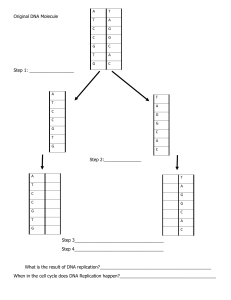
Biology Chapter 12/13 S-G DNA Replication Blue=The Scientists Green=DNA Replication Red=RNA and Protein Synthesis ● The Genetic Material ○ During the 20th century, scientists were determined to discover which part of a cell held the genetic material ○ Almost all scientists thought that the genetic material was proteins ● Griffth ○ In 1928, Griffith was attempting to develop a vaccine against pneumonia ○ Experiment ■ Griffith used 2 types of Bacteria ■ S strain bacteria ● Smooth and Harmful ■ R strain bacteria ● Rough and harmless ■ When Griffith gave the R strain bacteria the rat lived, as expected when he gave the S strain bacteria the mouse died ■ When Griffith gave the heat-killed S strain the mouse lived because the proteins denatured ■ However, when Griffith mixed the R strain bacteria with the heat-killed S strain the mouse died ■ Meaning that there was some form of genetic material left ○ Griffith was unable to identify the transformation that took place, so he was not able to identify the genetic material as DNA ○ Transformation ■ Process in which one strain of bacteria is changed by a gene or genes from another strain of bacteria ● Oswald Avery ○ 1944 ○ Researchers do not know whether the molecule that holds genetic information is, proteins, lipids, carbohydrates, RNA, or DNA ○ Avery performs Griffth’s experiment with a twist ○ Virus ■ A very tiny, simple nonliving particle that is made of protein and nucleic acids (DNA or RNA). Not visible to the naked eye only with a powerful electron microscope ○ Capsid ■ The protein coat that protects the genetic material and helps viruses attach and enter the host cell ○ 4 shapes of a virus ■ Crystals ■ Spheres ■ Cylinders ■ Spacecraft ○ In Avery’s experiments, they used identical extracts from heat-killed S cells, and they used hydraulic enzymes to kill either protein, DNA, or RNA. The treatments were then mixed with R cells. Using the process of elimination they were able to prove that DNA was the hereditary material. Furthermore, his colleagues were able to isolate DNA and show that it possessed the same transforming ability as the heat-treated extract ● Hershey and Chase ○ Used two different bacteriophages ○ Bacteriophages ■ Viruses that infect bacteria ○ Tagged the bacteriophages using a radioactive isotope (trackers) ■ Phosphate 32 to trace DNA ■ Sulfur 35 to trace proteins ○ When the viruses are attached to the bacterium. There was no radioactivity from the Sulfur 35 bacteriophage, meaning that the bacteriophage's genetic info was not proteins. However, when the Phosphate 32 bacteriophage was used, there was radioactivity in the bacterium meaning that the genetic information is DNA, not protein ○ Concluded, that DNA was the genetic material, proteins weren’t, and it is the genetic material for ALL LIVING CELLS ● DNA structure ○ Nucleotide ■ Monomer of nucleic acids made up of a 5-carbon sugar, a phosphate group, and a nitrogenous base ○ Nitrogenous bases ■ Adenine ■ Thymine (IN DNA ONLY) ■ Guanine ■ Cytosine ■ Uracil (ONLY IN RNA) ○ A-T, G-C, or A-U (IN RNA) ○ DNA has a sugar-phosphate backbone ● Chargaff’s Rule ○ If I have a certain number of Cytosines I will have the same number of Guanines. This applies to As and T’s ● Rosalind Franklin ○ 1950 ○ Used X-Ray Diffraction to take photos of DNA ○ Coiled (Forming Helix) ○ Double-Stranded ○ Nitrogenous bases are in the center ○ Photo-51 ○ However, her data was supposedly used without credit, and she was often ignored because she was a woman ● Watson and Crick ○ Francis Crick ■ British physicist ○ James Watson ■ American Biologist ○ Were building a 3D model of DNA ○ Used Franklin’s X-Ray without permission and realized that the structure was a Double Helix ○ Ended up winning the Nobel Prize ● DNA Replication ○ Unwinding ■ Parental DNA molecule is “unzipped” (the weak H bonds are broken b/w paired bases) ○ Complementary Base pairing ■ New complementary nucleotides are positioned by the rules of base pairing. (A =T, C = G) ○ Joining (Semi conservative model) ■ Complementary (matching) nucleotides join to form new strands. Each daughter DNA molecule contains an old strand and a new strand. ○ Unwinding ■ Enzymes unwind DNA ○ ○ ○ ○ ○ ■ Enzymes split “unzip” double helix ■ The enzyme, DNA polymerase, finds and attaches the corresponding Nitrogenous base ■ Each old/original strand serves as a template and is matched up with a new strand of DNA ■ New helixes wind back up DNA Structure ■ Antiparallel Strands ■ The 2 strands of DNA run in opposite directions (Antiparallel) ■ Nitrogenous bases connect at the center Basic Vocab ■ Origin of Replication ● Center of a Replication bubble ● The place where the DNA polymerase starts working ■ Replication Bubble ● A portion of the strand that is getting copied DNA Polymerase ■ Is only able to add nucleotides to the free 3’ end of a growing DNA strand (Hydroxyl Group) Leading Strand■ used by polymerases as a template for a continuous complementary strand working towards the replication fork unzipping it Lagging strand ■ Copied away from the replication fork in short segments called Okazaki fragments ○ DNA Polymerase and primers ■ The polymerase cannot initiate the synthesis of a nucleotide ● Can only add nucleotides to the end of an existing chain ■ To start a new chain requires a primer ● A short segment of RNA ○ Enzymes and important vocab ■ Topoisomerases ● Helps Supercoils/Winding (Prevents Tangling) ■ Helicase ● Unzips the double helix ■ Single Strand proteins ● Keeps the Hydrogen bonds separated ■ Ligase ● Joins DNA molecules together ■ Primases ● Put RNA primers where DNA polymerase is going to bind and make new strands ■