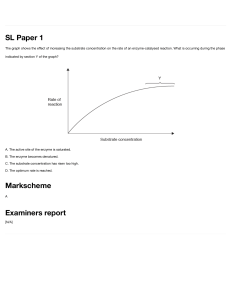
Enzymes 1 Enzymes – Classification Enzymes are proteins that act as catalysts. They increase the rate at which chemical reactions occur without being consumed or permanently altered themselves Enzymes are classified according to the reactions that they catalyze: 2 Enzymes – Classification 1. Oxidoreductases: they catalyze the transfer of electrons from one molecule, the reductant (electron donor) to another, the oxidant (electron acceptor). They can be oxidases or dehydrogenases • Oxidases are involved when oxygen acts as an acceptor of hydrogen or electrons • Dehydrogenases oxidize a substrate by transferring hydrogen to an acceptor 3 Enzymes – Classification 2. Transferases: they transfer a chemical group from one molecule to another. • • • • • • Acyltransferase Aminotransferase Glycosyltransferase Kinase Methyltransferase Nucleotidyltransferase… 4 Enzymes – Classification 3. Hydrolases: They split water and add it to molecules. They catalyze the hydrolytic cleavage of carbon-oxygen, carbon-nitrogen, and carbon-carbon bonds. • Phosphatase • Peptidase • Lipase • Hydrolase • Amylase 5 Enzymes – Classification 4. Lyases: they break double bonds trough the addition of water, ammonia, or carbon dioxide. They can also form double bonds by the removal of these groups from the substrate • • • • • Decarboxylase Synthase Aldolase Cyclase Endonuclease 6 Enzymes – Classification 5. Isomerases: they catalyze reactions that change or alter small parts of the substrate inducing intramolecular rearrangements • Epimerase • Mutase • Racemase 7 Enzymes – Classification 6. Ligases: also called Synthetase, they catalyze the joining of two molecules, using energy from ATP ADP • amino acid–RNA ligase: catalyzes the formation of a carbon-oxygen (C―O) bond between an amino acid & transfer RNA • amide synthetases & peptide synthetases: catalyzes the formation of Carbon– nitrogen (C―N) bonds 8 Enzymes – Nomenclature Usually ends in ase Identifies the reacting substrate; for example: • urease catalyzes the reaction of urea • lactase catalyzes the reaction of lactose Describes the function of the enzyme • Dehydrogenase • oxidase • decarboxylase Can be common name, with no direct relationship to substrate or reaction type, particularly for the digestive enzymes • Pepsin • Chymotrypsin • Trypsin 9 Enzymes – Energy of reactions An enzyme speeds a reaction by lowering the activation energy, changing the reaction pathway This provides a lower energy route for conversion of substrate to product Every chemical reaction is characterized by an equilibrium constant Keq, which is a reflection of the difference in energy between reactants aA, and products, bB 10 Enzymes – Activation energy As substrates are transformed into products during a chemical reaction, they go through an intermediate transition state The chemical reaction is at its highest energy at the transition state The difference between the energy level of the substrate and the energy level of the transition state is called the activation energy Activation energy is the energy needed to overcome the energy barrier of breaking and reforming bonds for a reaction to proceed. 11 Enzymes – Activation energy During a chemical reaction, substrates (A + BC) reach a transition state (A—B—C) before they are transformed into products (AB + C) Compared to an uncatalyzed reaction (left), enzymes lower the activation energy by stabilizing the transition state to an energetically favorable conformation (right) 12 Catalyzed & uncatalyzed reactions The rates of uncatalyzed reactions increase as the substrate concentration increases The rates of catalyzed reactions are limited by enzyme availability & show two stages: • The 1st stage is the formation of an enzyme-substrate complex • The 2nd stage is a slow conversion to product 13 Reactions & substrate concentration Increasing substrate concentration increases the frequency with which the enzyme & substrate collide As a result enzyme-substrate complexes form more quickly and the rate of reaction increases However, there is a limit as eventually there all the enzyme active sites are already occupied with substrate the enzyme active sites become saturated Any further increase in substrate concentration has no further effect on the reaction rate 14 Reactions & enzyme concentration Increased enzyme concentration increase the formation of enzyme-substrate complexes accelerate the reaction’s rate BUT, if more enzyme molecules present than substrate some enzymes won't have any substrate to bind to Increased enzyme concentration no further effect on the reaction’s rate If there is excess substrate 1st part of the curve can be approx. to a straight line 15 Enzyme-substrate complex The reversible enzyme-catalyzed reaction steps: • Formation of a temporary enzyme-substrate complex (E-S) when an enzyme comes into contact with its substrate conformational change when the substrate enters the active site (E-S is the transition state) • Formation of the enzyme-product complex (E-P) and when products are released, the enzyme is ready for another substrate 16 Enzyme-substrate complex details The part of the enzyme combining with the substrate is the active site Active sites characteristics include: • Pockets or clefts in the surface of the enzyme • R groups at active site are called catalytic groups • Shape of active site is complimentary to the shape of the substrate • The enzyme attracts & holds the substrate using weak non-covalent interactions • Conformation of the active site determines the specificity of the enzyme 17 The lock & key enzyme model In this model, the enzyme is assumed to be the lock and the substrate is the key and that both have fixed confirmation that lead to an easy fit • The enzyme and substrate have fixed conformations that lead to an easy fit • This model fails to take into account proteins conformational changes to accommodate a substrate molecule 18 The induced-fit enzyme model In this model, it is postulated that the enzyme’s active site is a flexible, not rigid, pocket • Upon exposure the shapes of the enzyme, active site, and substrate adjust to maximize the fit and form the E-S complex improving the catalytic reaction • There is s greater range of substrate 19 Enzyme-substrate complex specificity For an enzyme & substrate to react, surfaces of each must be complementary Enzyme specificity: the ability of an enzyme to bind only one, or a very few, substrates catalyzing only a single reaction Example: Urease is VERY Specific or has a HIGH DEGREE of Specificity Classes of enzyme specificity 1. Absolute – one enzyme acts only on one substrate (Ex. Urease decomposes only urea; arginase splits only arginine) 2. Group – one enzyme can catalyze one type of reaction for similar substrates (Ex. hexokinase adds a phosphate group to different hexoses) 3. Relative/linkage - one enzyme acts on different substrates which have the same bond type (Ex. pepsin splits different proteins) 4. Stereochemical – some enzymes can only catalyze the transformation of substrates which are in certain geometrical configuration (Ex. enantiomers) Cofactors & Coenzymes Active enzyme / Holoenzyme: • Polypeptide portion of enzyme (apoenzyme) • Nonprotein prosthetic group (cofactor) Cofactors are bound to the enzyme for it to maintain the correct configuration of the active site • Organometallic compounds • Metal ions • Organic compounds Cofactors & Coenzymes A large number of enzymes require an additional non-protein component to carry out its catalytic functions called as cofactors A coenzyme or metal ion is covalently bound to the enzyme protein is called prosthetic group Cofactors-two types: • Inorganic ions such as Fe2+, Mg2+, Mn2+, Zn2+ • A complex organic molecule called coenzyme Some enzymes require both a coenzyme & one or more metal ions for the activity Coenzymes function as transient carriers of specific functional groups Coenzymes They are essential for the biological activity of the enzyme and act as group transfer reagents (Hydrogen, electrons, or other groups can be transferred) They are low molecular weight organic substances, bound to the enzyme by weak interactions /hydrogen bonds, without which the enzyme can’t exhibit any reaction Coenzymes classification Metabolite coenzymes – synthetized from common metabolites and include nucleoside triphosphates Vitamin-derived coenzymes – derivatives of vitamins • vitamins are required for coenzyme synthesis & must be obtained from diet • Most vitamins must be enzymatically transformed to the coenzyme Water-soluble vitamins & their coenzymes 26 Environmental effects The environment surrounding an enzyme can have a direct effect on enzyme function Each enzyme exhibits peak activity at narrow pH range – optimum pH Enzymes contain R groups of aa with proper charges at optimum pH Extreme pH changes will denature the enzyme disrupt the tertiary structure destroy the catalytic activity • Pepsin (stomach) • Trypsin (small intestine) have different optimum pH Temperature (T°) effects An enzyme has an optimum T° associated with maximal function The rate of an uncatalyzed reaction will increase proportionally with T° increase Optimum T° is usually close to the T° at which the enzyme typically exists (37oC for humans) Excessive heat can denature a enzyme making it completely nonfunctional Measuring enzyme’s kinetics What is kinetics? – it is an approach to learn a reaction’s mechanism by studying factors that influence its rate How does this apply to enzymes? – The mystery with enzymes is to know what the enzyme is doing, when it will function, what is it doing, and how factors affect its rate More… – Does the enzyme reacts with the substrate directly? What concentration is needed? How should an enzyme be essayed? How does an inhibitor affect enzymatic activity? Kinetics ginve these answers and a whole lot more Substrate concentration & enzyme activity At low [sub], there is a steep increase in the rate of reaction with increasing [sub]. The catalytic site of the enzyme is empty, waiting for substrate to bind, and the rate at which product can be formed is limited by the [sub] which is available Substrate concentration & enzyme activity As the [sub] increases, the enzyme becomes saturated with substrate. When the catalytic site is empty, more substrate binds and undergoes reaction. The rate of formation of product depends on the activity of the enzyme itself, and adding more substrate will not affect the rate of the reaction to any significant effect Substrate concentration & enzyme activity Maximum rate of reaction (Vmax) - rate of reaction when the enzyme is saturated with substrate The relationship between Vmax and [sub] depends on the affinity of the enzyme for its substrate. This is usually expressed as the Km (Michaelis-Menten constant) of the enzyme, an inverse measure of affinity Km - concentration of substrate which permits the enzyme to achieve ½ Vmax An enzyme with a high Km has a low affinity for its substrate, and requires a greater concentration of substrate to achieve Vmax How to determine Km & Vmax Km and Vmax are determined by incubating the enzyme with varying concentrations of substrate; the results can be plotted as a graph of rate of reaction (v) against concentration of substrate [S] (a hyperbolic curve) The relationship is defined by the Michaelis-Menten equation: Vo = Vmax [S] / (Km + [S]) K , K , and K are specific rate constants k1 k2 E+S ES E+P k-1 1 -1 2 If assume [S] >>>>>[E] then ES will be fairly constant. The rate of rxn is determined by [ES] •Michaelis-Menten constant Km = (k2 + k-1) / k1 • When V0=1/2Vmax, Km=[S] The importance of Km & Vmax The Km permits to predict if the rate of formation of product will be affected by the availability of substrate Km reflects affinity of enzyme for its substrate Low Km enzyme normally saturated with substrate, will act at a constant rate, regardless of variations in the [sub] The importance of Km & Vmax High Km enzyme is not saturated with substrate, and its activity will vary as the [sub] varies the rate of formation of product depend on the availability of substrate Smaller Km enzyme has great affinity for its substrate Km indications - hexokinase In most tissues, the phosphorylation of glucose (Glc) to G6P is catalyzed by hexokinase It has a low Km (0.1 mM) high affinity for Glc operates at Vmax under physiological blood Glc of ~5 mM Activity does not change with blood Glc levels at fasting [Glc], hexokinase is at Vmax, but glucokinase activity varies according to [Glc] Glucokinase vs. Hexokinase Hexokinase has low Km efficiently use low levels of Glc. But quickly saturated Glucokinase is found in liver and β-cells of the pancreas Glucokinase allows liver to respond to blood Glc levels Glucokinase has a high Km, so it does not become saturated till very high levels of Glc are reached At low Glc levels, very little taken up by liver, so is spared for other tissues. Regulation of enzyme activity One of the major ways that enzymes differ from nonbiological catalysts is in the regulation of biological catalysts by cells How organisms regulate enzymatic activity? • Enzyme produced only when the substrate is present – common in bacteria • Allosteric enzymes • Feedback inhibition • Zymogens - Proteolytic enzymes are synthesized as inactive precursors, to prevent unwanted protein degradation • Protein modification 38 Allosteric enzymes Allosteric enzyme - enzyme that contains a region (2nd site) to which small, regulatory molecules "effectors" may bind in addition to and separate from the substrate binding site (1st site) and thereby affect the catalytic activity. • Positive allosteric speed up enzymatic activity • Negative allosteric slow down enzymatic activity Feedback inhibition Allosteric enzymes are the basis for feedback inhibition - a product late in a series of enzyme-catalyzed reactions serves as an inhibitor for a previous allosteric enzyme earlier in the series (enzyme’s activity is inhibited by the end product) This mechanism allows cells to regulate how much of an enzyme’s end product is produced Example - product F serves to inhibit the activity of enzyme E1 40 Zymogens - proenzymes A proenzyme - the inactive form of an enzyme Irreversible (Zymogen) or reversible (covalent modulation) Zymogenes are activated when one or more peptides are removed by proteolysis • The zymogen proinsulin is converted to its active form, insulin, by removing a small peptide chain 41 Proenzymes of the digestive tract Digestive enzymes are produced as zymogens, and are then activated when deeded Most of them are synthetized and stored in the pancreas, and then secreted into the small intestine, where they are activated by removal of small peptide sections They must be stored as zymogens otherwise they would damage the pancreas 42 Reversible covalent modulation In protein modification a chemical group is covalently added to or removed from the protein. This modification will activate or deactivate the enzyme A common example is phosphorylation of an enzyme by addition of a phosphate group to serine, threonine, or tyrosine mediated by a kinase The phosphorylation is reversible, and phosphatases typically catalyze the of the phosphate group removal 43 Inhibition of enzyme activity Chemicals can bind to enzymes and eliminate or drastically reduce catalytic activity Classify enzyme inhibitors on the basis of reversibility and competition • Irreversible inhibitors - bind tightly to the enzyme and thereby prevent formation of the E-S complex • Reversible competitive inhibitors - often structurally resemble the substrate and bind at the normal active site • Reversible noncompetitive inhibitors - usually bind at someplace other than the active site. Binding is weak and thus, inhibition is reversible 44 Irreversible inhibitors Irreversible enzyme inhibitors bind very tightly to the enzyme Binding of the inhibitor to one of the R groups of an aa in the active site • This binding blocks the active site the enzyme-substrate complex can’t form • The inhibitor interferes with the catalytic group of the active site eliminating catalysis Irreversible inhibitors include: • • • Arsenic Snake venom Nerve gas 45 Irreversible “Suicide” inhibitors Special type of irreversible inhibition of enzyme activity. Also known as mechanism based inactivation The inhibitor makes use of the enzyme’s own reaction mechanism to inactivate it In suicide inhibition, the structural analog binds to the active site of the enzyme and is converted to a more effective inhibitor with the help of the The substrate-like compound initially binds with the enzyme and the first few steps of the reaction are catalysed This new product irreversibly binds to the enzyme and inhibits further reactions 46 Suicide inhibitors - Example Aspirin acts as an acetylating agent where an acetyl group is covalently attached to a serine residue in the active site of the cyclooxygenase enzyme, rendering it inactive preventing inflammation, swelling, pain and fever 47 Suicide inhibitors - Example Clavulanic acid, which inhibits β-lactamase clavulanic acid covalently bonds to a serine reside in the active site of the β-lactamase, restructuring the clavulanic acid molecule, creating a much more reactive species that attacks another amino acid in the active site, permanently inactivating it, and thus inactivating the enzyme β-lactamase. 48 Reversible competitive inhibitors Reversible, competitive enzyme inhibitors are also called structural analogs • Molecules that resemble the structure and charge distribution of a natural substrate • Resemblance permits the inhibitor to occupy the enzyme active site • Once the inhibitor is at the active site, the enzyme activity is inhibited Inhibition is competitive because the inhibitor and the substrate compete for binding to the active site 49 Reversible competitive inhibitors Km increases 50 Reversible, noncompetitive inhibitors Reversible, noncompetitive enzyme inhibitors bind to R groups of amino acids or to the metal ion cofactors This binding is weak Enzyme activity is restored when the inhibitor dissociates from the enzymeinhibitor complex These inhibitors: • Do not bind to the active site No effect on Km • Do modify the shape of the active site once bound elsewhere in the structure 51 Comparison between competitive & noncompetitive inhibitors 52 Uses of enzymes in medicine Diagnostic – biomarker levels altered with disease • Heart attack: Lactate dehydrogenase, Creatine phosphate • Acute myocardial infarction: Creatine kinase, Myoglobin, Troponin I • Pancreatitis: Amylase, Lipase Analytical reagents – enzyme used to measure another substance • Urea converted to NH3 via urease: Blood urea nitrogen measured Replacement therapy • Administer genetically engineered β-glucocerebrosidase for Gaucher’s disease 53 Enzymes in diagnostic use 54 Isoenzymes Isoenzymes or isozymes - multiple forms of same enzyme (different structures) that catalyze the same chemical reaction Different chemical & physical properties: • Electrophoretic mobility • Kinetic properties • Amino acid sequence • Amino acid composition Example: lactate dehydrogenase is a tetramer with 5 isomers (different Km and Vmax) 55 Thank you 56