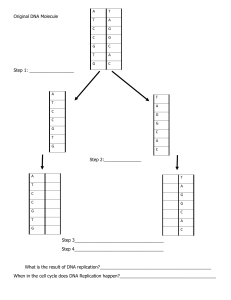
Overview General Features of DNA Replication Enzymology Detailed Mechanism GENERAL FEATURES General Features of DNA Replication Semiconservative Replication • Watson-Crick Model Two parental strands separate and that each then serves as a template for a new progeny strand Each daughter duplex has one parental strand and one new strand • Other potential mechanisms: Conservative replication Two parental strands stay together and somehow produce another daughter helix with two completely new strands Dispersive replication The DNA becomes fragmented so that new and old DNAs coexist in the same strand after replication General Features of DNA Replication Semidiscontinuous Replication • Semidiscontinuous Synthesis of one of the new strands is continuous; synthesis of the other is discontinuous, with the DNA being made in short pieces. o The lagging strand is replicated discontinuously as 1–2 kb Okazaki fragments in the opposite direction The discontinuity of synthesis of the lagging strand comes about because its direction of synthesis is opposite to the direction in which the replicating fork is moving Both strands grow in the 5’→3’ Other models: Continuous (a) o As the replicating fork moves to the right, both strands are replicated continuously in the same direction, left to right. o The top strand grows in the 3’→5’ direction, the bottom strand in the 5’→3’ direction. Discontinuous (c) o Both leading and lagging strands are made in short pieces (i.e., discontinuously); both strands grow in the 5’→3’ direction General Features of DNA Replication Priming of DNA Synthesis • Primer A piece of nucleic acid, 10–12 nt long, that the polymerase can “grab onto” and extend by adding nucleotides to its 3’end Not DNA, but a short piece of RNA Process of priming: 1. First, a replicating fork opens up 2. Next, short RNA primers are made 3. Next, DNA polymerase adds deoxyribonucleotides to these primers, forming DNA, as indicated by the arrows General Features of DNA Replication Bidirectional Replication • Replication bubble DNA replication begins with the creation of a “bubble”—a small region where the parental strands have separated and progeny DNA has been synthesized. As the bubble expands, the replicating DNA begins to take on the theta shape Contains two replicating forks that move in opposite directions away from the origin • Replicon DNA under the control of one origin of replication General Features of DNA Replication Rolling Circle Replication • Certain circular DNAs replicate, not by the θ mode, but by a mechanism called rolling circle replication • This mechanism is sometimes called the σ mode • One strand of a double-stranded DNA is nicked and the 3’-end is extended, using the intact DNA strand as template. • This displaces the 5’-end. ENZYMOLOGY Enzymology Three DNA Polymerases in E. coli • Pol I A single 102-kD polypeptide chain Three activities: 1. DNA polymerase 2. 3’ → 5’ exonuclease activity (proofreading) 3. 5’ → 3’ exonuclease activity Can be cleaved into two polypeptides 1. Large fragment (Klenow fragment) o polymerase and 3’ → 5’ exonuclease activity o α-helices: one is part of the “fingers” domain; the other is part of the “thumb” domain o β-pleated sheet: part of the “palm” domain 2. Small fragment - 5’ → 3’ exonuclease activity Enzymology Three DNA Polymerases in E. coli • Pol II and Pol III Gefter and colleagues used N-ethylmaleimide to knock out pol III so its activity could be measured as the difference between the activities in the presence and absence of the inhibitor o Three conclusions: 1. The dnaE gene encodes pol III. 2. The dnaE gene does not encode pol II, and pol II and pol III are therefore separate activities. 3. Because defects in the gene encoding pol III interfere with DNA replication, pol III is indispensable for DNA replication. • Of the three DNA polymerases in E. coli cells, pol I, pol II, and pol III, only pol III is required for DNA replication. Thus, this polymerase is the enzyme that replicates the bacterial DNA. Enzymology Three DNA Polymerases in E. coli • The Pol III Holoenzyme Carries out the elongation of primers to make both the leading and lagging strands of DNA Moves at a rate of 1000 nt/sec Multisubunit – has 10 polypeptides Core is composed of: 1. α – DNA polymerase 2. ε – 3’ → 5’ exonuclease activity 3. Θ - Stimulates ε exonuclease Enzymology Fidelity of Replication • Proofreading Fidelity is greatly increased by the proofreading mechanism of pol III (and pol I) Proofreading allows the polymerase to use another mechanism by which to get the base pairing right. Rate of error after the first and second pass: 𝟏𝟎−𝟏𝟎 − 𝟏𝟎−𝟏𝟏 o “Imperfect” fidelity allows for mutations, some of which help the organism to adapt to a changing environment through evolution. • RNA primers Primers are made with more errors, because their synthesis is not subject to proofreading Making primers out of RNA guarantees that they will be recognized, removed, and replaced with DNA by extending the neighboring Okazaki fragment. Enzymology Multiple Eukaryotic DNA Polymerases • Probable Roles of Some Eukaryotic DNA Polymerases Enzyme Probable role DNA polymerase α Priming of replication of both strands DNA polymerase δ Elongation of lagging strand DNA polymerase ε Elongation of leading strand DNA polymerase β DNA repair DNA polymerase γ Replication of mitochondrial DNA • Proliferating cell nuclear antigen (PCNA) Processivity factor Enhances the processivity of polymerase δ by a factor of 40. Enzymology Strand Separation • Helicase Harnesses the chemical energy of ATP to separate the two parental DNA strands at the replicating fork Without a functional helicase, the fork cannot move, and DNA synthesis must halt immediately. DnaB o Helicase that unwinds the DNA double helix during E. coli DNA replication o Encoded by E. coli dnaB gene o Stimulated by DnaG and SSB Single-Strand DNA-Binding Proteins (SSBs) • A class of proteins that also participate in DNA strand separation during replication by binding selectively to single-stranded DNA as soon as it forms and coat it so it cannot anneal to re-form a double helix • SSBs also protect DNA from degradation • Stimulate their homologous DNA polymerases • RF-A – human SSB Enzymology Topoisomerases • DNA gyrase introduce transient single- or double-stranded breaks into DNA and thereby allow it to change its shape, or topology. serves the swivel function; relaxes positive supercoils that form in replicating DNA ahead of the fork • The more you twist, the more supercoiling you will observe: one superhelical turn for every full twist you introduce. • Classification: 1. 2. Type I topoisomerases - introduce temporary singlestranded breaks, e.g. topoisomerase I of E. coli Type II - break and reseal both DNA strands, e.g. DNA gyrase of E. coli DETAILED MECHANISM Initiation Priming in E. coli • Primosome Collection of proteins needed to make primers for a given replicating DNA DnaB (helicase), DnaG (primase) Two functions: • oriC 1. 2. Operates repeatedly in priming Okazaki fragment synthesis to build the lagging strand Serves as the helicase that unwinds DNA to provide templates for both the leading and lagging strands Origin of replication 245 bp long Includes four “9-mers” with the consensus sequence TTATCCACA o Two of these are in one orientation, and two are in the opposite orientation o dnaA boxes: binding sites for the DnaA DnaA and DnaC - assists the binding of DnaB RNA polymerase - synthesize a short piece of RNA that creates an R loop HU protein - a small basic DNA-binding protein that can induce bending in doublestranded DNA Initiation Priming in Eukaryotes • More complex than bacterial replication • Origin of replication in SV40 DNA replication proceeded bidirectionally from the origin Ori sequence/ ori core o 64 bp long o Essential elements: 1. 17-bp region consisting only of A–T pairs - facilitates melting of the nearby palindrome region 2. Four pentamers (5’-GAGGC-3’) – the binding site for large T antigen (helicase activity unwinds the DNA and prepares the way for primer synthesis) 3. 15-bp palindrome - the earliest region melted during DNA replication The primase in eukaryotic cells associates with DNA polymerase 𝜶 Initiation Priming in Eukaryotes • Origin of replication in yeast Contained within autonomously replicating sequence (ARS1), w/ four important regions: 1. 2. 3. 4. A - 15 bp long and contains an 11-bp consensus sequence: B1 - binds with origin recognition complex B2 - melting of DNA occurs B3 - allow for an important DNA bend within ARS1 Elongation Speed of Replication • Pol III holoenzyme - the enzyme that carries out elongation in E. coli • In vivo – 1000 nt/sec • In vitro - 730 nt/sec • Highly processive Processivity - ability of the enzyme to stick to its job a long time without falling off and having to reinitiate The holoenzyme did not dissociate from the template throughout the process of elongation of the primer by at least 30 kb. Elongation The Pol III Holoenzyme and Processivity of Replication • Pol III Holoenzyme - E. coli The pol III core (αεθ) by itself is a very poor polymerase β-subunit/ β-clamp o “Sliding clamp” - confers processivity on the holoenzyme; that holds the enzyme on the template for a long time o Binds both the core complex and DNA o α is the core subunit to which β binds o PCNA – eukaryotic processivity factor γ complex o “Clamp loader”- helps the β-subunit join the preinitiation complex (core plus the DNA template) o Includes γ-, δ-, δ‘-, χ-, and ψ-subunits. o Forms the processive αδβ complex o ATP-dependent process o Also a clamp unloader Elongation The Pol III Holoenzyme and Processivity of Replication • Lagging Strand Synthesis One core of the pol III holoenzyme is responsible for continuous synthesis of the leading strand; other performs discontinuous synthesis of the lagging strand The γ complex loads the β clamp onto a primed DNA template. The γ complex, or clamp loader, dissociates from the β clamp. The core associates with the clamp. The core and clamp cooperate to processively synthesize an Okazaki fragment, leaving just a nick between two Okazaki fragments The polymerase core dissociates from the clamp. The γ complex reassociates with the β clamp. The γ complex acts as a clamp unloader, removing the β clamp from the template. Now it is free to repeat the process, recycling to another primer on the template. Termination • E. coli (circular) The two replication forks approach each other in the terminus region o Contains 22-bp terminator sites: Ter sites (TerA-TerF) Bind Tus proteins (terminus utilization substance) This leaves the two daughter duplexes entangled. • Eukaryotes (linear) - fill in the gaps left by removing primers at the 5’-ends of the linear chromosomes Decatenation: Disentangling Daughter DNAs • Catenanes - interlocking rings • Topo IV – decatenating enzyme • Topo II – eukaryotic decatenating enzyme Termination Termination Termination in Eukaryotes Termination Termination in Eukaryotes • Telomere maintenance Ends of eukaryotic chromosomes that are composed of repeats of short, GCrich sequences. o Tetrahymena: TTGGGG/AACCCC o Vertebrates (including humans): TTAGGG/AATCCC G-rich strand o Made by telomerase Ribonucleoprotein with essential RNA and protein subunits Telomerase RNA: serve as the template for telomere synthesis; acts as a reverse transcriptase o Added at the very 3’-ends of DNA strands C-rich strand o Priming then occur within these telomeres to make the C-rich strand Termination Termination in Eukaryotes • The Mammalian Telomere-Binding Proteins: Shelterin The group of telomere-binding proteins that “shelters” the telomere. Protect the telomeres from degradation, and also hide the telomere ends from the DNA damage factors that would otherwise recognize them as chromosome breaks 1. TRF1 - bind to the double-stranded telomeric repeats 2. TRF2 - bind to the double-stranded telomeric repeats 3. TIN2 - organizes shelterin by facilitating interaction between TRF1 and TRF2, and tethering TPP1/POT1 to TRF1 and TRF2. 4. POT1 - binds to the single-stranded 3’-tail of the telomere, beginning at a position just 2 nt away from the 5’-end of the other strand; 5. TPP1 – POT1-binding protein 6. RAP1 - binds to the telomere by interacting with TRF2 Termination Telomere Structure and Telomere-Binding Proteins in Lower Eukaryotes • Lower Eukaryotes Schizosaccharomyces pombe o Taz1 – like TRF; binds through Rap1 and Poz1 to a dimer of Tpz1 and Pot1 Saccharomyces cerevisiae 1. Rap1 – like TRF; partners with Rif1 and Rif2 2. Cdc13, Stn1, Ten1 - binds to the single-stranded telomeric tail Oxytricha 1. TEBPα and TEBPβ - evolutionarily related to POT1 and TPP1; bind to the singlestranded 3’-end of the organism’s telomeres and protect them from degradation Termination Telomere Structure and Telomere-Binding Proteins in Lower Eukaryotes • The Role of Shelterin in Suppressing Inappropriate Repair and Cell Cycle Arrest in Mammals DNA repair activities Function Repressed by 1. HDR (Homologous-directed repair) Promotes homologous recombination b/w telomeres on separate chromosomes, or between telomeres and other chromosomal regions TRF2 and POT1 2. NHEJ (Nonhomologous end-joining) Leads to chromosome fusion TRF2 in the G1 phase TRF2 and POT1 in the G2 phase 3. ATM (ataxia telangiectasia mutated) kinase pathway Responds directly to unprotected DNA ends TRF2 4. ATR (ataxia telangiectasia and Rad3 related) kinase pathway Responds to single-stranded DNA end that appears when one DNA strand at a chromosome break POT1 Pathways THANK YOU!