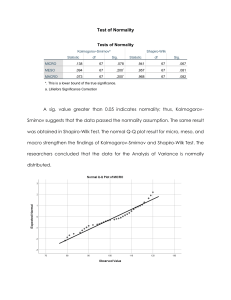
Normality Assumption, 1 Model Diagnostics: Normality Assumption Scenario Data and Instructions Download the Healthy_SCH23.xlsx subset of the Healthy Minds 2021-2022 data file from D2L. Upload the Healthy_SCH23.xlsx to SAS Studio. Upload your SAS code to D2L. Upload a Word document that contains ONLY the following: Task 2. The raw critical values for skewness and kurtosis. Task 3.The summary table for Flourishing. Task 7. The summary table for the residuals. Task 8. The summary table for the residuals. Your answers to the questions in Task 9. The portion of the APA results section that deals with descriptive statistics, Pearson correlations, and normality issues. Refer to the example in the Model Diagnostics notes. Hypothesis Feelings of loneliness can be predicted from a combination of depression, anxiety, and positive mental health feelings. Variable LoneSc Item Coding Sum of loneliness variables. Higher scores indicate greater feelings of loneliness. Depression Sum of depression variables. Higher scores indicate greater feelings of depression. Sum of anxiety variables. Higher scores indicate greater feelings of anxiety. Min = 3 Max = 9 Min = 9 Max = 36 Min = 7 Max = 28 Min = 8 Max = 56 Anxiety Flourishing Sum of positive mental health items. Higher scores indicate more positive mental health feelings. LAB 3: LAB 1: Check for impossible values. Explore variable transformations. Evaluate normality assumption of the initial regression model. Check for outliers. LAB 2: Evaluate linearity assumption of the regression model. Check for collinearity among the predictors in the regression model. Evaluate homoscedasticity assumption of the regression model. Re-evaluate normality assumption of the final regression model. Obtain bootstrap confidence intervals for the regression model. Normality Assumption, 2 Task 1. Bring in the Data and Create the UpdatedHealthy Data Set. Download the LoneSC – Data Management.sas file from D2L. Upload the LoneSC – Data Management.sas file to SAS Studio. Double-click on the LoneSC – Data Management.sas to edit it. Press F3 or the run icon to submit the code. This will create a data set called UpdatedHealthy. Task 2. Determine Skewness and Kurtosis Critical Values Download the Standardized Skewness and Kurtosis – Updated.xlsx file from D2L. Enter the sample size of 262 to obtain the critical values for skewness and kurtosis. We want to determine whether skewness and kurtosis values are significant. Fortunately, the sample size is 262 for all of the variables we are analyzing. This allows us to convert the Z score critical values to critical values for the raw scale. Once we obtain the values, we can compare all of the skewness and kurtosis values to the same raw scale values. This approach is NOT appropriate if there are different sample sizes for each variable. Write the raw scale critical values for skewness on the guidelines shown below. Negatively Skewed Symmetric -Critical Value 0 Positively Skewed +Critical Value Write the raw scale critical values for kurtosis on the guidelines shown below. Platykurtic -Critical Value Mesokurtic 0 Leptokurtic +Critical Value Normality Assumption, 3 Task 3. Evaluate the Flourishing Distribution ► Tasks and Utilities ► Tasks ►Statistics ►Distribution Analysis Data Tab Settings DATA: work.UpdatedHealthy Analysis Variables: Flourishing Options Tab Settings CHECKING FOR NORMALITY ●Histogram and goodness-of-fit tests ● Add inset statistics ● Normal quantile-quantile plot ● Add inset statistics INSET STATISTICS ● Number of Observations ● Goodness-of-fit test ● Mean ● Skewness ● Kurtosis Edit the Code ● Replace GoodnessOfFit with TestsForNormality. Then, press F3 or the run icon to submit the code. Summarize the distribution analysis for Flourishing in the table shown below. Since Flourishing does not have a normal distribution, variable transformations should be considered. ● Add the following variables to the Flourishing – Data Management.sas program. ● Press F3 or the run icon to submit the code. log_Flourishing = log(Flourishing); sqr_Flourishing = Flourishing**2; ●Save the updated Flourishing_Data Management.sas program. ● Repeat the distribution analysis for Log_Flourishing and sqr_Flourishing. Summarize the Results ● Complete the table shown below to summarize the distribution analyses you conducted. Flourishing Shapiro-Wilk W Shapiro-Wilk p value Conclusion Skewness Conclusion Kurtosis Conclusion Which representation is best? Log_Flourishing Sqr_Flourishing Normality Assumption, 4 Task 4. Evaluate the LoneSc Distribution The output needs to determine the best representation for LoneSc has been summarized below and the graphs are shown on the next page. You just need to review the summary table and the graphs to determine the best representation for LoneSc. The instructions are provided here to demonstrate all of the steps involved for a real research project. ► Tasks and Utilities ► Tasks ►Statistics ►Distribution Analysis Data Tab Settings DATA: work.UpdatedHealthy Analysis Variables: LoneSc Options Tab Settings CHECKING FOR NORMALITY ●Histogram and goodness-of-fit tests ● Add inset statistics ● Normal quantile-quantile plot ● Add inset statistics INSET STATISTICS ● Number of Observations ● Goodness-of-fit test ● Mean ● Skewness ● Kurtosis Edit the Code ● Replace GoodnessOfFit with TestsForNormality. Then, press F3 or the run icon to submit the code. The distribution analysis for LoneSc has been summarized in the table shown below. Since LoneSc does not have a normal distribution, variable transformations should be considered. ● Add the following variables to the LoneSc – Data Management.sas program. log_lonesc = log(lonesc); sqr_lonesc = lonesc**2; ● Press F3 or the run icon to submit the code. ● Repeat the distribution analysis for Log_LoneSc and sqr_lonesc. Summarize the Results ● Complete the table shown below to summarize the distribution analyses you conducted. LoneSc Log_LoneSc Sqr_LoneSc Shapiro-Wilk W Shapiro-Wilk p value Conclusion .919 < .0001 Nonnormal .903 < .0001 Nonnormal .894 < .0001 Nonnormal Skewness Conclusion .006 Symmetric -0.48 Negatively Skewed 0.43 Positively Skewed Kurtosis Conclusion -1.09 Platykurtic -0.83 Platykurtic -1.02 Platykurtic Which representation is best? Normality Assumption, 5 Normality Assumption, 6 Task 5. Evaluate the Depression Distribution The output needs to determine the best representation for Depression has been summarized below and the graphs are shown on the next page. You just need to review the summary table and the graphs to determine the best representation for depression. The instructions are provided here to demonstrate all of the steps involved for a real research project. ► Tasks and Utilities ► Tasks ►Statistics ►Distribution Analysis Data Tab Settings DATA: work.UpdatedHealthy Analysis Variables: Depression Options Tab Settings CHECKING FOR NORMALITY ●Histogram and goodness-of-fit tests ● Add inset statistics ● Normal quantile-quantile plot ● Add inset statistics INSET STATISTICS ● Number of Observations ● Goodness-of-fit test ● Mean ● Skewness ● Kurtosis Edit the Code ● Replace GoodnessOfFit with TestsForNormality. Then, press F3 or the run icon to submit the code. The distribution analysis for depression has been summarized in the table shown below. Since depression does not have a normal distribution, variable transformations should be considered. ● Add the following variables to the Depression – Data Management.sas program. log_Depression = log(Depression); sqr_Depression = Depression**2; ● Press F3 or the run icon to submit the code. ● Repeat the distribution analysis for Log_Depression and sqr_Depression. Summarize the Results ● Complete the table shown below to summarize the distribution analyses you conducted. Shapiro-Wilk W Shapiro-Wilk p value Conclusion Skewness Conclusion Kurtosis Conclusion Depression .953 < .0001 Nonnormal 0.58 Positively skewed -0.44 Mesokurtic Log_Depression .981 .0015 Nonnormal -0.08 Symmetric -0.72 Platykurtic Sqr_Depression .876 < .0001 Nonnormal 1.17 Positively skewed 0.68 Leptokurtic Which representation is best? I would use log depression, because the skewness is symmetric, and ketosis is only slightly platykurtic. Depression has a positively skewed Skewness value that’s slightly more intense (not sure how else to describe it) than the platykurtic value of log depression. Normality Assumption, 7 Normality Assumption, 8 Task 6. Evaluate the Anxiety Distribution The output needs to determine the best representation for anxiety has been summarized below and the graphs are shown on the next page. You just need to review the summary table and the graphs to determine the best representation for anxiety. The instructions are provided here to demonstrate all of the steps involved for a real research project. ► Tasks and Utilities ► Tasks ►Statistics ►Distribution Analysis Data Tab Settings DATA: work.UpdatedHealthy Analysis Variables: Anxiety Options Tab Settings CHECKING FOR NORMALITY ●Histogram and goodness-of-fit tests ● Add inset statistics ● Normal quantile-quantile plot ● Add inset statistics INSET STATISTICS ● Number of Observations ● Goodness-of-fit test ● Mean ● Skewness ● Kurtosis Edit the Code ● Replace GoodnessOfFit with TestsForNormality. Then, press F3 or the run icon to submit the code. The distribution analysis for Anxiety has been summarized in the table shown below. Since Anxiety does not have a normal distribution, variable transformations should be considered. ● Add the following variables to the Anxiety – Data Management.sas program. Then, press F3 or the run icon to submit the code. log_Anxiety = log(Anxiety); sqr_Anxiety = Anxiety**2; ● Repeat the distribution analysis for Log_Anxiety and sqr_Anxiety. Summarize the Results ● Complete the table shown below to summarize the distribution analyses you conducted. Shapiro-Wilk W Shapiro-Wilk p value Conclusion Skewness Conclusion Kurtosis Conclusion Anxiety .956 < .0001 Nonnormal 0.27 Symmetric -0.97 Platykurtic Log_Anxiety .955 < .0001 Nonnormal -0.37 Negatively Skewed -0.75 Platykurtic Sqr_Anxiety .903 < .0001 Nonnormal 0.79 Positively Skewed -0.455 Mesokurtic Which representation is best? I would use Anxiety, not any of the transformed data because Anxiety has the less severe variables that indicate skewness or kurtosis of the two that don’t have both non symmetrical/mesokurtic data. Normality Assumption, 9 Normality Assumption, 10 Task 7. Evaluate Residuals of Linear Regression Model Using Original Variables Obtain Residuals from the Regression Model: ► Tasks and Utilities ► Tasks ►Linear Models ►Linear Regression Data Tab Settings DATA: work.UpdatedHealthy Dependent Variable: LoneSc Continuous Variables: Flourishing, Depression, Anxiety Model Tab Settings Model Effects: Intercept, Flourishing, Depression, Anxiety Options Tab Settings You can use the default and just ignore the graphs or you can unselect all of the plots. Output Tab Settings ● Create observationwise statistics data set Data Set Name: work.Aresiduals ● Residual Press F3 or the run icon to submit the code. Analyze the Residuals from the Regression Model ► Tasks and Utilities ► Tasks ►Statistics ►Distribution Analysis Data Tab Settings DATA: work.Aresiduals Analysis Variables: r_ Options Tab Settings CHECKING FOR NORMALITY ●Histogram and goodness-of-fit tests ● Add inset statistics ● Normal quantile-quantile plot ● Add inset statistics INSET STATISTICS ● Number of Observations ● Goodness-of-fit test ● Mean ● Skewness ● Kurtosis Edit the Code ● Replace GoodnessOfFit with TestsForNormality. Press F3 or the run icon to submit the code. Complete the table below based on the output generated. Shapiro-Wilk W Shapiro-Wilk p value Conclusion Residuals using Original Variables .98 <.0001 Nonnormal Skewness Conclusion .49 Positively skewed Kurtosis Conclusion .06 Mesokurtic Normality Assumption, 11 Task 8. Evaluate the Residuals of the Linear Regression Model Using Transformed Variables Obtain Residuals from the Regression Model: ► Tasks and Utilities ► Tasks ►Linear Models ►Linear Regression Data Tab Settings Dependent Variable: LoneSc Continuous Variables: Sqr_Flourishing, Log_Depression, Anxiety Model Tab Settings Model Effects: Intercept, Sqr_Flourishing, Log_Depression, Anxiety Options Tab Settings You can use the default and just ignore the graphs or you can unselect all of the plots. Output Tab Settings ● Create observationwise statistics data set Data Set Name: work.Bresiduals ● Residual Press F3 or the run icon to submit the code. Analyze the Residuals from the Regression Model ► Tasks and Utilities ► Tasks ►Statistics ►Distribution Analysis Data Tab Settings DATA: work.Bresiduals Analysis Variables: r_ Options Tab Settings CHECKING FOR NORMALITY ●Histogram and goodness-of-fit tests ● Add inset statistics ● Normal quantile-quantile plot ● Add inset statistics INSET STATISTICS ● Number of Observations ● Goodness-of-fit test ● Mean ● Skewness ● Kurtosis Edit the Code ● Replace GoodnessOfFit with TestsForNormality. Press F3 or the run icon to submit the code. Complete the table below based on the output generated. Shapiro-Wilk W Shapiro-Wilk p value Conclusion Residuals using Transformed Variables .99 .02 Nonnormal Skewness Conclusion .45 Positively Skewed Kurtosis Conclusion .35 Mesokurtic Normality Assumption, 12 Task 9. Decide which model(s) will be used going forward (i.e., in lab 2 and lab 3). Which model should be used going forward (i.e., in lab 2 and lab 3)? Why? You want to use the original going forward because even after transformation, the subsequent analyses did not yield better results in some way or another so we should stick with the original. APA Results Section Results See Table 1 for descriptive statistics and Table 2 for Pearson correlations among the variables. Skewness, kurtosis, and Shapiro-Wilk’s W tests were evaluated to determine whether variable transformations should be considered (see Table 1). Flourishing was negatively skewed log_flourishing and sqr_florishing was created. Depression was positively skewed, so log_depression and Sqr_depression was created. The residual distribution from the regression model using the original scaling of all variables (Shapiro-Wilk’s W = 0.98, p <.0001, Skewness = .49, Kurtosis = .06) and the residual distribution from the regression model including the transformed variables (Shapiro-Wilk’s W = 0.99, p = .02, Skewness = 0.45, Kurtosis = .35) were similar, so the original scaling of variables was used to make interpretation easier. Table 1 Descriptive Statistics for All Variables (N = 262) Variable Shapiro-Wilk Test for Normality W p M SD Skewness Kurtosis Loneliness 6.07 1.96 .01 -1.09 .92 <.0001 Flourishing 42.54 9.73 -1.13 1.05 .91 <.0001 Depression 18.82 6.84 .58 -.44 .95 <.0001 Anxiety 18.02 7.10 .27 -.97 .96 <.0001 Table 2 Pearson Correlations for All Variables (N = 262) 1 1. Loneliness 2. Flourishing 3. Depression 4. Anxiety **p < .01. -- 2 3 4 -.61** <.0001 -- .60** <.0001 -.511** <.0001 -- .52** <.0001 -.40** <.0001 .76** <.0001 -- Normality Assumption, 13