con 259cl2 were performed as described [22]. To semi-automatically quantify the number of oocysts in photographs of dissected mosquito midguts, we used the “watershed segmentation” and “analyze particles” plugins of the ImageJ software after digital subtraction of the image background and smoothing of the signal. Oocyst counts obtained by this method are consistent with those obtained by manual counting of oocysts. A. gambiae lines and mosquito transgenesis Immunoblotting The TALEN-encoding vectors were inserted by ϕC31 integrase-mediated transgenesis [41] into the genome of A. gambiae lines X1 and X13, which are derived from laboratory strain G3. These two lines carry a PiggyBac transgene on chromosome II, containing an attP docking site. Individual transgenic larvae carrying the inserted left or right TALEN genes at the X1 or X13 attP site were identified by their red or yellow fluorescence, respectively. Resulting adults were crossed to their non-fluorescent parental line. In the F2 generation, fluorescent homozygous larvae were COPASselected [28] to establish stable TALEN-expressing lines. Whole-body mosquito extracts were obtained by grinding one adult female mosquito in 80 µl of protein sample buffer. The sample was denatured for 3 min at 95°C and centrifuged. 8 µl were loaded on 8% SDS-polyacrylamide gels. Hemolymph samples were prepared as described [42], 10 µl of the samples were loaded. Immunoblotting was performed using standard procedures [43] with rabbit polyclonal antibodies raised against the prophenoloxidase PPO2 [44] or against the C-terminal half of TEP1 [45]. Supporting Information Identification of TEP1 Mutants In putative mutants (mosquitoes arising from parents expressing both TALENs), we PCR amplified a region of TEP1 spanning the TALEN target site using Phusion or Phire Polymerases (Thermo, Fisher) and primer 5’TCAACTTGGACATCAACAAGAAGGCCGA-3’ in combination with either 5’-GCATATCTTTGTGCCACACTTT-3’ or 5’GCCACCGTAACGAATTTCCA-3’. The PCR product was digested with NcoI (Fermentas), the recognition site of which is centrally located in the sequence cut by the TALENs. PCR products corresponding to mutant TEP1 alleles were not digested by NcoI. These PCR products were sequenced PLOS ONE | www.plosone.org Text S1. The nucleotide sequences of plasmids and building blocks used to construct the TALEN-expressing transgenesis vectors are provided. Building blocks not listed here are published in references 5,7. (DOCX) Acknowledgements We thank the Boch and Bonas Laboratories (University of Halle, Germany) for sharing the TAL construction kit. 7 August 2013 | Volume 8 | Issue 8 | e74511 TALEN-induced TEP1 mutations in Anopheles gambiae Author Contributions Conceived and designed the experiments: EM AS. Performed the experiments: AS OT JS EM. Analyzed the data: AS EM EAL. Wrote the manuscript: EM AS EAL. References 1. WHO (2012) Malaria Report. 2. Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R et al. (2002) The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129-149. doi:10.1126/science.1076181. PubMed: 12364791. 3. Catteruccia F (2007) Malaria vector control in the third millennium: progress and perspectives of molecular approaches (in press). Pest Manag Sci. 4. Paton D, Underhill A, Meredith J, Eggleston P, Tripet F (2013) Contrasted Fitness Costs of Docking and Antibacterial Constructs in the EE and EVida3 Strains Validates Two-Phase Genetic Transformation System. PLOS ONE 8: e67364. doi:10.1371/ journal.pone.0067364. PubMed: 23840679. 5. Boch J, Scholze H, Schornack S, Landgraf A, Hahn S et al. (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326: 1509-1512. doi:10.1126/science.1178811. PubMed: 19933107. 6. Moscou MJ, Bogdanove AJ (2009) A simple cipher governs DNA recognition by TAL effectors. Science 326: 1501. doi:10.1126/science. 1178817. PubMed: 19933106. 7. Miller JC, Tan S, Qiao G, Barlow KA, Wang J et al. (2011) A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29: 143-148. doi:10.1038/nbt.1755. PubMed: 21179091. 8. Cermak T, Doyle EL, Christian M, Wang L, Zhang Y et al. (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res, 39: e82-. PubMed: 21493687. 9. Geissler R, Scholze H, Hahn S, Streubel J, Bonas U et al. (2011) Transcriptional activators of human genes with programmable DNAspecificity. PLOS ONE 6: e19509. doi:10.1371/journal.pone.0019509. PubMed: 21625585. 10. Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD et al. (2012) FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 30: 460-465. doi:10.1038/nbt.2170. PubMed: 22484455. 11. Chen S, Oikonomou G, Chiu CN, Niles BJ, Liu J et al. (2013) A largescale in vivo analysis reveals that TALENs are significantly more mutagenic than ZFNs generated using context-dependent assembly. Nucleic Acids Res. 12. (2011) Move over ZFNs. Nat Biotechnol 29: 681-684. doi:10.1038/nbt. 1935. PubMed: 21822235. 13. Huang P, Xiao A, Zhou M, Zhu Z, Lin S et al. (2011) Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol 29: 699-700. doi:10.1038/nbt.1939. PubMed: 21822242. 14. Liu J, Li C, Yu Z, Huang P, Wu H et al. (2012) Efficient and specific modifications of the Drosophila genome by means of an easy TALEN strategy. J Genet Genomics 39: 209-215. doi:10.1016/j.jgg. 2012.04.003. PubMed: 22624882. 15. Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C et al. (2012) Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci U S A 109: 17382-17387. doi:10.1073/pnas.1211446109. PubMed: 23027955. 16. Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG et al. (2012) In vivo genome editing using a high-efficiency TALEN system. Nature 491: 114-118. doi:10.1038/nature11537. PubMed: 23000899. 17. Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S et al. (2012) Highly efficient generation of heritable zebrafish gene mutations using homoand heterodimeric TALENs. Nucleic Acids Res 40: 8001-8010. doi: 10.1093/nar/gks518. PubMed: 22684503. 18. Watanabe T, Ochiai H, Sakuma T, Horch HW, Hamaguchi N et al. (2012) Non-transgenic genome modifications in a hemimetabolous insect using zinc-finger and TAL effector nucleases. Nat Commun 3: 1017. doi:10.1038/ncomms2020. PubMed: 22910363. 19. Sung YH, Baek IJ, Kim DH, Jeon J, Lee J et al. (2013) Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol 31: 23-24. doi:10.1038/nbt.2477. PubMed: 23302927. 20. Aryan A, Anderson MA, Myles KM, Adelman ZN (2013) TALEN-Based Gene Disruption in the Dengue Vector Aedes aegypti. PLOS ONE 8: e60082. doi:10.1371/journal.pone.0060082. PubMed: 23555893. PLOS ONE | www.plosone.org 21. Li T, Liu B, Spalding MH, Weeks DP, Yang B (2012) High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol 30: 390-392. doi:10.1038/nbt.2199. PubMed: 22565958. 22. Blandin S, Shiao SH, Moita LF, Janse CJ, Waters AP et al. (2004) Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector Anopheles gambiae. Cell 116: 661-670. doi:10.1016/ S0092-8674(04)00173-4. PubMed: 15006349. 23. Blandin SA, Wang-Sattler R, Lamacchia M, Gagneur J, Lycett G et al. (2009) Dissecting the genetic basis of resistance to malaria parasites in Anopheles gambiae. Science 326: 147-150. doi:10.1126/science. 1175241. PubMed: 19797663. 24. Dong Y, Aguilar R, Xi Z, Warr E, Mongin E et al. (2006) Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLOS Pathog 2: e52. doi:10.1371/journal.ppat.0020052. PubMed: 16789837. 25. Molina-Cruz A, Garver LS, Alabaster A, Bangiolo L, Haile A et al. (2013) The human malaria parasite Pfs47 gene mediates evasion of the mosquito immune system. Science 340: 984-987. doi:10.1126/ science.1235264. PubMed: 23661646. 26. Baxter RH, Chang CI, Chelliah Y, Blandin S, Levashina EA et al. (2007) Structural basis for conserved complement factor-like function in the antimalarial protein TEP1. Proc Natl Acad Sci U S A 104: 11615-11620. doi:10.1073/pnas.0704967104. PubMed: 17606907. 27. Papathanos PA, Windbichler N, Menichelli M, Burt A, Crisanti A (2009) The vasa regulatory region mediates germline expression and maternal transmission of proteins in the malaria mosquito Anopheles gambiae: a versatile tool for genetic control strategies. BMC Mol Biol 10: 65. doi: 10.1186/1471-2199-10-65. PubMed: 19573226. 28. Marois E, Scali C, Soichot J, Kappler C, Levashina EA et al. (2012) High-throughput sorting of mosquito larvae for laboratory studies and for future vector control interventions. Malar J 11: 302. doi: 10.1186/1475-2875-11-302. PubMed: 22929810. 29. Horn C, Wimmer EA (2000) A versatile vector set for animal transgenesis. Dev Genes Evol 210: 630-637. doi:10.1007/ s004270000110. PubMed: 11151300. 30. Szczepek M, Brondani V, Büchel J, Serrano L, Segal DJ et al. (2007) Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol 25: 786-793. doi: 10.1038/nbt1317. PubMed: 17603476. 31. Gupta A, Meng X, Zhu LJ, Lawson ND, Wolfe SA (2011) Zinc finger protein-dependent and -independent contributions to the in vivo offtarget activity of zinc finger nucleases. Nucleic Acids Res 39: 381-392. doi:10.1093/nar/gkq787. PubMed: 20843781. 32. Le BV, Williams M, Logarajah S, Baxter RH (2012) Molecular basis for genetic resistance of Anopheles gambiae to Plasmodium: structural analysis of TEP1 susceptible and resistant alleles. PLOS Pathog 8: e1002958. PubMed: 23055931. 33. Blandin S, Levashina EA (2004) Thioester-containing proteins and insect immunity. Mol Immunol 40: 903-908. doi:10.1016/j.molimm. 2003.10.010. PubMed: 14698229. 34. Catteruccia F, Levashina EA (2009) RNAi in the malaria vector, Anopheles gambiae. Methods Mol Biol 555: 63-75. doi: 10.1007/978-1-60327-295-7_5. PubMed: 19495688. 35. Nsango SE, Abate L, Thoma M, Pompon J, Fraiture M et al. (2012) Genetic clonality of Plasmodium falciparum affects the outcome of infection in Anopheles gambiae. Int J Parasitol 42: 589-595. doi: 10.1016/j.ijpara.2012.03.008. PubMed: 22554991. 36. Degennaro M, McBride CS, Seeholzer L, Nakagawa T, Dennis EJ et al. (2013) orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature. 37. Ghosh AK, Devenport M, Jethwaney D, Kalume DE, Pandey A et al. (2009) Malaria parasite invasion of the mosquito salivary gland requires interaction between the Plasmodium TRAP and the Anopheles saglin proteins. PLOS Pathog 5: e1000265. PubMed: 19148273. 38. Bhattacharyya MK, Kumar N (2001) Effect of xanthurenic acid on infectivity of Plasmodium falciparum to Anopheles stephensi. Int J Parasitol 31: 1129-1133. doi:10.1016/S0020-7519(01)00222-3. PubMed: 11429178. 8 August 2013 | Volume 8 | Issue 8 | e74511 TALEN-induced TEP1 mutations in Anopheles gambiae 39. Volz J, Müller HM, Zdanowicz A, Kafatos FC, Osta MA (2006) A genetic module regulates the melanization response of Anopheles to Plasmodium. Cell Microbiol 8: 1392-1405. doi:10.1111/j. 1462-5822.2006.00718.x. PubMed: 16922859. 40. Thailayil J, Magnusson K, Godfray HC, Crisanti A, Catteruccia F (2011) Spermless males elicit large-scale female responses to mating in the malaria mosquito Anopheles gambiae. Proc Natl Acad Sci U S A 108: 13677-13681. doi:10.1073/pnas.1104738108. PubMed: 21825136. 41. Meredith JM, Basu S, Nimmo DD, Larget-Thiery I, Warr EL et al. (2011) Site-specific integration and expression of an anti-malarial gene in transgenic Anopheles gambiae significantly reduces Plasmodium infections. PLOS ONE 6: e14587. doi:10.1371/journal.pone.0014587. PubMed: 21283619. 42. Rono MK, Whitten MM, Oulad-Abdelghani M, Levashina EA, Marois E (2010) The major yolk protein vitellogenin interferes with the anti- PLOS ONE | www.plosone.org plasmodium response in the malaria mosquito Anopheles gambiae. PLOS Biol 8: e1000434. PubMed: 20652016. 43. Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning A laboratory manual. Cold Spring Harbor: CSH Laboratory Press. 44. Fraiture M, Baxter RH, Steinert S, Chelliah Y, Frolet C et al. (2009) Two mosquito LRR proteins function as complement control factors in the TEP1-mediated killing of Plasmodium. Cell Host Microbe 5: 273-284. doi:10.1016/j.chom.2009.01.005. PubMed: 19286136. 45. Levashina EA, Moita LF, Blandin S, Vriend G, Lagueux M et al. (2001) Conserved Role of a Complement-like Protein in Phagocytosis Revealed by dsRNA Knockout in Cultured Cells of the Mosquito, Anopheles gambiae. Cell 104: 709-718. doi:10.1016/ S0092-8674(01)00267-7. PubMed: 11257225. 9 August 2013 | Volume 8 | Issue 8 | e74511