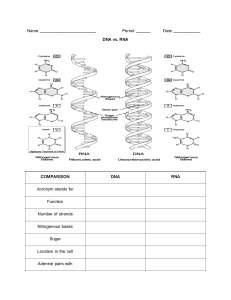
MODULE 2│PHARMBIOSCI 3 BIOCHEMISTRY BIOCHEMISTRY • II. AA with uncharged Polar groups generally, deals with the physical and chemical properties of compounds that make up the smallest unit of life and how these compounds undergo processes and relate it with how it affects the daily function of human beings. R–H PROTEINS • constitute 70% of the organic matter of cell. The simplest unit is amino acid, and proteins are polymers of these repeating units linked together With OH Glycine Gly G Serine Ser S Threonine Thr T Tyrosine Tyr Y Asparagine Asn N Glutamine Gln Q Cysteine Cys C AMINO ACIDS • organic molecule containing both carboxyl and amino functional groups • there are about 300 amino acids occurring in nature, but only about 20 are commonly occurring and are constituents of proteins. Except for proline, an imino acid, all 19 are alpha amino acid with the structure shown below: With amide • • Sulfur containing • Cysteine and Methionine Glycine • Acts as inhibitory neurotransmitter; antagonized by strychnine With SH CLASSIFICATION OF AMINO ACIDS I. AA with Non-polar R-groups Alanine Ala A Valine Val V Leucine Leu L Isoleucine Ile I Proline Pro P Phenylalanine Phe F III. AA with charged Polar groups Glutamic Glu Acid Alkyl R group E Aspartic Acid Asp D Lysine Lys K Histidine His H Arginine Arg R Alkyl R group A. PROTEIN STRUCTURES • Aromatic group Tryptophan Trp W Structurally organized into four levels (with each level having its proper specificity): PRIMARY, SECONDARY, TERTIARY, and QUATERNARY 1. Primary Structure • S– containing • Methionine Met M • • • * Proline is an alpha imino acid, it doesn’t contain a free amino group Module 2 – Biochemistry Page 1 of 19 The simplest level of structural organization composed of the amino acid resides linked through peptide bonds The sequence is written from left to right (N–Terminal to C–Terminal amino acid In an electric field Positively charge proteins →Cathode (-) Negatively charged proteins → Anode (+) Proteins at the isoelectric Ph → no migration since net charge is 0 RJAV 2022 • 2. Secondary Structure • α-Helix • A helical configuration of the polypeptide chain formed by the Hydrogen bonding between the peptide groups of every first and fourth AA residues. • β-Sheets • Pleated sheet structure resembling an accordion • Can be in a form of a parallel or an antiparallel chain • Examples are Keratin, Collagen, and Fibroin • Creutzfeldt – Jakob Disease • Caused by the transmission of Prions (a proteinaceous infectious agent that causes neurodegenerative diseases) that results to the misfolding of the normal prion protein found abundantly in the brain • Prions causes Mad Cow Disease • Replacement of the normal α-helical arrangement of the normal prion protein with βpleated sheet • Dementia and Involuntary Jerking Movements (Startle Myoclonus) 3.Tertiary Structure • • • • Refers to the spatial arrangement of the polypeptide chain Can either be Fibrous or Globular Different bonds contribute to the stabilization of the structure • Covalent bond: Disulfide Bond • Polar bonds: Hydrogen bonding, Ionic bonding • Non-Polar/ van der-Waals Chaperones • Proteins that assist in the folding and unfolding of the polypeptide to correctly form the tertiary structure • Example: Heat Shock Proteins 4. Quaternary Structure • Spatial arrangement of proteins made up of several polypeptide chains, with each peptide chain having a tertiary structure • Referred to as Oligomers • Example: Hemoglobin Denaturation (Denativation) of proteins happen when certain agents such as heat and urea destroy the higher Structural levels of protein without destroying the peptide bonds Ehlers – Danlos Syndrome • A group of connective tissue disorders that are generally characterized by hyperextensible skin, joint hypermobility, and defects in large blood vessels • 3. Insulin • A polypeptide hormone produced by the beta – cells of the pancreas • Synthesis of Preproinsulin in the rough ER → Proinsulin is cleaved in the secretory granules → C – peptide is released → the remaining molecule forms Insulin C. QUALITATIVE TESTS FOR PROTEINS AND AMINO ACIDS 1. Biuret Test • Identifies the presence of proteins • Uses 1% NaOH and a solution of Copper (II) sulfate • Positive Result: Violet color 2. Ninhydrin Test • Uses Ninhydrin to detect the presence of amines • Positive Result: Purple (Ruehlmanns’ Purple) = Amino Acids with a free amin group, Yellow = Imino Acids (Proline & Hydroxyproline) 3. Xanthoproteic Test • For Tyrosine, Tryptophan • Uses conc. HNO3 and 40% NaOH • Positive Result: Yellow Color 4. Millon’s test • Specific for Tyrosine • Uses Mercury dissolved in conc. HNO3 • Positive Result: Yellow Coloration 5. Hopkin’s – Cole Test • Specific for Tyrosine (Indole group) • Uses Glyoxylic Acid in Glacial Acid and Sulfuric Acid • Positive Result: Purple Color on the surface 6. Nitroprusside Test • Specific for Cysteine • Uses nitroprusside in alkaline solution • Positive Result: Red Coloration 7. Sakaguchi Test • Specific for Arginine (guanido group) • Uses NaOH, α-naphthol, and Bromine solution ENZYMES Renaturation/(Renativation), on the other hand, is the recovery of the protein from its denatured state • B. MEDICALLY IMPORTANT PROTEINS 1. Hemoglobin • Globular Transport Protein for Oxygen • Contains Heme which is a complex of porphyrin ring and a Ferrous • Sickle Cell Anemia • Caused by a mutation in hemoglobin where the Glutamate at position six in the beta–chain of hemoglobin is replaced by Valine • Has higher affinity to CO than CO2 2. Collagen • Found mainly in the bones and cartilage • Mainly contains Glycine, Proline, Hydroxyproline • Types: • I – Found in skin and bones • Osteogenesis Imperfecta is a disorder in the synthesis of Type I collagen characterized by a distinctive blue sclera and predisposed multiple childhood fracture • II – Found in the Cartilage • III – Found in the arterial walls • IV – found in the basal lamina • V – Found in the hair and placenta Module 2 – Biochemistry • • • These are biological catalysts of protein nature which catalyzes energetically feasible reaction without altering the reaction route. Like other nonbiological catalysts, enzymes are not consumed during a reaction and decreases the Ea of a reaction Enzymes are highly specific for their substrates and products with most enzymes recognizing only 1 compound as substrate. The velocity of a reaction is directly proportional with temperature • However, as the temperature goes higher than 37°, enzymes start to degrade • Changes in pH can cause changers in reaction and may cause enzymes to denature • Pepsin: optimal pH is 2 Many enzymes require cofactors or coenzymes to catalyzes reactions. These coenzymes are commonly derived from vitamins and metal ions. A. NOMENCLATURE 1. Oxidoreductase • Catalyzes REDOX reactions • Also called as dehydrogenases or reductases 2. Transferases • Catalyzes reactions involving the transfer of different groups from the substrate to another • Amino Transferases, Methyl transferases Page 2 of 19 RJAV 2022 3. Hydrolases • Catalyzes the substrate bond cleavage by adding water • Amylase, Saccharase Pyrimidine 4. Lyases • Catalyze bond – cleaving reactions without oxidation or addition of water • Pyruvate decarboxylase 5. Isomerases • Catalyzes structural rearrangements • Triose isomerase 6. Ligases • Also called as synthetases • Catalyzes the addition of 2 molecules using ATP as energy source • DNA – Ligase B. DNA DOUBLE HELIX • • B. INHIBITION • Inhibitors are agents capable of exerting a specific deterrent action on the activity of an enzyme 1. Competitive • An inhibitor that is structurally related to the substrate binds to the active center preventing the formation of the enzyme – substrate complex • • • • • 2. Noncompetitive • An inhibitor binds to the enzymes or enzyme – substrate complex which leaves the active center free and may induce conformational hinders the formation of enzyme – substrate complex. C. DENATURATION 3. Irreversible • An inactivator bonds covalently to the enzyme and inactivate it • • Also known as isozymes These are enzymes that may differ in amino sequences and physical properties but catalyze the same reaction. • A. DNA STRUCTURE • • Upon heating, DNA strands separate the base pairs reform once the temperature is slowly decreased E. HYBRIDIZATION DNA & RNA • Separation of the DNA strands due to heat or alkali exposure without breaking the phosphodiester bond D. RENATURATION C. ISOENZYMES • • Proposed by Watsons and Crick in 1953 The secondary structure of DNA is formed by the pairing two polynucleotide chains that are antiparallel. • One chain rubs 5’ – 3’ and the other 3’ – 5’ • The base sequences of the two strands are complementary • Adenine and Thymine via 2 hydrogen bonds (major groove) • Guanine and Cytosine via 3 hydrogen bonds (minor groove) One full turn of DNA helix contains nucleotides Has 3 different conformations: B form: Right-handed Z form: Left-handed A form: dehydrated and compact; Right-handed A single strand of DNA or RNA pairs with a complementary base sequence on another strand of DNA or RNA F. HISTONES Contains nucleosides that is made up of a nitrogenous base, deoxyribose, and phosphate The nitrogenous bases are classified into Purine and Pyrimidines The primary structure of DNA and RNA is linear polynucleotide chain made up of mononucleotides which are linked by a 3’,5’ phosphodiester bond (3’ C of one sugar to the 5’ C of the next sugar) • • These are small of DNA small, basic proteins rich in Arg and Lys; not present in prokaryotes Eukaryotic chromatins consist of a DNA complexed with histones G. RNA vs. DNA The Nucleotides of DNA • • RNA contains ribose as sugar instead of deoxyribose Uracil replaces Thymine • • Single stranded with extensive base pairing RNA can sometimes act as catalysts and enzymes (ribonucleases, Peptidyl transferase) Purines H. TYPES OF RNA 1. Messenger (mRNA) • Contains a cap consisting of a methylated guanine triphosphate attached to the hydroxyl group on the ribose at the 5’ end • The poly(A) tail contains up to 200 Adenine nucleotides 2. Ribosomal (rRNA) • Aids in the formation of ribosomes • Prokaryotic: 70s (Large subunit = 50s; Small subunit = 30s) • Eukaryotic: 80s (Large subunit = 60s; Small subunit = 40s) Module 2 – Biochemistry Page 3 of 19 RJAV 2022 • 3. Transfer (tRNA) • Has a characteristic cloverleaf structure DNA REPLICATION • • Known as the transfer of genetic formation within single class of nucleic acids, i.e., from DNA to DNA or, as in certain viruses from RNA to RNA. DNA Replication is: • Bidirectional • Replication begins at a specific origin and simultaneously moves out in both directions • Prokaryotes have one site of origin while eukaryotes can have multiple sites of origin • Semiconservative • The resulting daughter DNA contains one intact parenteral strand and newly synthesized complementary strand. b. c. d. e. f. Promoters contain a sequence TATAAT known as the Pribbenow box or TATA BOX • In eukaryotes, Hogness box (TATA Box) has a sequence of TATATAA An RNA Polymerase – closed promoter complex is formed The DNA unwinds at promoter and forms the open promoter complex RNA Polymerase initiates the mRNA synthesis with a purine ribonucleoside triphosphate RNA Polymerase holoenzyme catalyzes the elongation of mRNA by about 4 more units Upon reaching about 10 nucleosides, the sigma – subunit dissociates and will bind to another RNA Polymerase 2. Termination a. Intrinsic (Rho – independent Transcription Termination) • Controlled by specific sequences called termination sites wherein the sequences form a hairpin loop structure that allows the RNA Polymerase to detach from the DNA template strand b. Rho – dependent Transcription Termination • Involves the Rho – Protein • The rho –protein binds to the RNA and chases the polymerase until it reaches the termination site to facilitate the dissociation of the RNA Polymerase from the DNA Template Strand. A. STEPS IN REPLICATION 1. Initiation a. The parenteral DNA is separated and uncoiled by helicases where the uncoiled DNA strand serves as the template b. As the strands separate. An active site for synthesis known as replication fork is formed where each separated strand is stabilized by a single – stranded protein c. As the strands separate, supercoiling happens which is removed by DNA Topoisomerase • Etoposide inhibits topoisomerase d. The primase aids in the production of the RNA primer in a 5’ – 3’ direction C. EUKARYOTIC TRANSCRIPTION 1. RNA Polymerase (Promoters) 2. Elongation & Termination a. From the initiation site, DNA Polymerase III adds deoxyribonucleotides at the 3’ OH end of the primer • DNA polymerase can only copy as 3’ – 5’ direction and produce the daughter strand in a 5’ – 3’ direction • DNA polymerase always requires primers and cannot initiate the formation of new strands b. DNA Polymerase III forms continuous strands in a 3’ – 5’ direction forming the leading strand c. In the 5’ – 3’ strand, a discontinuous formation of DNA happens forming smaller fragments known as Okazaki fragments. These fragments are joined together by DNA ligase d. Once complete, RNA primer is removed by the exonuclease activity of DNA Polymerase I e. Gaps are filled with the complementary bases f. Replication is terminated, forming 2 daughter DNA molecules. a. RNA Polymerase I • Found in the nucleolus and synthesizes the precursors of ribosomal RNA b. RNA Polymerase II • Found in the nucleoplasm and synthesizes mRNA precursors • α-Amanitin (a toxin from the mushroom Amanita phalloides) binds and inhibits RNA Polymerase II which halts mRNA synthesizes resulting to severe GIT symptoms, liver toxicity, and death. c. RNA Polymerase III • Found in the nucleoplasm and synthesizes the tRNA precursors 2. Steps in Eukaryotic Transcription a. RNA TRANSCRIPTION • • • • • The process of making an RNA form a DNA template is known as transcription. Is the major control point in gene expression and protein production Catalyzed by DNA – dependent DNA polymerase No primer is needed Uses one strand of DNA as template A. RNA POLYMERASE • • • Can initiate synthesis of new chains without primer Copies the DNA template in a 3’ – 5’ direction and resulting RNA elongates in a 5’ – 3’ direction Precursors: Ribonucleoside Triphosphate (ATP, UTP, GTP, and CTP) 3. Reverse Transcription • The process of transcribing the single stranded RNA into a double – stranded DNA • Enzyme: Reverse Transcriptase (RNA – dependent DNA Polymerase) • Retrovirus contain RNA as genetic material RNA TRANSLATIN AND PROTEIN SYNTHESIS B. STEPS IN PROKARYOTIC TRANSCRITION A. GENETIC CODE • 1. Initiation and Elongation • Rifampin inhibits the beta subunit of bacterial dependent RNA Polymerase • Actinomycin D binds to DNA and inhibits the elongation or RNA Transcription by RNA Polymerase a. mRNA Synthesis 1. RNA Polymerase II produces a heterogenous nuclear RNA (hnRNA) containing exons and introns • Exons are sequences appearing in the mature mRNA • Introns are the non-coding region and are removed during splicing 2. The primary transcript (hnRNA) is capped at the 5’ end 3. A poly(A) tail with a nucleotide length range of 20 – 200 is added at the 3’ end of the transcript 4. The introns are removed and the exons are connected to form the mature mRNA through splicing • A collection of codons that specify all the amino acids found in proteins Characteristics: • It is degenerate (Redundant; Many amino acids have numerous codons) • It is non – overlapping RNA polymerase is directed by the sigma factor to bind to the promoter Module 2 – Biochemistry Page 4 of 19 RJAV 2022 • Begins with the start codon AUG (Methionine) near the 5’ end of the mRNA which determines the reading frame • Ends with a stop codon (UGA, UAA, UAG) near the 3’ end • The code is comma less (no markers to differentiate one codon from one another) • The code is nearly universal Codons • A sequence of 3 bases in mRNA that specifies a particular Amino acid • g. 3. Termination a. b. • • Mutations in DNA are carried over into the mRNA that causes changes in the encoded protein • • • 1. Point mutations • A base in the DNA is replaced by another which alters the codon in the MRNA a. b. The stop codon is recognized by a release factor The newly synthesized protein is released and the synthesizing complex dissociates CARBOHYDRATES B. MUTATIONS • The steps are repeated until the ribosome encounters a stop codon Silent • The codon containing the changed base codes for the same amino acid Missense • The codon containing the changed base codes for a different amino acid The most widespread compounds involved in the buildup functions of the cell Presence of a carbonyl (Aldo or Keto group) and at least two hydroxyl groups Stereoisomers: Same chemical formula but differs in the position of hydroxyl groups Enantiomers: Stereoisomers that are mirror images Epimers: Stereoisomers that differ in the position of the hydroxyl group at only one asymmetric carbon. A. MONOSACCHARIDES • • • Nonsense • The codon containing the changed base codes for a stop codon 2. Insertions • Occur when a base or a number of bases are added to the DNA These are simple carbohydrates that are named depending on the number of Carbon atoms and the specific carbonyl group present They can either be Dextrorotatory or Levorotatory Examples: D – Galactose, L - Fructose c. B. OLIGOSACCHARIDES • • • • • • • 3. Deletions • Occur when a base or a number of bases are deleted to the DNA 4. Frameshift • Occurs when the number of bases added or deleted is not a multiple of 3 which shifts the reading frame to a completely different set of codons • C. STEPS IN EUKARYOTIC TRANSLATION • C. POLYSACCHARIDES 1. Initiation • a. b. c. The 40s ribosomal subunit binds close to the 5’ cap until it recognizes the start codon (AUG). • The ribosomes recognize the AUG in the correct context known as the Kozak Sequence (ACCAUGG) • In prokaryotes, Streptomycin binds to the 30s subunit which hinders initiation A special initiator tRNA recognizes the start codon carriers a non formylated Methionine The 60s ribosomal subunit is recruited from the 80s initiation complex. c. d. e. f. The ribosome moves from 5’ to 3’ direction The amino acid containing tRNA (aminoacyl-tRNA’s) forms a complex with elongation factor and enters the empty A-site The anticodon of the aminoacyl-tRNA is matched against the mRNA codon in the A-site When the correct aminoacyl-tRNA enters the A-site, the polypeptide chain in the P-site is linked to the new amino acid in the A-site via a peptide bond • The reaction is catalyzed by peptidyl transferase • Chloramphenicol inhibits prokaryotic peptidyl transferase • The new peptidyl-tRNA is left in the A site The peptidyl-tRNA is translocated from the A-site to the Psite facilitated by an elongation factor • Erythromycin binds to a site on the 50s subunit which inhibits translocation A new aminoacyl-tRNA in the A-Site is made available for the next codon in the mRNA Module 2 – Biochemistry These are high – molecular carbohydrates with more than ten monosaccharide units linked by glycosidic bonds Neutral Polysaccharides such as Starch (Plants) and Glycogen (human and animals) are found inside the cells as reserve material Acidic polysaccharides such as Chondroitin sulfate and Hyaluronic acid are found extracellularly D. DIGESTION OF CARBOHYDRATES • • • • 2. Elongation a. b. Composed of 2 to 12 monosaccharide units linked by glycosidic bond Sucrose Table Sugar Glucose + Fructose Lactose Milk Sugar Galactose – linked β-1,4 to glucose Salivary α-amylase, which is present in the saliva, cleaves starch by breaking the α-1,4 linkage between glucose residues Sucrase converts sucrose to glucose and fructose Lactase coverts lactose to glucose and galactose α-glucosidase inhibitors work in the intestine to slow down the digestion of carbohydrates to facilitate better post digestive blood glucose control E. QUALITATIVE TESTS FOR CARBOHYDRATES 1. Molisch Test • General test for Carbohydrates • Monosaccharides give the most rapid result • Uses α-naphthol and sulfuric acid • (+) result: purple ring 2. Benedict’s test • Tests for the presence of reducing sugars • All monosaccharides give a positive result • Uses Copper (III) sulfate, Sodium citrate and Sodium carbonate in a mildly basic solution • (+) result: red to orange precipitate 3. Barfoed’s Test • Uses Copper (III) in a slightly acidic medium • Used to differentiate a reducing monosaccharide from a reducing disaccharide Page 5 of 19 RJAV 2022 • (+) result: red ppt (within 3 minutes = Monosaccharide, longer than 3 minutes = disaccharide) Anabolism External trigger 4. Bial’s Test • Test for differentiating pentose and hexose monosaccharides • Used concentration HCl and orcinol + ferric chloride • (+) result: Pentoses = bluish to green, Hexoses + brownish to gray Endocrine gland Negative feedback Hormone 5. Seliwanoff’s Test • Test for differentiating aldoses from ketoses • Uses 6M HCl and resorcinol • (+) result: Ketoses = Cherry red, Aldoses = bluish-green to peach Target organ TOPICS ON HUMAN PLANT AND METABOLISM • INTRODUCTION What is metabolism? • • Metabolism is the sum of all chemical reactions in the body (the reactions are catalyzed by specific enzymes) A series of metabolic steps with a specified end-product is called a metabolic pathway • Metabolic pathways are linear or cyclic • Metabolic pathways are either catabolic or anabolic • Each pathway usually has an irreversible reaction to dictate the direction of the process A↔B↔C→D Next question: How do we know if the catabolic or anabolic process will dominate? • Answer: control the irreversible steps (often by negative feedback). When one process is on, the opposite must be off Why do we need metabolism? • • To generate energy Humans are thermodynamically open systems, and the second law states that entropy (disorder) must increase. Humans that don’t produce energy will give up to entropy, their bodies will lose homeostasis, and they die. Entropy is counteracted it performs. We don’t die as result ATP will serve as the primary energy currency. Other cofactors can also be equated to ATP: • NADH (reduced NAD+) = 2.5 ATP • FADH (reduced FAD+) = 1.5 ATP • Other triphosphates (ex. GTP) = 1 ATP *NADH and FADH2 will only become ATP after going through the electron transport chain • • • • “______lysis” “______oxidation” Catabolism Smaller molecules Larger molecules OVERVIEW Energy • • Anabolism Respiration involves most biomolecules They converge at acetyl-CoA, which ultimately enters the citric cycle Carbohydrate Protein Fat “______genesis” “______synthesis” Digestion and absorption Catabolism – breaking down Anabolism – building up • • • • Catabolism and anabolism, by direction, are opposites Do you think the same exact enzymes can perform the opposite processes? Answer: NO Different enzymes must be used because each pathway has at least one irreversible step. If there is no irreversible step, a “futile cycle” will result (pathways moving around without direction and achieving totally nothing) Catabolism Simple sugars (mainly glucose) Fatty acids + glycerol Amino acids Catabolism Smaller molecules Acetyl-CoA Larger molecules Energy ETC Module 2 – Biochemistry Page 6 of 19 RJAV 2022 INTRODUCTION • • • • • Cell respiration and carbohydrate metabolism are not one and the same! Yet, they are discussed together because carbohydrates are commonly used materials in respiration Familiar processes: • Glycolysis • Krebs’ Cycle • Electron Transport Chain Classification equation for cell respiration C6H12O6 + 6O2 → 6CO2 + 6H2O + ATP Complete set of processes: • Glycolysis and gluconeogenesis • Glycogen metabolism (glycogenesis and glycogenolysis) • Pentose phosphate pathway • Metabolic effects of insulin and glucagon • Krebs’ Cycle • Electron Transport Chain • • • CARBOHYDRATE METABOLISM • Digestion first! Carbohydrates from food intake DIGESTION Pentose phosphate pathway GLYCOGENOLYSIS Glycogen Glyceraldehyde-3phosphate (GAP) will proceed to payoff Dihydroxyacetone phosphate (DHAP) will not poceed) For DHAP to proceed it must isomerize into GAP (on top of the original GAP) Gives a total of TWO GAP molecules → everything in the payoff phase must be multiplied by TWO! Pentoses + CO2 Glucose ↑↓ Glucose 6-phosphate GLYCOGENESIS GLUCONEOGENESIS GLYCOLYSIS Pyruvate Lactate 1. GLYCOLYSIS • • • • • • • • • aka Embden-Meyerof-Parnas pathway breakdown of glucose to two molecules of pyruvate Cytosolic Energy-producing Consists of two phases • Energy investment (steps 1-5) • Energy payoff (steps 6-10) ATP yield? • 2 ATP produced (7 ATP if NADH is processed) Effect on blood sugar? • Lowers blood sugar • Works during fed states (after eating) • Stimulated by insulin Irreversible steps? • 3 steps: #1, #3, & #10 “The committed step”? • Rate-limiting step – step 3 Module 2 – Biochemistry Page 7 of 19 RJAV 2022 Pyruvate • end product of glycolysis can be converted back to glucose. Glucose Synthesis of glycogen Glucose-6phosphate Glycogen Pentose phosphate pathway Ribose, NADPH Degradation of glycogen Glycolysis Gluconeogenesis Pyruvate • G6P bridges many pathways in carb metabolism • Just because G6P cannot go back to glucose, it doesn’t mean it fully commits to glycolysis • Step 1 can NOT be the committed step The next irreversible step (Step 3) is • The enzyme for step 3, PFX, is highly regulated • 2. FATES OF PYRUVATE Pyruvate Aerobic conditions in humans, animals, and microorganisms ACETYL CoA Anaerobic conditions in humans, animals, and microorganisms Anaerobic conditions in some microorganisms LACTATE ETHANOL NEW ENZYMES (only 4): • • • To reverse step 10: • Pyruvate carboxylase* • PEP carboxykinase To reverse step 3: • Fructose-1,6-biphosphatase To reverse step 1: • Glucose-6-phosphatase (G6Pase) • G6Pase is only in the liver *Do not confuse with pyruvate decarboxylase! liver – 90% kidney – minor; more likely 10% Amino Acids, Glycerol • • Amino acids – through the citric acid cycle Glycerol – via the following reaction 3. GLUCONEOGENESIS (GNG) • • • • • Conversion of non-carbohydrates into glucose Substrates include pyruvate, amino acids, glycerol, and lactate Similar to reverse of glycolysis • Irreversible steps require different enzymes Timing: fasted state (which release of glucagon) Effect: increased blood glucose Module 2 – Biochemistry Page 8 of 19 RJAV 2022 Uridine diphosphate glucose (UDPGlc). I. GLYCOGENESIS • • • • Lactate • • • To reverse step 1: • Glucose-6-phosphatase (G6Pase) • G6Pase is only in the liver • Thus, ~90% of GNG occurs in the liver The GNG of lactate is cyclic. • Cori Cycle • Synthesis of Glycogen Anabolic Requires formation of Contains α1, 4 and α1,6 bonds Stimulated by: • Insulin Effect: • reduce blood glucose Timing: • Fed state The biosynthesis of glycogen. The mechanism of branching as revealed by feeding 14C-labeled glucose and examining liver glycogen at intervals Steps: 1. 2. 3. 4. FATES OF GLUCOSE-6-PHOSPHATE A. GLYCOGEN METABOLISM Glycogen • A branched homopolysaccharide • Contains α1, 4 (linearity) and α1,6 (branching) glycosidic bonds G6P to G1P by phosphoglucomutase (PGM) G1P + UTP → UDP-glucose UDP-glucose is added to a glycogen molecule (Glcn) by Glycogen synthase • After step 3, glycogen becomes longer (Glcn+1) • The branching enzyme uses part of the chain to make α1,6 bonds II. GLYCOGENOLYSIS (GGL) • • • • • • Breakdown of glycogen Catabolic Requires hydrolysis of α1, 4 and α1,6 bonds Stimulated by: • Glucagon Effect: • Increase blood glucose Timing: • Fasted state Steps: 1. Glycogen phosphorylase cleaves a glucose molecule from glycogen into G1P 2. G1P → G6P by PGM 3. G6P → glucose by G6Pase • The glucose goes to the blood and to the organs that need it • Any branches block phosphorylase, and must be removed by the debranching enzyme III. GLYCOGEN STORAGE DISEASES • • Diseases in the metabolism of glycogen Commonly lead to hypoglycemia, hyperlipidemia, and/ or hepatomegaly; some are fatal. Pathways of glycogenesis and of glycogenolysis in the liver. , Stimulation; Module 2 – Biochemistry , inhibition.). Page 9 of 19 RJAV 2022 GSD TYPE (common name) 0 1 (Von Gierke’s) 2 (Pompe’s) 3 (Cori’s) 4 (Andersen’s) 5 (McArdle’s) 6 (Hers’) 7 (Tarui’s) ENZYME DEFICIENT Glycogen synthase G6Pase Lysosomal alpha-glucosidase Debranching enzyme Branching enzyme Muscle phosphorylase Liver phosphorylase PFK B. PENTOSE PHOSPHATE PATHWAY • • • • *All reactions are irreversible *Step 1 is the major rate-limiting step Oxidative phase is controlled by level of NADP+ NADPH is not NADH! Uses of NADPH: • Lipid biosynthesis • Detoxification (in conjugation with glutathione (GSH)) THE THING CALLED GLUTATHIONE • aka HEXOSE MONOPHOSPHATE SHUNT) Another pathway for G6P An alternate pathway of glycolysis in the cytosol Consists of two phases I. Oxidative II. Non-oxidative • • Essential for protection of the cell against oxidative and chemical insults GSH is oxidized to the disulfide form by help of glutathione peroxidase to counteract oxidative stress Most of the NADPH in the RBC is used by glutathione reductase to maintain GSH in the reduced state • G6PD is a key enzyme in the conversion of NADP to NADPH (and therefore, is also key in glutathione activity) • Imagine the sequence of events that follow if there is G6PD deficiency *G6PD is deficient if there is minimal amount of NADPH, there is also minimal amount of active glutathione. If there is minimal amount of active glutathione, the we will not be able to detoxify Hydrogen peroxide which leads to ↑ = damage → cell death; RBCs: hemolysis • • In G6PD-deficient people, their cells (especially RBC) are susceptible to oxidative stress When triggered by drugs or reactive oxygen species (ROS), causes acute hemolysis (and related consequences jaundice and dark urine) II. NON-OXIDATIVE PHASE • • Leads to synthesis of other sugar phosphates Merely a shuffling of carbons between pairs of sugar I. OXIDATIVE PHASE • • • Involves three steps Riboluse-5-phosphate serves as the final product Steps 1 and 3 produce NADPH *All reactions are reversible *Non-oxidative state phase is controlled by the requirements of pentoses • • Module 2 – Biochemistry Page 10 of 19 Can simply lead back to glycolysis or lead to Ribose-5Phosphate for nucleotide synthesis PPP and glycolysis work simultaneously RJAV 2022 • • BUT any excess demand for R5P or NADPH puts priority on PPP over glycolysis Otherwise, glycolysis is usually dominant Series of electron transfers that generates a proton gradient to fuel the synthesis of ATP “Series of electron transfers…” 5. CITRIC ACID CYCLE • • • • aka Krebs cycle*/ tricarboxylic acid (TCA) cycle Cycle that converts acetyl CoA to two molecules of CO2 The “central hub” Takes place at the mitochondrial matrix Acetyl-CoA → 2CO2 *There is another Krebs cycle, in fact, the citric acid cycle is the second cycle that Hans Krebs discovered! • • There is a certain flow, and each electron acceptor is immediately followed by another Oxygen is the last electron acceptor • In fact, ~90% of oxygen we breathe is used in the ETC • (the remaining oxygen is used by WBC to fend off pathogens) “that generates a proton gradient…” • • • • 3 NADH 1 FADH2 1 GTP The Citric Acid Cycle acid is amphibolic: (Catabolic or Anabolic) • Citrate – fatty acid synthesis • α-kg – amino acid catabolism • Succinyl-CoA – heme synthesis • OAA – GNG and nucleotide synthesis Depends on balance of body acetyl-CoA and OAA levels (to be discussed more in lipid metabolism) • • “…to fuel the synthesis of ATP.” Complex I: NADH-CoQ reductase Complex II: Succinate-CoQ reductase Complex III: CoQ-cytochrome c reductase Complex IV: Cytochrome c oxidase • • Oxidative Phosphorylation • • NADH and FADH2 are oxidized to NAD+ and FAD, which allows the ETC to happen in the first place (and ultimately result to ATP synthesis) Very different from other types of phosphorylation • Substrate-level phosphorylation • Photophosphorylation (in photosynthesis) But they can get flux back to the matrix using complex V Complex V actually uses proton influx to create ATP • Proton gradient generation and ATP synthesis are different events, but they are coupled by complex V (chemiosmotic coupling) Coupling them is their way of being regulated: • • • • • Module 2 – Biochemistry When the protons leave the matrix into the IMS, they can’t get back using any complex from I to IV Page 11 of 19 The ETC will work only when ATP levels are low, and will produce ATP as a response The ETC will slow down when ATP levels are high Some cells in the body have two capacities to disconnect the two processes using uncouplers (ex. thermogenin, 2,4-DNP) • The ETC (and proton gradient) goes on without creating ATP • If the ETC goes on with creating ATP, the energy resulting from the process manifests as heat • Serves as heat-generating mechanism for some animals All complexes must form a continuous flow Blocking the complexes can lead to severe complications! RJAV 2022 We need some computations, too LIPID METABOLISM Lipids with Fatty Acids (Saponifiable) • TAGs • Phospholipids Lipids without Fatty Acids (Non-saponifiable) • Terpenes • Sterols SUMMARY ON CELL RESPIRATION (with ATP count) 1. LIPOGENESIS Citric Acid Cycle • FATTY ACID SYNTHESIS Energy yield per acetyl-CoA: OLD CONVERSION (NADH = 3 ATP │ FADH = 2 ATP) 3 NADH 1 FADH2 1 GTP TOTAL CURRENT CONVERSION (NADH = 2.5 ATP │ FADH = 1.5 ATP) 9 2 1 12 ATP/ acetyl 2.5 1.5 1 10 ATP/ acetyl • • Modern ATP Computation (aerobic respiration) PROCESS Glycolysis 2 pyruvate to 2 acetyl 2 x acetyl to 2 CO2 (TCA cycle) • MOLECULES PRODUCED 2 ATP 2 NADH x 2.5 = 5 ATP Minus two if GP shuttle is used 2 NADH x 2.5 = 5 ATP ATP YIELD 5–7 3 NADH x 2.5 = 7.5 ATP 1 FADH x 1.5 = 1.5 ATP 1 GTP x 1 = 1 ATP Total of 10 ATP/ acetyl 20 • Occurs in the cytosol RLS: Conversion of acetyl-CoA to malonyl-CoA by acetylCoA carboxylase (ACC) When synthesized, fatty acids are esterified into phospholipids and triglycerides Timing: Fed state (stimulated by insulin) 5 30 – 32 Overall Old ATP Computation (aerobic respiration) PROCESS Glycolysis 2 pyruvate to 2 acetyl 2 x acetyl to 2 CO2 (TCA cycle) MOLECULES PRODUCED 2 ATP 2 NADH x 3 = 6 ATP Minus two if GP shuttle is used 2 NADH x 3 = 6 ATP ATP YIELD 6–8 3 NADH x 3 = 9 ATP 1 FADH x 2 = 2 ATP 1 GTP x 1 = 1 ATP Total of 12 ATP/ acetyl 24 Overall Module 2 – Biochemistry 6 36 – 38 Page 12 of 19 RJAV 2022 2. LIPOLYSIS A. DIGESTION AND GENERAL LIPOLYSIS Digestion in GIT: • A small number of TAGS are digested by lipases (can be found in intestines) • A larger amount is converted into micelles by bile acids – emulsification → further absorption Breakdown in fat: • Triggered by the fasted state, with glucagon release or insulin inhibition • Initial hydrolysis of triglycerides by hormone-sensitive lipase (HSL) into glycerol and fatty acids. How about Phospholipids? • • • • • • B. FATTY ACID OXIDATION aka beta-oxidation Nearly complete reverse of fatty acid synthesis Timing: Fasted state (stimulated by glucagon) Successive breakdown of fatty acids by 2 carbons at a time (acetyl-CoA) • All acetyl-CoA will be oxidized in the TCA cycle Occurs in the mitochondrial matrix Requires transport of fatty acids to the matrix! “Nearly complete reverse of fatty acid synthesis” Module 2 – Biochemistry Page 13 of 19 RJAV 2022 What is the next ATP production for the complete oxidation of lauric acid, C12 saturated fatty acid, to CO2 and H2O? Cycles: 5 x 4 ATP = Acetyls: 6 x 10 ATP = “Successive breakdown of fatty acids by 2 carbons at a time” 20 60 (-2) = 78 Shortcuts: #ac = #C / 2 #cy = #ac – 1 3. FATES OF HMG-CoA (hydroxymethyl glutaryl – CoA) 3x acetyl-CoA “Requires transport of fatty acids to the matrix” A. MEVALONATE PATHWAY • • • • • • Module 2 – Biochemistry Page 14 of 19 Pathway for cytoplasmic synthesis of sterols and terpenes* Stimulated in the fed state (by insulin) Building block: acetyl-CoA *In non-animals like plants Most are also under terpenes Terpenes are made up of isoprene units (right) Gives rise to many odors given off by plants RJAV 2022 VOLATILE OILS • Most are also under terpenes • Give rise to many odors given off by plants • Isoprene 5C TYPE Monoterpene Diterpene Sesquiterpene Triterpene Tetraterpene # ISOPRENES 2 4 3 6 8 # CARBON 10 20 15 30 40 Sunlight + Liver + Kidney = Good bones B. KETOGENESIS • • • • • *Happens in cytosol Production of ketone bodies in the mitochondrion • Acetone • Acetoacetate • Beta-hydroxybutyrate Timing: Fasting/ Starvation • Accompanies β-oxidation due to acetyl-CoA overflow • TCA cycle saturates → excess acetyl-CoA become HMG-CoA, and then acted upon by HMG-CoA lyase Our tissues normally shift from using sugars to using fats in the fasted state The brain cannot use the fats due to the blood-brain barrier Ketone bodies serve as the emergency fuel of the brain in these cases High blood Glucose (after meal) Low blood Glucose (between meals) Liver Adipose tissue Module 2 – Biochemistry Page 15 of 19 RJAV 2022 • Skeletal Muscle • • Ketoacidosis – excessively elevated ketone bodies, causing acidification of the blood Ketogenic diet – weight loss; induced Ketosis (but was used for what before?) ↑fats, ↓carbs, regular protein Diabetic Ketoacidosis Heart NUCLEOTIDE METABOLISM • • Brain Purine and pyrimidine nucleotides are synthesized separately Often synthesized using de novo (“of new” or “from scratch”) pathways or salvage (from readily-made nucleotides) I) PYRIMIDINE METABOLISM SYNTHESIS AND UTILIZATION OF KB’s • OMP (Orotidine monophosphate) – parent pyrimidine FOLATE II) PURINE METABOLISM • Ketosis – shift from dependence on glucose to dependence on ketone bodies • Ketonemia – elevated KB in blood (20mg/100mL) • Ketonuria – elevated KB in urine (70mg/100mL) Module 2 – Biochemistry Page 16 of 19 RJAV 2022 4. AMINO ACID DERIVATIVES • Amino acids can be used as precursor for many different compounds, including: • Neurotransmitters • Hormones/ Autacoids • Pigments • Cofactors/ Carriers • Secondary plant metabolites (alkaloids, flavonoids, tannins) A. FROM TYROSINE • • • • Catecholamines • DA, NE, E • Adrenergic NTs Thyroid hormones • T3, T4 Melanin – enzyme Tyrosinase • Primary skin pigment Phenylpropanoids* D. FROM ARGININE • • Nitric oxide (NO) • Vasodilatory substance • Related to MoA of some direct vasodilators Urea • Final product of protein catabolism E. FROM SERINE • • • Sphingosine • Formed by condensation of serine with palmitic acid Ethanolamine • Found in phosphatidylethanolamine Choline • Found in phosphatidylcholine/ acetylcholine B. FROM TRYPTOPHAN • • Serotonin • Autacoids and Neurotransmitters Melatonin • Sleep-wake cycle F. MISCELLANEOUS • • • S-adenosylmethionine (SAM) • From methionine • One-carbon transport GABA • From glutamic acid L-carnitine • From lysine • Tranexamic acid C. FROM HISTIDINE • • Histamine • Contributes to allergic reactions • • Module 2 – Biochemistry Page 17 of 19 6-aminopenicillanic acid • From cysteine and valine Glutathione • Tripeptide of E, C and G • Endogenous antioxidant REMEMBER: all pituitary and hypothalamic hormones are also peptides! RJAV 2022 G. PORPHYRIN METABOLISM • • • • • • • Porphyrins – chelating macrocycles • Starts from the condensation of glycine and succinylCoA • Includes heme and chlorophyll Heme: • Heme is incorporated to RBCs as hemoglobin • When the RBCs die, the hemoglobin releases heme and is metabolized in the liver to waste product bilirubin • Hyperbilirubinemia can lead to jaundice • Other derivatives include stercobilin (feces) and urobilin (urine) Carbon monoxide (CO) made from biliverdin production is toxic (it competes with oxygen in hemoglobin binding) Conversion of heme all the way to bilirubin happens in the spleen, but after, is delivered to the liver. 3 MAJOR PROCESSES: A. Transamination B. Oxidative deamination C. Urea cycle A. AMINO ACID CATABOLISM: TRANSAMINATION • • • • • Glutamate collects amino groups in most tissues and is converted to glutamine Pyruvate collects amino groups in muscles and is converted to alanine Alanine goes to the liver and is converted back to pyruvate, but aKg is converted to glutamate The liver therefore collects all amino groups for further processing All above processes are transamination processes, where pyridoxal phosphate (PLP) is the main cofactor Bilirubin has to be glucuronidated first before being ready for excretion Treatment for neonatal jaundice: Phototherapy Before: Phenobarbital (enzyme inducer) 5. AMINO ACID CATABOLISM • • • • Excess amino acids are not stored Nitrogen balance • Positive: intake > output • Negative: outtake > input Metabolized to ammonia by different tissues in the body Converted to urea in the liver • Urea is the final form B. AMINO ACID CATABOLISM: [O] DEAMINATION • • In the liver, deamination of glutamate will lead to release of ammonia (ionized: ammonium) Deamination is accompanied b replacement of a keto group → “oxidative deamination” C. THE UREA CYCLE • • • • Module 2 – Biochemistry Page 18 of 19 Aka: Krebs-Henseleit cycle Ammonia is so far the product from deamination Ammonia is toxic to the body Ex. when it accumulates in the brain, it can result to hepatic encephalopathy RJAV 2022 • • Therefore, ammonia in the liver is further converted to urea (final product of nitrogen disposal) Involves arginine and nonstandard amino acids citrulline and ornithine Name AA Affected Alkaptonuria Tyrosine Albinism Tyrosine Homocystinuria Methionine Phenylketonuria (PKU) Maple syrup urine disease (MSUD) Phenylalanine Branched-chain amino acids Enzyme Deficient Homogentisate oxidase Tyrosinase Cystathionine synthetase Phenylalanine hydroxylase Branched-chain keto acid dehydrogenase Effects Crippling arthritis Light complexion photophobia Ectopia lentis, osteoporosis Retardation, ketonuria, diet restrictions COMMON INBORN ERRORS OF METABOLISM (IEM)/ AMINOACIDOPATHIES Module 2 – Biochemistry Page 19 of 19 RJAV 2022