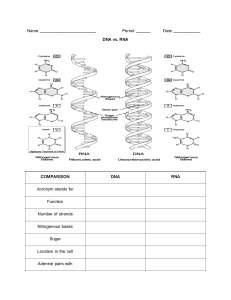
UNIT 1: EUKARYOTIC CELLS NUCLEUS AND MITOCHONDRIA CELL THEORY: 3 principles 1. all living organisms are composed of one or more cell cells 2. Cell is basic unit of life 3. All cells arise from pre-existing cells 4. NEW: genetic information is encoded by DNA in the cell NOTE: viruses might have RNA instead of DNA and they aren’t organisms as they aren’t composed of cells WHAT DEFINES A EUKARYOTIC CELL? - Eu = true - Karyote = kernel o Describing true nucleus within eukaryotic cell - EUKARYOTIC CELL: Cells that contain membrane bound nucleus and organelles - Capable of undergo mitosis - Some exceptions o Mature red blood cells (RBCs) have no nucleus or DNA but are born with one (just expel it during maturation) EUKARYOTIC CELL - Plasma membrane - Rough ER - Smooth ER - Nucleus - Mitochondria - Lysosome - Golgi Apparatus - Up to dozens of linear chromosomes PROKARYOTIC CELL - Cells of unicellular organisms, bacteria, archaea - DO NOT contain membrane bound nuclei and organelles - 1 circular DNA chromosome - Chromosome (DNA) - Cell wall - Cell membrane - Flagellum DNA is found in chromosomes DNA CHROMOSOMES are composed of CHROMATIN, NUCLEAR ENVELOPE: chromatin is contained within double membrane which are 2 sets of phospholipid bilayers - Studded with NUCLEAR PORES: protein complexes embedded in nuclear membrane that allows selective passage of larger molecules like RNA in/out of nucleus NUCLEOLI: little nucleus; where ribosome assembly occurs (synthesizing ribosomal RNA and packaging it with protein to form ribosomes) RIBOSOMES: complexes within cell that synthesize proteins - - Everything else in cell is incorporated in cell cytoplasm which are all suspended in CYTOSOL: gel like fluid o Ex. Proteins, organelles, cytoskeletal fibers In cytoplasm, subcellular components are trafficked from one location of a cell to destination METABOLISM: converting energy sources into form of usable energy (ex. ATP) to drive all cellular functions MITOCHONDRIA - Do what they want when they want, powerhouse of cell o Replicates independently of cell cycle via binary fission - HISTORY: Bacteria are good at aerobic metabolism converting energy sources into usable forms of energy in presence of oxygen BINARY FISSION: asexual reproduction where DNA is replicated and cell splits into 2 ENDOSYMBIOTIC THEORY: Awhile, another cell engulfed a bacterium and took advantage of its ability to make ATP efficiently in exchange for a place to reproduce safely inside the cell - First eukaryotic cell was born - Mitochondria arose when bacterium was engulfed by another cell; used as internal battery and to speak by host cell - Mitochondria look and act a lot like bacteria - Average human cell may contain 100 or 1000s of mitochondria o Have their own mitochondrial DNA (mtDNA) that has circular structure (derived from ancestral bacterial DNA - Most proteins that function in mito. Are derived from nuclear genes - Deleterious mutations to mitochondrial DNA can cause many disorders o Are all maternally inherited as we obtain mito from our mothers Mito has inner membrane and outer membrane In between spaces is INTERMEMBRANE SPACE Within inner membrane is MITOCHONDRIAL MATRIX where Krebs’s Cycles Occur - Where complexes of ETC and ATP synthase are imbedded - Invaginates as CRISTAE which greatly increases available surface area of membrane ENDOPLASMIC RETICULUM ENDOPLASMIC RETICULUM: net like organelle that’s continuous with cellular membrane - Folds and modifies newly synthesized proteins - Enclosed double membrane which invaginates to form cisternae - Has rough ER and smooth ER - Proteins can be modified by addition of carbohydrate groups (GLYCOSYLATION), proteolytic cleavage, disulfide bridges or addition of other functional groups ROUGH ER: tough surface created by ribosomes on it RIBOSOMES: protein RNA complexes that are responsible for synthesizing proteins - Human cells have 10 mil ribosomes - Ribosomes bound to rough ER feed newly synthesized proteins directly into rough ER where proteins are folded in 3D shape with help of chaperone proteins - - Proteins synthesized on membrane bound ribosomes are destined for 1. Synthesized secretion outside of cell 2. Synthesized membrane embedded proteins FREE RIBOSOMES float freely in cytoplasm synthesize proteins that have intracellular destination (e.g., cytoplasmic proteins involved in cellular metabolism) SMOOTH ER: important role in lipid metabolism, steroid hormone synthesis, detoxification - Ex. Liver have extensive smooth ER as it is the largest detoxification site in the body - As part of LIPID METABOLISM: smooth ER produces phospholipid components of cellular membranes - Smooth ER muscle cells are specialized to store calcium in muscles for muscle contraction - SARCOPLASMIC RETICULUM: muscles in smooth ER GOLGI APPARATUS - Once proteins are synthesized, they are packaged into vesicles to go to Golgi apparatus - Post office of the cell - GOLGI: composed of stack like chambers known as cisternae o Receives protein packed vesicles at the CIS FACE, which modifies them internally and packages them into membrane bound vesicles that bud off from TRANS FACE o Sent to PLASMA MEMBRANE for secretion outside of cell or other cellular membrane LYSOSOMES LYSOSOME: membrane bound vesicles that pinch off from Golgi; house hydrolytic enzymes that chop and digest everything - Garbage disposal system of the cell - Extracellular stuff that needs to be broken down enters the cell through endocytosis ENDOCYTOSIS: vesicle pinches off from cell membrane and vesicle fuses with lysosome inside cell AUTOPHAGY: Intracellular debris in cytoplasm can become enclosed within a vesicle that then fuses with lysosome for degradation - if lysosome releases digestive enzyme into cellular (neutral) cytoplasm = enzymes won’t do much lysosomal enzyme’s function BEST at acidic pH o lysosome interior is kept at pH of 4.5 to 5 PEROXISOMES PEROXISOMES: accumulate and neutralize peroxides (e.g., hydrogen peroxide) - protects against oxidative stress - critical role in breaking down long chains of fatty acids via beta oxidation CYTOSKELETON AND ITS COMPONENTS - cell has skeletal network of fibres that serve as architecture within the cell and acts as highway system for trafficking molecules and vesicles 3 Major Components of Cytoskeleton 1. Microfilaments o Smallest o Double helix (intertwined strands) of actin polymers o G-ACTIN: single actin monomer that is globular protein that polymerizes with other actin monomers o F-ACTIN: form microfilament strand o Play role in cell motility/movement and PHAGOCYTOSIS: engulfment of a cell o Contribute to formation of CLEAVAGE FURROR that splits a cell into 2 o Actin (smallest with diameter of 6nm) components of microfilament interact with protein MYOSIN to initiate muscle contraction 2. Intermediate filaments o Intermediate in size (10nm) o Melting pot of number of different polymers that provide structural support to cell o Involved in cell-to-cell adhesion processes o Ex. Lamin is intermediate filament that provides structural support to nucleus in form of nuclear lamina o Ex. Keratin is intermediate filament that makes up hair, skin, and nails. 3. Microtubules o Wider than microfilaments (25nm) o Polymers of alpha-tubulin and beta-tubulin (TUBULIN DIMERS) o Maintain structural integrity of a cell and act as highways along where motor proteins can traffic intracellular vesicles o Motor proteins use energy derived from ATP to walk down microtubule filament o TWO MOTOR PROTEINS THAT WALK DOWN MICROTUBULE ARE: KINESIN Moves in anterograde fashion (centre to edge of cell such as vesicle being moved by neurotransmitters being delivered down axon to axon terminal) Motor ATPase Moves from minus to plus DYNEIN Moves in retrograde fashion (moves towards centre of the cell such as movement of endocytosed materials from plasma membrane to lysosomes) Motor ATPase Moves from plus to minus o Also form MITOTIC SPINDLES which are structures that separate chromosomes during cell division o Cell undergoing division has two centrosomes (one at either pole of the cell) Each centrosome is composed of two CENTRIOLES which are cylindrical structures made up of nine sets of microtubule triplets o CENTROSOMES are main microtubule organizing centres (MTOC) o Microtubules extend into cell, latch onto chromosomes, and pull them to either side of cell during cell division o Can also see microtubules in EUKARYOTIC FLAGELLA OR CILIA which are whip-like structures involved in cell motility In euk, both cilia and flagella characterized by 9 + 2 STRUCTURE where outer ring of 9 pairs of microtubules surround inner ring of 2 microtubules o FLAGELLA: longer, thinner, tail like appendages that protrude from cell to propel it forward (e.g., sperm cells) Can be found in prokaryotes but the flagella are structurally different EUKARYOTIC FLAGELLA PROKARYOTIC FLAGELLA - Flap back and forth - Use rotary motion - Movement powered by ATP - Powered by proton gradient - Composed of protein called FLAGELLIN, NOT microtubules o CILIA: shorter projections that help move substances along cell surface Ex. Respiratory tract where cilia help clear mucus out of lungs Ex. Line fallopian tube which helps send egg from ovaries to uterus NOTE: both microfilaments and microtubules have POLARITY or directionality to them - Label one end + and other – - Filaments polymerize or grow at the + end o Depolymerize or shrink at – end - TRENDMILLING: when actin/tubulin monomers added at one end, while simultaneously being removed from other end o Moving across cell without changing very much in length - Polymerization can be halted by CAPPING OF PROTEINS, that bind or cap either end to stabilize polymer. LESSON 2: GENETIC CODE AND DNA NUCLEIC ACIDS - Uses 4 nucleotides (A,C,G,T) responsible for all messages to build proteins and cells among other things - DNA: deoxyribonucleic acid, double helix with sugar phosphate backbone held together by phosphodiester bonds – stores and transmits genetic information - RNA: ribonucleic acids, uses uracil and contains ribose as its pentose sugar, more found in single stranded state – decodes DNA to make proteins - Nucleic acids: long strands of DNA sequences made up of various combinations of the 4 nucleotides and have 3 parts: nitrogenous base, a pentose or 5 carbon sugar and phosphate group - Nucleoside: pentose sugar and base SUGARS RNA uses five carbon sugar called ribose and DNA uses deoxyribose phosphate group has negative charge due to large presence of electrons distributed among 4 oxygen atoms Nucleoside diphosphate: 2 phosphates Nucleoside triphosphate: 3 phosphates (seen in ATP) - cyclic AMP is chemical signaling model inside cell. NITROGENOUS BASE Have cyclic structures with many nitrogens Pyrimidines = 3 rings (DNA = C,T) Purines = 2 rings (DNA = A,G) Base pairs held with hydrogen bonds Experience hydrophobic interactions with adjacent bases stacked on top of eachother – enhances stability of nucleic acids individual nucleotides joined in linear string by sugar phosphate backbone (alternating units of pentose sugar and phosphate groups connected by phosphodiester linkages) Phosphate group links 2 nucleotides together by binding 3prime carbon on sugar of one nucleotide and binding 5 prime sugar carbon of second nucleotides Gives directionality 5’ – 3’ and - 3’ – 5’ in opposite direction Complementary strand has to be complementary and antiparallel, running in 3’ to 5’ direction EX. 5’ACTG 3’ - Complementary is 3’TGAC 5’ so 5’ end starts with C - So it should be 5’ CAGT 3’ - If we wanted to separate 2 strands of double stranded DNA, could use heat or chemical denaturants (ex. urea) to separate the 2 strands, called melting the DNA - Melting temperature (Tm): 50% of DNA strands are denatured o When denaturing DNA, are breaking hydrogen bonds that hold the base pairs together and the stacking action o Reversible reassociate with each other called annealing - Higher melting temperature = more GC bonds - Double helix is called right handed - B-DNA is the most common form of DNA – 10.5 base pairs per turn of double helix and every 10 base pairs is around 34 Angstroms (3.4x10-9m) - A-DNA is dehydrated form of B-DNA that can be formed by DNA-RNA hybrid helices, tigher, more condensed structure. - Z-DNA is left handed and looser and less condensed CENTRAL DOGMA AND GENETIC CODE - DNA RNA Protein = Central Dogma of Molecular Biology o DNA serves as template for RNA - DNA is source of heritable genetic information - mRNA stores genetic material that is inherited maternally - reverse transcriptase reverse transcribes RNA back into DNA which virus propgate in host MESSENGER RNA (mRNA) - messenger template to synthesize protein - communicates info about genetic code to translational machinery in cytoplasm to make the right proteins - involves directly in gene expression - read in group of 3 nucleotides at the time (codons) so there are 64 codons - genetic code = degenerate: > 1 codon can encode for 1 AA - AUG = START CODON - UAA, UAG, UGA = STOP CODON - Wobble hypothesis: third nucleotide in codon has wobble room HETEROGENOUS NUCLEAR RNA (hnRNA) - precursor to mRNA that’s spliced and modified to become mature mRNA TRANSFER RNA tRNA - small molecule that transfers AA to growing polypeptide chain - cloverleaf structure that recognizes codons and sequences and add them to polypeptide chain during translation on ribosomes RIBOSOMAL RNA rRNA - synthesized in nucleoli and plays role in translation, making up to 50% of ribosome’s weight - some molecules act as ribozymes - siRNA and miRNA (micro RNA) inhibited gene expression, interfering and destruction of corresponding mRNA DNA REPLICATION THEORIES OF DNA REPLICATION CONSERVATIVE MECHANISM entire DNA molecule completely duplicated to create new DNA molecule (conserving entire original molecule DISPERSIVE DNA backbone broken in multiple places as two strands unwind and new and old DNA would have sections of new and old DNA SEMI CONSERVATIVE two DNA strands separate and serve as template for synthesis of a new strand – new and old DNA molecules would contain both one old and one new strand - 1950s: Meselson and Stahl conducted experiment where E.coli bacteria were grown in medium containing nitrogen 15 isotope (heavier than nitrogen 14 isotope) - - - After many generations, heavier nitrogen isotope was almost completely incorporated into bacterial DNA Bacteria then transferred to medium with nitrogen 14 isotoppe and replicated, DNA then extracted and centrifuged on salt gradient to determine density o If was done with conservative mechanism after one round of replication, would expect 2 separate bands (one older, heavier DNA and one lighter nitrogen 14 containing DNA) o All DNA actually had intermediate density suggesting new DNA molecules contain parts of old and new DNA which first dispersive or semi conservative After 2 rounds of replication, two equal bands appeared in centrifuge tube, one intermediate and one lighter o Rules out dispersive method as it would expect all new DNA molecules to contain nitrogen 15 containing DNA and some newer nitrogen 14 DNA o Best explanation two strands of DNA were separated and used as template Meselson Stahl Experiment o Supported semi-conservative replication (one strand of parent DNA is in every new DNA molecule) PROKARYOTE REPLICATION - One origin of replication – a specific sequence recognized by pre-replication complex, replication proceeds in both directions along circular bacterial chromosome EUKARYOTE REPLICATION. - Multiple origins of replication – DNA replication produces identical sister chromatid connected to original DNA molecule centromere of original chromosome - Unwinding of DNA helix done by helicase enzyme which unwinds 2 strands of DNA at replication fork o Single stranded binding proteins – proteins that latch on separated strands and prevent them from coming back together o Topoisomerase (DNA gyrase) – alleviates supercoiling created by helicase by making incisions in one of DNA strands and rotating cut strand around the other and rejoining then to relax DNA o Primase – synthesizes short RNA primer that has free 3’ hydroxul group used as start point for synthesis of new strand o DNA polymerase – performs DNA synthesis; travels along one of the separated DNA strands and adds nucleotides sequentially after RNA primer in 5’ to 3’ direction – since strands run anti-parallel, DNA polymerase reads DNA in 3’ to 5’ direction, while synthesizing DNA in 5’ to 3’ o Leading strand: moves in direction of helicase, 3’ to 5’ direction o Lagging strand: moves in 5’ to 3’ direction; moving away from helicase and misses new DNA as it’s exposed behind it (solution = new DNA polymerase to latch on as more DNA is exposed and catch up to new strand and as replication fork goes further and another polymerase is added and synthesizes etc.) o Okazaki Fragments: fragments o DNA ligase: attaches Okazaki fragments together by bridging the gaps forming phosphodiester bonds between sugar carbon backbone PROKAYOTIC DNA POLYMERASES ENZYME FUNCTION DNA Polymerase I removes RNA primer, replaces primer with DNA, repairs DNA DNA Polymerase II Repairs DNA DNA Polymerase III Synthesizes new DNA and proofreads DNA via 3’ to 5’ exonuclease activity EUKARYOTIC DNA POLYMERASE ENZYME DNA POLYMERASE α DNA POLYMERASE δ DNA POLYMERASE ε DNA POLYMERASE β DNA POLYMERASE γ OTHER ENZYMES ENZYME REVERSE TRANSCSRIPTASE TELOMERASE FUNCTION Initiates DNA synthesis Synthesize new DNA and replaces RNA primer with DNA Extends leading strand and repairs DNA Repairs DNA Replicates mitochondrial DNA FUNCTION Synthesizes DNA from RNA template – used by retroviruses (HIV) to be integrated into genome of host Extends telomeres (repetitive sequences at ends of eukaryotic chromosomes) in stem/cancer cells LESSON: DNA PACKING AND ORGANIZATION CHROMOSOMES - Human genome is organized into 23 pairs of linear chromosomes, making 46 individual chromosomes total in diploid cell - 23 pairs are autosomes – homologous pairs of chromosomes (22 inherited paternally, 22 inherited maternally, 23rd pair represented by sex chromosomes, X and Y) o Female: XX o Male: XY - DNA is organized into nucleosomes – microscopic structures that contain 200 base pairs of DNA wrapped twice around a spool of 8 histone proteins and H1 histone linker protein o Nucleosomes connected by linker DNA (looks like beads on a string) o First level of organization for DNA packaging - DNA chromosomes are further condensed into chromatin fiber (30 nm diameter) and later packed into loop domains (300 nm in length) and later, further into a highly condensed chromosome CHROMOSOME ANATOMY - 2 identical sister chromatids joined together at central centromere. Each sister chromatid has 2 arms, a p arm and q arm. - - - Chromatin that is highly packaged is called heterochromatin, which is tightly coiled dense form of chromatin seen in cell division Euchromatin (true chromatin) is loose, like spaghetti like configuration and allows DNA to be readily replication and/transcribed Charge attractions between DNA and histone proteins o Nucleotide have nitrogenous base, pentose sugar and phosphate component DNA has negative charge + positive charge from histone [on histone tail] (electrostatic interactions) Regulated by enzymes that add/remove acetyl groups from + charged AA lysine on histone tails Acetylation (addition of acetyl group_ mask this charge by reducing interactions between DNA and histones promoting looser configuration (euchromatin) to allow transcription of genes ex. Histone acetyltransferase acetylation euchromatin (looser) o Histone acetylation – adds acetyl group to histone protein that makes them less + charge; removes positive charge on histone proteins o Histone deacetylation – reintroduces positive charges to histone protein Removal of acetyl group and tightening of interactions between histone and DNA = formation of tightly coiled heterochromatin o Catalyzed by histone deacetylation enzyme. SINGLE COPY AND REPETITIVE DNA - 98.8% of genome is non-coding (don’t code for proteins) (junk DNA) o 44% transposons o 24% introns and regulatory sequence o 15% unique noncoding DNA o 15% repetitive DNA - 1.2% of genome actually contains actual genes (single copy genes) - Among 3 billion base are 20 000 single copy, protein encoding genes – can produce around 2 million unique proteins (alternative splicing of mRNA, post transcription/translational modifications) NON CODING DNA - 1/3 of repetitive DNA is genome of short repeats (higher mutation rate) o Ex. VNTP (Variable Number Tandem Repeats) which are repeats of short DNA sequences (around 10-100 BP) and number of repeats varies by person – polymorphism or genetic differences in number of repeats is done in fingerprinting o Ex. if Maria has 70 BP and Tom has 130 BP and 130 was found at the crime scene, more likely it was Tom. - STRs (Short Tandem Repeats) is similar to VNTP but is shorter (~2 to 6 nucleotide per repeat) - SNIP (Single Nucleotide Polymorphism) – 1 nucleotide in length, between people, stretches of DNA in people may differ in one nucleotide at a specific location, single nucleotide substituions at specific genetic locus which results in different AA sub at SNP’s position o Source of genetic variation throughout DNA can be used as biological marker o Ex. at specific site on q arm of chromosome 19, Maria may have C and Tom has T, if DNA found at crime scene has T, then Murphy is more implicated o NOTE: single nucleotide found that that location must be shared by at least 1% of the population - Transposons: mobile genetic elements known as jumping genes as they can jump to other parts of genomes; around 50% of human genome and has 2 classes o Class I Transposons (Copy and Paste): relocates to other parts of genome by coping and pasting. First transcribed into RNA and code a reverse transcriptase enzyme that uses RNA template to synthesize DNA copy of transposon sequence that can insert itself wherever o Class II Transposons (Cut and Paste): splices transposons sequence out of DNA and allows it to put its roots elsewhere. o When they accidentally insert themselves into coding DNA, can disrupt gene function in deleterious ways. SITES OF REPTITIVE DNA - Centromere is central region of chromosome that unites 2 sister chromatids – contain blocks of repetitive DNA sequences that are rich in GC base pairs and tightly packaged as heterochromatin which maintains structural integrity for association of kinetochore proteins and microtubule spindle fibers during mitosis - Telomeres: end part of chromosome arms, RNA dependent DNA polymerase that binds RNA sequence, complementary to telomere repeat and then DNA polymerase fills in complementary strand – repetitive sequence of non-coding DNA made up of repeat TTAGGG which cap the ends of chromosomes - When replication fork reaches end of chromosome, DNA polymerase has no trouble replicating leading strand but lagging stand is more challenging to replicate because if we replace an RNA primer at the end to synthesize a final Okazaki fragment, have no way to replace RNA primer with DNA as initiating DNA synthesis requires free 3’ hydroxyl group from RNA primer o Left over with short single strand overhand o After many replications, becomes problem as telomeric ends keep shortening o Ex. stem cells, cancer cells, germ cells, exhibit telomerase which bring telomeres to run risk of telomeric shortening cutting into actual genetic sequences LESSON: GENETIC INHERITANCE MENDELIAN GENETICS - Gregor Mendel = father of genetics using peas - Phenotype: physical characteristics (individual’s outwards appearance) - Genotype: actual genetic make up - Gene: DNA sequence that codes for protein that produces given trait, can be found at specific location on chromosome (locus) - Alleles: Variations in the same genes; can be dominant or recessive o Dominant: one copy required to express trait (use capital letters) o Recessive: two copies required to express trait (lowercase letters) o Homozygous: individual with 2 copies of same allele Homozygous dominant (BB) Homozygous recessive (bb) o Heterozygous: individual with 2 different alleles (Bb) carriers of recessive allele o Hemizygous: one copy of allele is present - Wildtype: default genotype/phenotype in population; encodes most common variants of trait (w+w+) - Mutant: result from mutated alleles (ww) LOSS OF FUNCTION MUTATION - Mutations tend to be recessive that aren’t noticed unless they affect both alleles GAIN OF FUNCTION MUTATIONS - Protein gain’s new function – dominant or recessive? - Ex. enzyme that is switchable (turned on or off) and has gain of function mutation that makes the enzyme always turned on, are mostly dominant. EXAMPLES Punnett Squares - F1 generation: RR x rr = all circular peas - F2 generation: Rr x Rr = 75% round, 25% wrinkled peas - Genotypic ratio: 1:2:1 (RR: 25%, Rr: 50%, rr: 25%) - Phenotypic ratio: 3:1 (RR or Rr: 75%, rr: 25%) Test Cross: dominant phenotype of unknown genotype is crossed with a homozygous recessive organism. (Ex. RR vs rr) would result in all heterozygous offspring (dominant round phenotype) Backcrossing: when hybrid is crossed back with its parents or an organism genetically like the parent. Goal is to obtain offspring similar to parents. DIHYBRID CROSS - phenotypic ratio: 9:3:3:1 9/16 dominant for traits yellow and round 3/16 dominant for one trait round but green 3/16 dominant for yellow but wrinkled 1/16 are recessive for both traits (wrinkled and green) GENETIC PRINCIPLES - Exceptions (deviations from simple dichotomies) TYPES OF DOMINANCE - Complete dominance – one dominant allele masks the effect of recessive allele - Codominance – two dominant alleles can be expressed simultaneously o Ex. ABO blood typing system where red blood cell can express A surface antigen, B surface antigen, both or neither) o If individual has both A and B alleles, one doesn’t mask presence of another but both are dominant and expressed (AB blood type) - Incomplete dominance – heterozygote exhibits blended phenotype that’s intermediate between 2 homozygous phenotype o Ex. snapdragon flower where homozygous flowers are completely red or white but heterozygous are blended of pink o Associated with gradient phenotype PENETRANCE AND EXPRESSIVITY - Penetrance: likelihood that an individual with a given phenotype will manifest corresponding phenotype (display associate phenotype) o Is whether or not gene is expressed o ex. females that have mutation in BRCA1 gene has 80% risk of developing breast cancer in lifetime – exhibits 80% penetrance. o Isn’t 100% due to influence of other genes or environmental factors o Like yes or no - Expressivity: intensity or extent of a variation in a phenotype. o Ex. even if 10 people expressed the same phenotype prone to congestive heart failure, severity of condition may differ between individual o Ejection fraction (volume of blood heart can pump) might be 40% in one and 20% in another o Shades of grey - NOTE: environmental conditions and other factors that can influence an individual’s phenotype INHERITANCE PATTERNS - Genetic pedigree: specific type of family tree; males are squares, females are circles and those affected by the condition are shaded in (phenotype) AUTOSOMAL PATTERNS OF INHERITANCE - Autosomal dominant: Traits inherited in autosomal fashion, where allele in question is dominant and someone with one copy of that allele will express dominant phenotype (ex. dominant black hair) - Autosomal recessive: individual must inherit two copies of recessive allele to express the recessive phenotype (ex. recessive red hair) possible for recessive traits/genes to skip generations One parent with one copy of the recessive allele can be a carrier who won’t manifest recessive phenotype but child may be affected if they inherit one copy from each parent A child with a condition inherited in autosomal dominant fashion must have one parent with the condition unless the mutant allele arose spontaneously (de novo) SEX LINKED INHERITANCE - Y chromo is smaller, less genes than X, all sex linked disorders are x-linked conditions: asymmetry in the sex of affected individuals o X linked recessive disorders more common than sex linked dominant disorders X LINKED RECESSIVE PATTERN OF INHERITANCE - Recessive phenotype expressed in females who have the recessive allele present on both X chromosomes. - Males express phenotype if they have only one copy of the recessive allele as they only have 1 x chromosome. - Ex. hemophilia - More prevalent in males, inherited by mothers GENETIC LINKAGE LAW SEGREGATION INDEPENDENT ASSORTMENT DOMINANCE DEF Allele pairs segregate randomly from each other into gametes (during meiosis from M and P) Alleles for separate traits are independently inherited (ex. maternal and paternal chromosomes on metaphase plate) Recessive alleles will be masked to some degree by dominant alleles PARENT CROSS Rr x Rr Round x round OFFSPRING 75% round (RR or Rr) 25% wrinkled (rr) RrYy x RrYy Round, yellow x round, yellow 9/16 round, yellow 3/16 round, green 3/16 wrinkled yellow 1/16 wrinkled green RR x rr Round x wrinkled 100% round (Rr) INDEPENDENT ASSORTMENT - Consider 2 genes on the same chromosome – are going to be dragged to same pole of cell and same daughter gamete but not always inherited together. - During prophase 1, homologous chromosomes experience crossing over between 2 sister chromatids, swapping paternal and material genetics, called genetic recombination. - Chiasmata: crossing over points, form at random locations between 2 sister chromatids. o less distance between them less likely to cross over and undergo recombination and more likely to be inherited together GENETIC LINKAGE centimorgans: distance associated with 1% increments/change in recombination frequency. o Ex. 2 genes with 5 centimorgans between them recombine around 5% of the time, 30 centimorgans recombine 30% of the time Two genes with recombination frequency of ~50% = unlinked. (either on separate chromosomes or separate enough that they recombine as much as if they were on separate chromosome) SINGLE VS DOUBLE CROSS OVER recombination occurs at any odd number of cross over points between 2 genes POPULATION GENETICS AND EVOLUTION PRINCIPLES OF EVOLUTION Evolutionary System - Variation – genotypic and phenotypic variation - Reproduction – sexual and asexual forms if reproduction - Differential reproduction due to selective pressures – must be more able to reproduce more than unfavourable variations. - Natural selection: favourable traits are favoured in population over time – selects favourable traits over time, differential survival and reproduction GROUP SELECTION - Natural selection applied to group level - Inclusive fitness: traits passed on that promote survival of the group (altruism, empathy) - if you help close relatives to survive and vice versa, you share many of those alleles and more prevalent in future generations if passed on (J.B.S Haldane) – selfish genes. HARDY-WEINBERG EQUILIBRIUM THE GENE POOL - allele = alternative forms of genes - evolutionary success indicated by how popular certain alleles are in gene pool. - Hardy Weinberg Equilibrium: mathematical relationship between allele and phenotype frequencies – used to model stable gene pools - Criteria for Stable Genepool o No mutation o Random matin g o No gene flow (cannot leave and enter randomly) o Large population size o No natural selection - p+q = 100% of alleles in population - (p+q)2=12 p2+2pq +q2 = 1 o p2 (AA) +2pq (Aa) +q2 (aa) = 1 - T REX EXAMPLE: 16% of t-rex are short armed (q2=0.16, q=0.4) p2+2pq +q2 = 1 - q2=0.16 q = 0.4 p+q=1 p+0.4=1 p = 0.6 looking for # of t rex’s that are carriers (homozygous) which is 2pq (Aa) - knowing that p=0.6 and q=0.4 - 2pq = 2(0.6)(0.4) - 2pq = 0.48 or 48% that are heterozygous SELECTION AND GENETIC CHANGE - Selective pressures work on spectrum for non-categorical traits (height and weight) - Traits are often polygenic: determined by more than 1 gene STABILIZING SELECTION - Favours intermediate phenotypes and selects against extreme phenotypes. - Ex. Muscle strength in Huskies as they must go through snowy terrains but not too large to sink into slow. DIRECTIONAL SELECTION - Favours an extreme phenotype in one direction. - Ex. During ice age, black bears in Europe decreased in size during warmer periods and increased in size during cold periods. DISRUPTIVE SELECTION - Inverse of stabilizing selection – favours extreme phenotypes over intermediate phenotypes. - Ex. Darwin’s island finches – had small or large beaks but no medium beaks as it was because of the size of seeds available (small or large) so medium beaks provided no advantage. MECHANISMS WHERE GENE POOLS MAY CHANGE - Natural selection - Chance - Genetic drift – change in gene pool due to random chance, often in smaller populations whereby certain alleles are produced often but not due to any selective pressures. EXAMPLES OF GENETIC DRIFT: BOTTLENECK EFFECT -Bottleneck effect: where genetic drift occurs due to some external event, ex. Natural disaster, that greatly reduces the population size. -Artifically increases/decreases allele frequency -Ex. Using a light to get rid of bacteria but not thorough, leaving some bacteria behind. Many of surviving bacteria is resistant to anti-biotics by change, so the genes are more prevalent in population due to radiation induced natural disaster. FOUNDERS EFFECT - - When a small group breaks off from the larger population and founds a new colony Reduction in genetic diversity due to smaller group and has variation and genes may be non-representative of the original population. Allele frequencies may shift but not due to selective pressures. Random errors in the genome accumulate at a fixed rate – extent to where 2 genomes differ serve as proxy for how long ago they had a shared common ancestor. - Molecular clock can be applied to help solve questions about mystery diseases (ex. How HIV originated and spread. o can track evolution of specific viral strain. Ex. Analyzing difference in evolutionary history of 2 similar viral genomes to tell us how closely related they are, which may provide clues about where it came from. o Can determine how long ago two different species diverged from a common ancestor. (when speciation events occurred) GENE FLOW - Gene flow: movement of alleles due migration between population, which may increase the genetic diversity of the population. - Ex. Lions from 2 prides mating with eachother. FACTORS INFLUENCING EVOLUTION - Gene flow - Random chance - Migration - Mutation - Selective pressures SPECIATION AND EVOLUTION - Species: group of organisms that can reproduce to produce fertile offspring and reproductively isolated from other groups (biological species concept) o Doesn’t account for asexual reproduction, hybridization and gene sharing between 2 species REPRODUCTIVE ISOLATION - prezygotic barriers: prevents zygote from forming - postzygotic barriers: prevent formed zygote from being viable or fertile - hybrids: offspring produced by 2 different species (ex. Mule = horse and donkey, is viable but not fertile, called hybrid sterility) - hybrid breakdown: first generation is fertile but offspring of that hybrid are infertile. INBREEDING - inbreeding: breeding between individuals that are closely genetically related. - carries risk of greater phenotypic expression of deleterious recessive mutations within population. OUTBREEDING - outbreeding: breeding with and passing alleles between unrelated members of the same species, genetically distant members of species - can be beneficial for genetic diversity, especially in inbred populations. - - - - divergent evolution: when species diverge from common ancestor to become more genetically and phenotypically different. o ex. Human fit vs chimp foot convergent evolution: when two different species independently evolve the same or similar trait and become more phenotypically similar to adapt to similar environments. o Ex. Echolocation in bats and dolphins which are derived from the same genetic mutation but not inherited by common ancestor but evolved independently Parallel evolution: when 2 closely related species share a common ancestor and evolve similar traits but independent of one another o ex. butterfly species developing similar colouration patterns. Coevolution: evolution of 2 species in response to each other o Common in symbiotic relationships: between 2 or more organisms that benefits, harms, or has no effect on one or both parties. o Mutualism: both organisms benefit (ex. Humans and intestinal bacteria which synthesize vitamins, break down food in exchange for nutrients) o Commensalism: benefits one party without affecting the other (ex. Human mites that feed on dead hair follicles) o Parasitism: when parasite derives benefit from the host which is harmed in the process (ex. Tapeworms in human intestines.) CELL CYCLE - - - - - - Average cell cycle lasts around 24 hours Makes 2 daughter cells G0: resting phase, makes proteins but not actively dividing (last indefinitely) – can be called nonprolific or non mitotic cells (ex. Neurons) M phase: mitosis or meiosis where cell division occurs Interphase: takes 90% of cell cycle, cell growth, organelle duplication, protein synthesis, DNA replication occurs o can be broken down into G1, S (synthesis), G2 growth phase o most of cell is loosely packed as euchromatin to allow transcription and replication to occur o EXCEPTION: heterochromatin which has tightly packed DNA in centromere G1: cell size and volume of cytoplasm increases as cell grows, actively synthesizing proteins and molecules to prepare for division. o Must pass G1 checkpoint (restriction point) where cell decides whether to commit to cell division. o if cell is damage free, conditions are right (nutrients and growth factors) and is large enough to divide, allowed to proceed to S phase and if not, enters G0 S phase: DNA synthesis/replication occurs – all 46 human chromosomes can be replicated to form identical sister chromatids. o NOTE: while actual DNA content doubles, number of chromosomes remain the same G2: undergoes growth again – must pass G2 checkpoint before going to mitosis o If DNA not properly replicated or complete, or DNA damage present, cell may not be able to proceed (or pause until damage is fixed) M phase: when cell division occurs through mitosis (cell divides into 2 identical copies ot itself) or meiosis (sexual reproduction used to produce gametes) o When cell transitions from interphase to m phase, rest of DNA condenses more densely into chromatin (heterochromatin) making it easier for cells to line up and separate chromosomes. o Must pass M checkpoint/Spindle checkpoint which ensures chromosomes are correctly attached to microtubule fibres that will pull them to either side of the pole of the cell. CYCLIN - Present at every stage of cell cycle and each cyclin specialized in particular phase. - Cyclins levels are cyclical, so levels of any given cyclin are low during most of cell cycle. - When levels of particular cyclin are high, bind and activate cyclin dependent kinases (CDKs) which phosphorylate and activate target proteins that promote phases of certain cell cycle until next cyclin comes - Kinases: enzymes that phosphorylate molecules MITOSIS - Mitosis: cell division that results in one cell dividing into 2 identical copies of a cell - When mitosis goes unchecked, can result in cancer, wounds would never heal ex. - Occurs in somatic cells – all the cells in the body except germ cells (sperm and egg cells) - 4 phases: prophase, metaphase, anaphase, telophase (PMAT) - Preceded by interphase (cell grows and replicates DNA), mitosis divides all content equally between 2 daughter cells. - Some cells go through mitosis regularly (ex. Epithelial cell in digestive tract) - When cell division goes unchecked, can spiral out of control and result in cancer PROPHASE - Nuclear envelop disintegrates, nucleoli disappears, and DNA condenses into tightly packed chromatin - DNA has already been replicated during S phase, making 2 identical copies of a chromosome called sister chromatids joined together at centromere. o NOTE: homologous chromosomes are pairs of maternal and paternal copies of same chromosomes, can contain different alleles of same gene o Kinetochore: complex of proteins assembles on centromere on chromosome o Centromeres are on the poles of the cells which are structures composed of 2 cylindrical centrioles, which are made of microtubule fibers o Microtubule fibers/spindle fibers extend from both centrosome at either pole of the cell forming mitotic spindle, and then, attaches to kinetochore on chromosome which allows them to pull apart sister chromatids. METAPHASE - Spindle fibers organized 45 human chromosomes in a line down the cell, called metaphase plate. - Cell must pass M checkpoint during cell cycle. - - Nondisjunction: failure of one or more pairs of homologous chromosomes or sister chromatids to separate normally during cell division, usually resulting in abnormal distribution of chromosomes in daughter cell. Aneuploidy: abnormal number of chromosomes ANAPHASE - Pull spindle fibers from metaphase plate to either side - Microtubule fibers attached to centromeres pull apart two identical sister chromatid of each chromosome so that 1 sister chromatid of chromosome 1 is placed in 1 daughter cell and the other chromatid is placed in the other daughter cell TELOPHASE - Have separated chromosomes into 2 identical teams on either side of cell - Nuclear envelope and nucleoli reappear but for each daughter cell - Cytokinesis: cytoplasmic division of a cell where cytoplasm is split into 2 so 2 identical cells are created o Takes place when actin and myosin filaments form contractile ring down middle of cell creating cleavage furrow on cell surface that splits cell membrane so they can split. MEIOSIS - Form of sexual reproduction that generates haploid gametes (sperm and egg cells) that can combine with other gametes via fertilization to form single celled diploid zygotes which grow to embryo fetus. - Greatly enhances genetic variability in offspring and confers evolutionary advantage (offspring may be better adapted to environment) - Only occurs in germ cells found in testes and ovaries o Germ cells undergo meiosis to produce ova and sperm via oogenesis and spermatogenesis. - 2 rounds of division – contain 4 phases of mitosis (PMAT) o Meiosis I o Meiosis II MEIOSIS I Prophase I Maternal and paternal copies of each chromosome pair up with each other in process called synapsis, which forms structures called tetrads (4 sister chromatids total in form of 2 pairs of homologous chromosomes) Each pair is referred to as homologous pair because they represent same chromosomes (genes the same but alleles differ, such as pat = green eyes, mat = brown) Adjacent chromatids from homologous chromosomes exchange genetic information with eachother by crossing over Chiasmata: points of crossover where mat and pat exchange chunks of genetic sequence, which results in recombinant DNA Metaphase I - Chromosome line sup in 23 pairs (each homologous pair is a tetrad linked at centromere) in random arrangement which explains independent assortment o Independent assortment: during meiosis, individual genes on different chromosomes assort into daughter cells independently of each other, allowing different combinations of traits to appear in offspring. Anaphase I - Chromosomes pulled apart to different poles of cell. - Two chromosomes in homologous pairs are separated to opposite poles. - Each daughter cell contains half the amount of chromosomes (23) but with 2 sister chromatids still attached. Telophase I - Splits into 2 - Parent cells go from diploid haploid. - Daughter cells become haploid at end of meiosis one - Chromosomes will be paired with their identical copies. MEIOSIS II Prophase II - Nuclear envelope breaks down Metaphase II - 23 Chromosomes line up at metaphase plate in single file line. Anaphase II - Sister chromatids are pulled apart to either pole of cell Telophase II - Nuclear envelope reforms and two cells bud off, causing 4 haploid daughter cells MITOSIS - Somatic cells - Takes cell and turns it into 2 identical copies (2N) - Divides into 2 so both daughter cells have exactly half of genetic material. - 46 chromosomes line up single file - Sister chromatids on same chromosome pulled apart in mitosis - Genetically identical daughter cells - 2 diploid daughter cells MEIOSIS - Germ cells - Generates daughter cells that are genetically variable - 4 daughter cells that have different alleles in genetic makeup - Half as many chromosomes as parent cells (parent = diploid 2N with 46 chromosomes, each daughter is haploid 1N with 23 chromosomes) - Daughter cells are gametes - - Meiosis I: maternal and paternal chromosomes in homologous pair form tetrads and crossing over occurs between adjacent chromatids Meiosis I: 23 pairs of chromosomes line up Meiosis I: homologous chromosomes are pulled apart during anaphase I Genetically different haploiddaughter cells MICROBIOLOGY ARCHAEA AND BACTERIA PROKARYOTES No distinct nucleus and no membrane bound organelles. ARCHEA - Unicellular prokaryotic organisms - Many are extremophiles. o Thrive in extreme temperatures, pH or salt conditions. - Use whatever energy source they can get on flagella (organic compounds, ammonia, metal ions, photosynthetic pathways.) BACTERIA - Human body has almost same bacteria as cells - Symbiotic relationships: o Mutualistic: beneficial to both parties. o Commensal: beneficial to one, no harm to the other. o Parasitic: beneficial to one, harmful to host o Pathogens: bacteria that are harmful to the body (E Coli, MRSA) - Most bacteria in human body is commensal o Bacteria that has positive effect on body are mutualistic (ex. Bacteria that synthesize Vitamin K and B7 in large intestine) - Pathogenic Bacteria: harm their hosts through different mechanisms o Reproduce intracellularly, outside of host cells or can secrete damaging toxins o Normally treated with anti-biotics but rise of antibiotic resistance. o Multidrug resistance is responsible for MRSA. TYPES OF BACTERIA - Categorized by shape o Coccus = sphere shaped o Bacillus = rod shaped o Spirillum = spiral shaped. - Oxygen requirements (aerobic) o Obligate aerobes: oxygen required for metabolism. o Anaerobes: oxygen not required for metabolism. Obligate anaerobes: cannot survive in presence of oxygen Aerotolerant anaerobe: tolerate oxygen Facultative anaerobes: aerobic metabolism when oxygen is present; anerobic metabolism when oxygen is lacking. PROKARYOTIC CELL STRUCTURE STRUCTURAL FEATURES - Lack of membrane bound nucleus and organelles. - Bacterial DNA is located in nucleoid region of cytoplasm. - Genetic material of bacteria is single circular chromosome composed of double stranded DNA - DNA transcription and translation can occur simultaneously in prokaryotes. - Plasmids: smaller, circular DNA fragments of non-essential genes o can confer antibiotic resistance or can code for virulence factors that enhance bacteria’s virulence. Virulence: ability to spread and harm it’s host. BACTERIUM STRUCTURE - Encased in cell wall which provides structural support and extra layer of defense. o Wall is rich in rigid polysaccharide peptidoglycan (takes up substance called gram stain) GRAM STAINING - Technique used to separate bacteria into 2 categories based on structure of their cell walls, can also identify bacteria species. - Gram Positive Bacteria: thick peptidoglycan rich cell wall traps stain within cross linked polymers, turning them into a deep shade of purple. - Gram Negative Bacteria: thin layer of peptidoglycan surrounded by a lipopolysaccharide. o when cell is washed, strips outer membrane of gram negative bacteria, and washes away any exposed crystal violet complexes. o Cell walls do not retain stain so after washing, counterstain safranin is applied, which turns gram negative bacteria a shade of pink, so it’s visible. - o Likely E.Coli. o Difficult to treat. o Lipopolysaccharide is antigenic and capable of inducing innate immune response in humans. During gram staining, crustal violet dye is applied to bacteria, then iodine solution is added, forming insoluble complex within dye, then cells are washed with ethanol or acetone. BACTERIAL RESPIRATION - ETC lines inner mitochondrial membrane, ETC is located on cell membrane. - Endosymbiotic theory: mitochondria originated when cyanobacterium was engulfed by early eukaryotic cell. Since then, coexist in symbiotic relationship (eukaryotic mitochondria) RIBOSOMES IN BACTERIA - Prokaryotic ribosome has 30S and 50S subunits, forming 70S ribosome. - Eukaryotic ribosome has 60S and 40S subunits, forming 80S ribosome. - Svedberg Units: describe sedimentation rates – how long it takes particle to sediment in test tube during centrifugation (related to mass and shape) o Aren’t additive. - Structure differences make prokaryotic ribosomes target for certain antibiotics which try to disrupt bacterial protein synthesis without affecting function of eukaryotic cell. FLAGELLA - Eukaryotic cells are composed of specific arrangement of microtubule filaments that whips back and forth to propel cell in environment. - Prokaryotic cells rotate to propel cell forward and has 3 parts 1. Filaments - made of flagellin) 2. Basal body - structure embedded in membrane responsible for rotation) 3. Hook – connects filament and basal body. PROKARYOTIC REPRODUCTION AND GENE TRANSFER - Bacteria reproduces by binary fission, where single cell divides into 2 identical daughter cells. 1. Prokaryotic parent cell 2. Circular chromosome is replicated. 3. Cell growth occurs. 4. New cell wall grows and segregates between 2 daughter cells. 5. Two chromosomes pulled towards either half of the cell 6. Parent cell splits into 2 daughter cells. - Takes around 20 minutes but growth is constrained by nutrient resources available, which results in characteristic growth pattern with 4 phases: 1. Lag phase – bacteria introduced and must adapt to new environment so growth lags in this stage. 2. Exponential phase – population multiplies rapidly, y axis of growth curve is logarithmic to show growth rate. 3. Stationary phase – growth is stalled as maximum stable population is reached, growth is limited by nutrients and space. 4. Death phase – resources are depleted, and bacteria will start to die off. HORIZONTAL GENE TRANSFER - Bacteria’s means of achieving genetic variability despite asexual reproduction. - NOT reproduction (no new organisms are created) - Transfer of genes to existing bacteria. 3 Mechanisms for sharing genes 1. Transformation – bacteria absorbs DNA from external environment. a. Demonstrated by Fred Griffith (1928), Avery McCarthy 2. Transduction – gene transfer mediated by viruses that infect bacteria (bacteriophages) a. During infection, bacteriophages incorporate part of bacterial genome from host, and store it until they can infect next bacterial cell, which will typically be integrated into new host genome. b. Helps increase microbiological diversity c. Implication for biotechnology 3. Conjugation – special plasmid (fertility factor) is transferred between 2 cells a. F+ Bacterium has fertility factor, with mates with b. F- Bacterium: lacks fertility factor c. F+ and F- bacterium mate by extended sex pilus and forming bridge between 2 cells. Plasmid is replicated and transferred to latter bacterium, which becomes F+ bacterium. d. Good for genetic diversity but mechanism of antibiotic resistance. VIRUSES, PRIONS, VIROIDS, PARASITES VIRUSES - Viruses always pathogenic, wanting to infect cells and hijack machinery in order to multiply and spread. - Most scientists consider viruses as “non living” VIRUS STRUCTURE - - - virion: fully assembled, infectious virus. Genetic material is housed within capsid. Bacteriophages: viruses that infect bacteria has more complex structure with a head, tail and tail fibers attaches to cells by tail fibers and injects its genome into host cell. In some viruses that infection humans and animals, capsid is enclosed in lipid envelope mostly made of phospholipids and proteins. Enveloped virsues are sentivie to environmental factors (light and heat) Enveloped viruses must be transmitted via bodily fluids (HIV) Non-enveloped viruses can survive outside the virus (ex. Rotavirus) Viruses are smaller than pro and euk cells. o Human cells = 10-15mm in diameter o Bacteria = 0.5-5mm in length o Viruses = 20 to 300/400 nm in diameter. Genomes vary in size with 4-1000 protein encoding genes. VIRAL GENOMES 1. Single stranded DNA (ssDNA) 2. Double stranded DNA (dsDNA) 3. Single stranded RNA (ssRNA) a. Considered positive sense if they contain mRNA that can be directly translated into protein b. Considered negative sense of RNA is complementary to mRNA and must be copied into mRNA by viral enzyme RNA replicase prior to translation. 4. Double stranded RNA (dsRNA) VIRAL INFECTION - When RNA virus infects host cell with DNA genome, genetic material must be reverse transcribed into DNA, if goal is to integrate into host genome. - Retroviruses (single stranded RNA virus) contain enzyme reverse transcriptase, that synthesizes DNA from viral RNA so it can be integrated into host genome and integrated into host DNA. o Reverse transcriptase: used in reverse transcriptase PCR, where RNA template is reverse transcribed into complementary DNA for PCR amplification. Ex. HIV, responsible for AIDs (responsible for death of ~35m) and hard to treat as a retrovirus, it’s genetic content becomes integrated into host genome, required death of host cell to destroy the virus. Treated with antiretroviral drugs that target reverse transcriptase. o Retrotransposons: mobile elements that make up 40% of human genome. VIRUS REPLICATION - STEPS o Infect cell with its genetic material. o Hijack host cellular machinery to replicate. o Assemble number of new virions o Exit host cell. o Spread to new ones. - Replication process depends on wither it’s a DNA or RNA viral genome. - DNA Viral Genome: translocates to nucleus, transcribed by host cell’s RNA polymerase. - RNA Viral Genome: immediately translated into protein in host cytoplasm OR reverse transcribed by reverse transcriptase (retrovirus) - Release of virions from the host cell is called extrusion, which leaves host cell intact as viral particles move onto their next target. BACTERIOPHAGE REPLICATION - - Viruses that affect bacteria - Initiate one of two life cycles: - Lytic cycle: when bacteriophage replicates rapidly in cytoplasm, eventually producing so many virions that host cell lyses and releases army of virion into its environment. - Lysogenic Cycle: marked by integration of viral genetic material into the host genome. Virus, now called prophage or provirus, lies dormant until triggered by environmental signal, where viral sequences are transcribed into RNA, resuming lytic cycle. Host cell is going to experience cell lysis. ANTIVIRAL MECHANISM - Life cycle makes viable targets for antiviral medication. Along with meds that target reverse transcriptase, others target attachment of virus to host cell, interfering with nucleotide and protein synthesis, and blocking reassembly of virions. VIROIDS - Smaller, simple RNA particles that infect plants. - Single stranded RNA sequences that lack capsid or envelope - Bind to complementary RNA sequences in plants to silence gene expression. PRIONS - Misfolded proteins that cause other proteins to misfold and aggregate together, harming cellular function. - Responsible for Creutzfeldt-Jakob disease, related to mad cow disease. - Responsible for fatal familial insomnia and kuru. OTHER MICROORGANISMS - Non-harmful fungi (mycobiome) - Pathogenic fungi (yeast infection, ring worm, other fungal diseases) - Parasitic organisms o Protozoa: single celled parasites (ex. Protozoan carried by mosquitos that cause malaria) o Helminths: multicellular worms (tapeworm, roundworms) o Ectoparasites: multicellular parasites that live outside the host. (ex. Fleas, lice) o Immune system mediates these infections primarily by eosinophils and IgE antibodies. o Are eukaryotes. BIOTECHNOLOGY RECOMBINANT DNA RESTRICTION ENZYMES/ENDONUCLEASES - Prokaryotic DNAses that evolved in bacteria and archaea as an internal defense system against viruses. (and bacteriophages) - Some are non-specific and will cleave DNA when they can. - Recognize and cleave highly specific recognition sites on DNA o 4-8 base pairs long o Palindromic sequences: symmetric, inverted repeats where 5’ to 3’ sequence on one strand is identical to 5’ to 3’ sequence on complementary strand. - When restriction enzymes cleave DNA at restriction sites, can create blunt (2nd picture) or frayed “sticky” ends. (1st picture) o Sticky ends preferred as they can be ligated together more effectively by DNA ligase. PLASMIDS - Insert sequence/gene into vector, smaller DNA molecule, that transfers DNA into target cells, hijack replication machinery and can integrate into host genome. - 2 types of vectors - Plasmids: circular DNA molecules that bacterial readily uptakes in environment and that replicates independently of bacterial chromosome. o gene sequence or “insert” is synthesized that contains 2 restriction sites, matching the restriction sites on a plasmid vector. o Vector may contain other functional sequences (promotor, origin of replication) to enable transcription of target gene. o Vector may also have antibiotic resistance gene (ampicillin resistance cassette) so bacteria that has incorporated plasmid can be selected for, with an antibiotic that wipes out any bacteria lacking resistance gene. o Main goal = allow gene of interest to replicate within the cell - Insert and vector are digested with restriction enzymes generating complementary sticky ends that can be ligated together. o Should first eliminate any excess DNA to improve efficiency where insert and vector ligate together. o To do this, the 2 mixtures are run on gen, electrophoresis, to separate fragment by size, fragments of interest are cut out of gel, combined and ligated together by DNA ligase. - Then, need to treat bacteria with plasmid mixture o Bacteria readily uptakes external DNA by transformation. - - - Need system to differentiate from bacteria transformed with recombinant plasmids and non-recombinant plasmids (no gene of interest) or those that did not take up DNA o Solution = engineering plasmids with antibiotic resistance genes o When we treat plate of bacteria with antibiotic, only those with antibiotic resistance gene will survive o some of these bacteria might contain non-recombinant plasmid, lacking novel gene sequence. Plasmids also constructed with reporter gene: which does for a protein that elicits some visible phenotypic change (ex. Colour), where insert is spliced into reporter gene on vector interrupting protein synthesis and resulting phenotypic change o Ex. Gene of interest is inserted in middle of gene for beta-galactosidase, which deactivates this enzyme and prevents it from cleaving substrate. o If cleavage of substrate turns bacterial colonies blue, blue colonies must contain an intact beta-galactosidase gene (intact lacZ) o white colonies must contain recombinant plasmids with split beta-galactosidase gene (split lacZ) 2-4kb in length BACTERIOPHAGES - 4-25kb - Viruses that infect bacteria with their DNA, which enables uptake of much larger DNA sequences than what is normal by bacterial transformation. RNA AND DNA - Same techniques can be used for RNA, but need to convert it into DNA first with reverse transcriptase to turn RNA into complementary DNA or cDNA (complementary to template RNA) - cDNA can be ligated into a plasmid and replicated in host cell o cDNA: contains exons but no introns o advantage: eukaryotic genes are usually interrupted by noncoding sequences which are removed by splicing in mRNA o genomic DNA and cDNA fragments have been used to construct DNA libraries hat allow researchers to study structures and functions of genes throughout organism’s genome. BIOTECHNOLOGY APPLICATIONS AND ETHICS GOALS OF GENETIC ENGINEERING 1. mass protein production a. recombinant expression vectors in bacterial systems ideal for amplifying genes of interest and generating large amounts of synthetic protein by hijacking replication and protein synthesis machinery of prokaryotes b. use recombinant bacterial plasmids to mass produce insulin, vaccine components, interferon, tumor necrosis factor 2. alterations to genetic code a. can be used to edit genes. b. recombinant plasmids are frequently used to modify and inactivate genes in cell culture or animal model. c. Transgenic organisms: organisms whose genomes have been modified (introducing novel gene, modifying existing gene via plasmid or bacteriophage mediated gene transfer) d. Knockout organisms: organisms who have had one or more genes deleted by swapping out target gene to change the genome, with a mutant or defunct gene copy. i. HOW? Under right conditions, vector will undergo homologous recombination with target DNA sequence, exchanging DNA between vector and host genome. ii. Similar genes used to treat humans such as cystic fibrosis, a genetic condition affecting lungs caused by 1 mutation that causes CFTR gene non-functional. By delivering functional copy of this gene by plasmid, to patient’s lungs epithelial cell, normal allele should replace non-functional allele and help recover lung function. GMO - Genetically modified organisms - Genetically engineered strains of crops developed that are resistant to insects, pesticides and many environmental conditions. - Some strains modified to have certain nutrients (beta-carotene in golden rice) RESPIRATORY SYSTEM RESPIRATORY SYSTEM ANATOMY - Air enters oral cavity/nostrils (nares) nasal cavity has mucous membrane covered with tiny hairs called vibrissae that help out filter particulate matter (first line immune defense) air passes pharynx (back of the mouth) path diverges into trachea and esophagus epiglottis: flag of cartilaginous tissue that covers and protects trachea from food, shunting food towards esophagus opens up to allow air to pass into trachea during breathing. Air passes to larynx which contains vocal cords, which vibrate when air moves over them, that is consciously controlled during speaking and singing. TRACHEA - Air continues down trachea with is linked with ciliated epithelial cells and mucous producing goblet cells. o Mucous traps stray bacteria/particular matter and thrust upwards by cilia towards oral cavity to be expelled/swallowed as phlegm. BRONCHI AND BRONCHIOLE - Trachea is branched into two main bronchi, which also split off. o Bronchitis: bronchi infected. o Bronchiolitis: bronchioles infected. ALVEOLI - Smallest bronchioles are terminated in air sacs known as alveoli. - Alveoli make up large surface area of the lungs, as they are tightly packed so total surface area available to gas exchange. - Surface tension of liquid lining alveoli normally cause them to collapse on themselves BUT they are coated in layer of surfactant that breaks up and reduces surface tension. - Neonatal Respiratory Distress Syndrome – insufficient surfactant production in fetuses. o Fixed artificially by administering artificial surfactants until lungs grow large enough to produce their own. o Pneumonia – when alveoli become infected. GAS EXCHANGE - Alveoli specialized for gas exchange. - Alveolar wall is 1 cell thick, oxygen from oxygen-rich air diffuses readily across the membrane into alveolar capillaries. - Blood from alveolar capillaries have just returned from systemic circulation (oxygen poor, CO2 rich) - As O2 diffuses from alveoli down concentration gradient into capillaries, CO2 follows its ow concentration gradient out of capillaries and into alveoli expelled out by exhalation. HEMOGLOBIN - Oxygen entering the blood latches onto hemoglobin, oxygen carrier of circulatory system, as oxygen diffuses poorly in blood. LUNGS - Lungs enclosed by ribcage within thoracic cavity. - Lungs encased in two serous membranes, the parietal pleura: lines the thoracic wall, and pulmonary pleurae: adheres to the lungs. o Are continuous with eachother. - Space between parietal pleura and pulmonary pleurae is called pleural cavity. o During inhalation, muscular diaphragm beneath lungs contract moving downwards and causing thoracic cavity to expand resulting in reduced pressure in pleural cavity. o As thoracic cavity expands, lungs must expand. o Gases like to equalize pressures between regions of space to maintain equilibrium. - Negative Pressure Breathing: Volume and pressure have inverse relationship, increased lung volume reduces alveolar pressure which causes air to flow to lungs to maintain equilibrium with ambient air pressure. o To breath in, ↑ volume, ↓ pressure, causing air to be sucked in. BREATHING - Can be voluntary or involuntary, controlled by respiratory center located in medulla oblongata of brain stem. - Inhalation is an active process as downwards contraction of diaphragm requires energy - Exhalation is a passive process as diaphragm relaxes and air is expelled from the lungs, without input of energy. o Can be active with internal intercostal muscles between ribs and abdominal muscles can produce more forceful exhalation (ex. exercise.) RESPIRATORY LUNG VOLUME - - Tidal volume (TV): During normal breathing, volume of air in each breath Inspiratory reserve volume (IRV): any additional air that we can possibly inhale. Expiratory Reserve Volume (ERV): any additional air that we can possibly exhale. Total Lung Capacity (TLC): Taking the deepest breath possible, total volume of air our lungs can hold. Vital Capacity: Maximum volume of that air from TLC that we can exhale. o Max volume of air that’s dynamic and can move into and out of lungs is vital compacity. Residual Volume: remaining air in the lungs that cannot be exhaled. o Air that remains in the alveoli so lungs don’t collapse completely. NOTE: at max volume, lungs contain more air that can be exhaled. RESPIRATORY SYSTEM PHYSIOLOGY IMMUNE DEFENSE - Hairs in nasal cavity and mucus from respiratory tract stop pathogens, trapping microbes to get them expelled by sneezing, or cilia pushing mucus upwards to be coughed out or swallowed. - Respiratory tract produces own antimicrobial proteins called defensins, that have antimicrobial activity against bacteria, fungi, viruses. o Ex. loss of cilia from extensive exposure to cigarette smoke increases risk of infection. THERMOREGULATION - Regulation of body temperatures. o Ex. vessels in nasal cavity and trachea dilate in hot conditions to increase surface area to environment. They constrict in cold conditions to retain heat as blood passes through. - o Capillary beds are sensitive as injury and even conditions that cause mucous membrane in nasal cavity to crack resulting in nosebleeds. Rely on other mechanisms like sweating, but mammals cannot sweat so they reply on panting or rapid breathing. Gas exchange – oxygen diffuses from alveoli in the lungs into alveolar capillaries and CO2 moves into opposite direction of the blood and into the alveoli. ELECTRON TRANSPORT CHAIN - Oxygen is the final electron acceptor to ATP production via aerobic respiration. o Oxygen transported via carrier protein, hemoglobin. - Carbon Dioxide is a by-product of the Krebs’s Cycle so it can be considered waste product. BICARBONATE BUFFER - Carbon Dioxide participates in equilibrium with carbonic acid and bicarbonate ions, and largely travels through bloodstream in form of bicarbonate. o CO2 reacts with H2O to form carbonic acid, which exists in equilibrium with bicarbonate. - Elevated carbon dioxide production directly translate to increased concentration of H+ which acidifies blood and decreases pH. o This is buffer system that helps maintain the blood pH at 7.4, o Acidemia: blood pH lower than 7.4, is too acidic. o Alkalemia: blood higher than 7.4, is too basic. Need to increase concentration of protons to lower pH back down. Can include breathing more slowly, hypoventilation, will promote CO2 in our blood. - Chemoreceptors in CNS and PNS detect O2, CO2, and H+ concentrations (and pH in plasma) in blood. o If we hold breath for too long, it depletes O2 while allowing CO2 and H+ to build up. Our respiratory rate speeds up to remove CO2 and replenish O2 until blood gases and pH return equilibrium levels. - Wants to maintain homeostasis. CIRCULATORY SYSTEM BLOOD BLOOD STRUCTURE - Plasma – makes up fluid component, makes up 55% of volume, carries ions, proteins, nutrients, and gases. - Buffy coat – small white layer in test tube, contains white blood cells (leukocytes) and platelets, makes up 1% of blood. - Red Blood Cells (erythrocytes): 45% of blood, carry gases, specifically for O2, CO2 in blood. o Hematocrit: proportion of blood that is comprised of red blood cells. o RBC carry hemoglobin molecules which are O2 carriers. o Produced in red bone marrow of flat and long bones (like WBC) o Body releases erythropoietin (EPO) from kidneys when it wants to stimulate more erythrocyte production. o Just about when they mature, they eject nuclei and other organelles (due to them being energy efficient but don’t live long) Live for around ~120 days before being sent to spleen for destruction. GLYCOPROTEINS – RBC - Express special glycoproteins (sugar coated proteins) on cell surface. - ABO Blood Typing is based on which ABO glycoproteins are expressed on person’s RBC. - Depends on blood type o A antigen = blood type A o B antigen = blood type B o blood A and B antigens = blood type AB o expresses neither antigens = blood type O universal donor blood type but cannot receive any blood other than O type. - - blood type depends on which alleles they have. o Ex. person with blood type A might have two A alleles or one functional A allele and one allele that isn’t expressed. o Ex. blood type AB demonstrates codominance, which demonstrates that both alleles can be expressed at the same time. Immune system creates antibodies against the ABO antigens that we don’t produce. o Ex. Blood type A will produce anti-B antibodies o Ex. Blood type B will produce anti-A antibodies. o Ex. Blood type O will produce anti-A and anti-B antibodies. Rh FACTOR – RBC - Rh Factor: antigen that can be expressed on RBC. - Either have it (Rh+) or don’t (Rh-) o A+ = RBC have A antigen and Rh factor antigen - Ex. mother is Rh- but mother is Rh+, and mother’s immune system will make anti-Rh antibodies that might attach fetus. o Mothers will administer anti-rH antibodies that prevent her immune system from needing to produce own antibodies. PLATELETS – BUFFY COAT - Platelets are cell fragments broken off from special cells called megakaryocytes and essential to coagulation (blood clotting) o no nucleus. - Ex. body lets her bleed out or creates biological band aid, called coagulation cascade. - Coagulation Cascade: involves clotting factors that lead to a molecule, called prothrombin being converted into thrombin, which then converts molecule fibrinogen into fibrin. o Fibrin: fibrous molecule that forms skeleton of blood clot, and platelets form on top to form a platelet plug. o Clot will disintegrate as wound heals. THROMBOSIS AND THROMBOEMBOLISM – BUFFY COAT - Blood clot in heart (heart attack) or brain (stroke) - Hemophilia: missing one of the factors required to form blood clots, more susceptible to excessive bleeding CARDIOVASCULAR ANATOMY SYSEMTIC CIRCULATION - Takes oxygen rich blood, delivers it to body’s tissues and then returned the blood, now depleted of oxygen, back to the heart. - When blood returns to the heart, it has spent all its O2 so it has to refuel on O2 by going though pulmonary circulation. PULMONARY CIRCULATION - Pulmonary circulation: circulation system through the lungs where blood flow through vessels adjacent to the air sacs (alveoli) of lungs. - Oxygen from the lungs flows across into blood vessel while CO2 flows across to lungs to be exhaled. - now that blood is filled with capacity with oxygen, it’s ready to return to the heart, which has power to pump the blood back to systemic circulation. ARTERIES - arteries: vessels with thick, muscular walls that carry blood away from the heart. - Are adaptable. o Ex. running marathon and it’s hot day, sympathetic NS notices we are running and flight in fight or flight can pump more blood in tissues and constrict arteries to our digestive tract. o Vasoconstriction: narrowing (constriction) of blood vessels by small muscles in their walls. o Vasodilation: widening of blood vessels by small muscles in their walls. - Arteries in systemic circulation carry oxygenated blood to the body - Pulmonary artery carry deoxygenated blood from heart to lungs - Umbilical artery in cord carries deoxygenated blood from fetus to placenta, where blood becomes oxygenated before returning to fetal circulation. - Arteries branch off into smaller arteries called arterioles. - Eventually arterioles lead to capillaries, which are tiny, thin vessels (walls are 1 cell thick) and RBC go through one by one. o Ideal environment for gas exchange to occur. o Flow rate and blood rate is slower. VEINS - Veins carry blood towards the heart. - Veins branch off as smaller venules from capillary beds and then merge to form larger veins that connect directly to heart. - Blood returns to heart through veins, which has thinner, less muscular walls than arteries do. o BP in veins is low. - Working against gravity (lower part to higher) o Uses two-fold, where veins are one-way valves (prevent backflow) and rely on skeletal muscles to squeeze vein to help blood return to heart. BLOOD VESSELS - Arteries and veins have outer tunica externa, an outer external layer. o tunica media: have middle layer made of smooth muscle, o Tunica intima: thin layer of endothelial cells - Capillaries have 1 cell thick layer of endothelial cells. HEART - Atherosclerosis: when fatty plaque builds up in heart valves - - - - o Plaque can narrow blood vessels and make it harder for O2 to reach body tissues. o Can lead to heart attack. Human heart has 4 chambers. o Atria: collecting chambers of the heart, where blood enters heart from systemic and pulmonary system o Atria funnels blood into venctricles, which have thick, muscular walls that contract to eject blood out of heart, Right atrium: collects deoxygenated blood from systemic circulation Right ventricle: pumps deoxygenated blood through pulmonary artery into the lungs, where blood becomes oxygenated. Oxygenated blood returns through pulmonary veins back to left atrium of heart. Funnels blood into left ventricle, which pumps blood into aorta to the rest of the body. Blood in systemic circulation will eventually be returned into right atrium via superior and inferior vena cavae. (BP lowest here) Ventricles are kept separate by valves. o Atrioventricular valve: controls blood flow between atria and ventricles o Tricuspid valve: on right side of heart o Bicuspid/mitral valve: on left side of the heart. Valves between ventricles and major arteries. o Aortic valve o Pulmonary valve When heart (ventricles) contract, is called systole (contract) and BP increases. When heart relaxes, it is called diastole, (relax), BP drops BP measures as systole (peak)/diastole(lower) o Normal is ~ <120/80mm. CARDIOVASCULAR PHYSIOLOGY ELECTRICAL SYSTEM - Heart has own electrical system, starting with sinoatrial (SA) node in right atrium where group of pacemaker cells live. - Pacemaker cells fire off electrical signals that instruct the heart to contract 60-100 times per minute. - Also send out AP into atria, causing atria to contract and push blood into ventricle. - All cardiac muscle fibers in atria contract simultaneously because of gap junctions. GAP JUNCTIONS - Gap junctions: membrane ion channels that allow ions to flow between individual muscle cells, so that all fibres depolarize and contract at the same time. - located in intercalated discs, which are structures that connect neighbouring cardiac muscle cells CONDUCTING SYSTEM OF THE HEART - Sinoatrial node (SA) - - Atrioventricular (AV) node – sits at junction between atria and ventricles to help transmit the signal to the ventricles. o Electrical stimulus sent out to bundle of his and Purkinje fibers which then contract to push out blood of the ventricles and into systemic and pulmonary circulation. Bundle of his (atrioventricular bundle) Right and left bundle branches Purkinje fibres NOTE: PNS slows down HR and SNS speeds up HR - Heart rate can also be increased by epinephrine and norepinephrine from adrenal medulla. HEMOGLOBIN AND IT’S ROLE IN GAS TRANSPORT - Ex. marathon runner is in race, and all sorts of leg muscles are contracting but what does muscle contraction require? ATP - How do we get ATP? Mitochondria - What does Mitochondria need to make ATP? Oxygen from lungs HEMOGLOBIN - Hemoglobin: O2 binding molecule inside red blood cells, a metalloprotein with 4 subunits (2 alpha, 2 beta) - Each subunit has heme group which has heterocyclic porphyrin ring with Fe2+ ion in center. o Iron binds O2 in red blood cells - Each hemoglobin can hold 4 O2 molecules at a time - When hemoglobin circulates back to lungs after delivering O2, can bind to CO2 from tissue and transport it to lungs for expiration. - Oxygen binding to hemoglobin is cooperative – interaction in which binding of ligand at one site of molecule influences binding at another site. o The number of O2 molecules bound to an individual hemoglobin molecule at given time affects how likely hemoglobin is to bind to more O2 molecules. o In high O2 environments, when 1 O2 binds to 1 of hemoglobin subunits, makes hemoglobin more likely to bind a second O2, … ex. o In low O2 environments, removal of 1 O2 makes it more likely for 2nd to disassociate and so on. o Due to hemoglobin changing conformation – in low O2, exists in the T (taut) state, which has low affinity for oxygen. o When hemoglobin binds, it shifts to R (relaxed) state, which has high affinity for oxygen. o Results in hemoglobin binding being favoured when large amounts of O2 present (lungs) and O2 disassociation being favoured when low amounts present (tissue) HEMOGLOBIN OXYGEN DISASSOCIATION CURVE - sigmoidal shape due to cooperative binding. - At low O2 concentration, oxygen disassociates from hemoglobin (t-state) - In R state, likes O2. - Curve can be affected by chemical environment. - Ex. marathon runner will be creating more CO2 and more lactic acid, which makes blood more acidic. Generating lots of heat, causing graph to shift from the right (curve down) - 2-3 BPG – by-product of glycolysis - Curve for fetal hemoglobin is shifted to left as well (greater affinity for O2 than mother). SICKLE CELL ANEMIA - If one AA in hemoglobin protein was mutated from glutamate to valine, hemoglobin will aggregate together in low O2 environments, causing sickle cell anemia. - Dangerous b/c misshapen blood cells can clog capillaries and become destroyed more easily. - Those with one copy (instead of 2) have sickle cell trait, that is milder form of disease and has some protection against malaria. MYOGLOBIN - Oxygen storing in muscle, has only one subunit and doesn’t exhibit cooperative binding. - Mostly, myoglobin has higher affinity for O2 than hemoglobin. - In tissue, they play tug of war for O2 and myoglobin steals O2 for tissues. DIGESTIVE SYSTEM ANATOMY OF DIGESTION FOOD - 3 biomolecules (carbohydrates, protein, lipids) - Salivary glands begin to produce enzyme rich saliva in anticipation of food. - Saliva lubricates food and contains enzymes that begin to digest carbohydrates and lipids. o Salivary Amylase breaks down starch into smaller oligosaccharides and disaccharides. o Lingual Lipase: begins to process digestion of lipids (fats) BUT protein digestion doesn’t begin in the mouth. o Lysozyme: antimicrobial enzyme that is strong enough to skill some bacteria but not all. - Bolus: ball of food after interaction with salivary glands, goes down digestive pipeline through esophagus o Moves through esophagus by wavelength contractions of smooth muscle lining, called peristalsis. - Esophagus: fibromuscular tube which food passes through stomach. Esophagus runs behind trachea and pierces through diaphragm en route to stomach. - Epiglottis covers trachea to prevent food from travelling down “wrong pipe” - Bolus then emptied into stomach passing through lower esophageal sphincter (cardiac sphincter), which acts as a gate which food can pass from esophagus into stomach but blocks contents of stomach from esophagus (GERD) - Stomach is highly acidic chamber which breaks down micro molecules o Parietal Cells of stomach secrete gastric acid, which is composed of hydrochloric acid and various salts o Keeps stomach pH between 1.5 and 3.5. o Chief Cells of stomach screen enzyme, pepsin, which does most of protein digestion and secrete biologically inactive precursor, pepsinogen, which is cleaved in acidic conditions to pepsin. o Zymogens: inactive precursor enzymes. o Intrinsic Factor: necessary to absorb vitamin B12 and water to dilute bolus. o Mucous Epithelial Cells produced by carbonate rich mucus that generate gastric acid at lining of stomach, providing protection against acidic interior. o Acid neutralized bacteria that has come into stomach. o EXCEPTION: ½ of world’s stomachs contain H. Pylori Bacteria, which strive in acidic conditions and never experience symptoms but can erode stomach lining and cause painful peptic ulcers - NOTE: o pylorus = bottom of stomach o fundus = top of stomach o body = middle of stomach GERD - Gastroesophageal Reflux Disease, caused by acidic contents of stomach going into esophagus, causing burning sensation, which can occur when lower esophageal sphincter is weakened. ANATOMY OF ABSORPTION - Bolus of food enters stomach from the esophagus, then stomach churns and begins to digest the bolus into chyme, an acidic mixture of semi digested food and gastric juices. - Chyme passes through pyloric sphincter of stomach into small intestine. - In small intestine, waves of smooth muscle contraction push food content through small intestine. Avg is inch in diameter and 22ft long. o Surface of small intestine invaginates into series of folds, which are composed of microscopic villi that project into the lumen. Entero comes from Greek word intestine. o Enterocytes: intestinal epithelial cells that line intestine. o Each has hundreds of finger-like projects of plasma membrane called microvilli, which are densely packed and mimic brush bristles, that inner lining of small intestine is called brush border. Microvilli at brush border greatly increase absorptive surface area of small intestine. - Small intestine is broken down into short duodenum, jejunum, ileum. o Chyme (from stomach) is emptied in duodenum, which triggers release of hormones that aid in digestion. Secretin: stimulates the secretion of bicarbonate to neutralize acidic chyme, maintaining the small intestine at slightly basic pH of 6-7. Cholecystokinin (CCK) release the digestive enzyme from pancreas and bile of gallbladder. LIVER, GALLBLADDER, AND PANCREAS - Gallstones accumulate painfully in gallbladder but can be removed surgically - Gallbladder is small sac responsible for storing bile. - Bile: produced in the liver, yellow-green fluid that contains bile salts, which facilitate fat absorption; are amphipathic (containing polar and non polar regions) o Non-polar regions associate triglycerides and polar regions associate with water, forming spherical micelles, that emulsify lipids in aqueous environment. - Lipid Emulsification breaks up lipids, exposing greatest amount of surface area to water soluble lipase enzyme, also facilitates absorption by enterocytes in small intestine,. o BILE SALTS ARE NOT ENZYMES, just break up lipids to allow digestive enzymes to work. LIVER - Secretes bile - Detoxifies and processes compounds absorbed from SI prior to entry into systemic circulation - o Capillaries of SI drain into hepatic portal vein, which enters hepatic portal system of liver o Portal System: system of blood vessels with capillary bed at each end, akin to hypophyseal portal system in hypothalamus. Capillaries of digestive tract pick up nutrients, and drugs from digestive organs and compounds are sent into secondary capillary system. Metabolizes drugs and medication Stores glycogen and triglycerides Mobilizes glucose and fatty acids. PANCREAS - Secretes alkaline fluid that contains digestive enzymes into pancreatic duct, which drains to duodenum of small intestine - Enzymes include o Pancreatic amylase – digest sugar into disaccharides o Pancreatic lipase – digests triglycerides into fatty acids and monoglycerides o Proteases – break protein into small peptides and AA o WORK BEST at more alkaline pH found in small intestine. - Has exocrine (secretes enzymes through pancreatic duct into duodenum) and endocrine (releases hormones into bloodstream, ex. insulin) functions. SMALL INTESTINE CON’T - SI receives partially digested food from the stomach, and communicates via hormonal signals to the accessory digestive organs that it needs help (calls for digestive enzymes, bicarbonate from pancreas and bile from gallbladder) - Enterocytes also secrete enzymes at brush border, called brush border enzymes, including: o Disaccharidases – that cleave disaccharide sugars into monosaccharides (ex. glucose) for absorption. o Peptidases – hydrolyze peptides into shorter peptides and AA. - Once carbohydrates, lipids, and proteins in ingested food have become digested into smallest units, intestinal epithelial cells will absorb resulting monosaccharides, fatty acids, and AA. o Fats are initially absorbed in the lacteals and transported by the lymphatic system before entering venous circulation. - Any remaining, undigested material passes onto large intestine. LARGE INTESTINE - Divided into cecum, colon, rectum. - Cecum: pouch connected to the ileum of SI through ileocecal sphincter. o Appendix: attached to cecum; thought of as vestigial organ (organ that serves no biological purpose), but might be reservoir for gut bacteria. - Colon: long tube that arcs over SI in order from ascending colon to transverse colon to descending colon to sigmoid colon, which terminates at rectum. - - Rectum: stores feces prior to excretion, pass through anal sphincter and through anus. Main Function: absorbs water from chyme converting it into solid feces. Laxatives: affect large intestine by interfering with water absorption BUT doesn’t affect nutrient absorption in the small intestine. o Eases constipation but not good for weight loss. o Nutrient absorption occurs to limited extent in large intestine o has largest community of bacteria (gut bacteria such as gut flora, microbiota) synthesize necessary vitamins such as: vitamin B7 (biotin) and vitamin K (blood coagulation) which are absorbed in the large intestine. After passing through large intestine and rectum, food is fully transformed into feces FECES - Contain water and indigestible material, mostly cellulose as humans lack enzymes to digest cellulose form plants o Cellulose rich foods (celery) are low calorie as it cannot get nutritional value from it. - Through pathology, feces can show things related to digestion and absorption of nutrients o Ex. liver dysfunction – can affect bile production and lipid digestion, causing more fat in feces, called steatorrhea. HORMONAL AND NERVOUS CONTROL OF DIGESTION ENDOCRINE SYSTEM - Regulates digestion, can be categorized as those that affect appetite, those that stimulate digestion and those that halt digestion. - Leptin: hormone secreted by sat cells (adipocytes) that help suppress appetite. - Ghrelin: hormone secreted by specialized cells in pancreas and upper stomach when stomach is empty and stimulates appetite. o Gastric bypass surgery: removal of cells in upper stomach that secrete ghrelin to dampen hunger signals. DIGESTION SIGNALS - Digestion stimulated by signals that indicate food has been ingested. - G cells secrete hormone, gastrin, which tells parietal cells when to produce gastric acid. - When acidic chyme reaches SI, S cells in duodenum release hormone secretin, which stimulates release of bicarbonate from pancreas to SI to maintain alkaline pH. - Cholecystokinin (CCK) is released from SI, stimulates secretion of digestive enzymes from pancreas and bile from gallbladder into SI. o Can inhibit appetite. SOMATOSTATIN - Somatostatin halts pro-digestion hormones o Gastrin o CCK - o Secretin Stalls stomach emptying Halts release of pancreatic hormones (insulin and glucagon) Known as growth hormone inhibiting hormone as it inhibits the release of growth hormones. ENTERIC NERVOUS SYSTEM - Branch of ANS (not under conscious control) - PNS and SNS have important effects on GI tract. o Since SNS inhibits fight or flight, it can contract blood vessels of digestive tract. o PNS (rest and digest) dilate blood vessels of digestive tract to promote digestion. DIGESTION AND ABSORPTION OF BIOMOLECULES LIPIDS - Maintain cellular membrane - Energy storage AMINO ACIDS - Synthesize new proteins CARBOHYDRATES - Harvest glucose as energy source CARBOHYDRATES - Trace digestion of carbohydrates, proteins, and lipid macromolecules starting in the mouth where food will need mastication (chewing) and lubrication from saliva in oral cavity. o Contains salivary amylase, an enzyme that digests starches into tri and disaccharides (ex. maltose) - Bolus then passes down esophagus via peristalsis, and goes into stomach. - Stomach doesn’t process carbs enzymatically but churning actions mechanically breaks them down, increasing surface area to hydrolytic enzymes in SI. o Acidic chyme: slurry of food particles and stomach acid, enters duodenum and triggers release of CCK, which induces pancreatic secretion into SI. o Pancreatic amylase: is induced in pancreatic secretion and hydrolyzes polysaccharides into tri and disaccharides. - brush border disaccharidases: completes digesting the sugars; enzymes created by enterocytes lining the SI that digest disaccharides into monosaccharides at brush border, which include: o sucrase – digest sucrose into glucose and fructose o maltase – digests maltose into 2 glucose units o lactase – digests lactose into glucose and galactose. - Monosaccharides that were liberated at brush border are taken up by intestinal epithelial cells for passage into bloodstream. o Requires coordination between transporters. o Sodium potassium pump uses ATP to pump 3 Na into bloodstream and pumping 2 K into cell, which maintains electrochemical gradient where Na concentration low inside cell. Na can flow down concentration gradient into cell from intestinal lumen. Occurs via Na+ glucose symporter (driven by high extracellular Na+), which allows 1 glucose molecule come for every 2 Na ions it transports inside the cell. Glucose then travels via facilitated diffusion through glucose uniporter out of cell and into circulatory system. PROTEIN - Isn’t digested until stomach, where stomach acid cleaves pepsinogen into active form of pepsin, which hydrolyzes peptide bonds in protein. o Pepsin target sites between hydrophobic or aromatic AA, so it has enough specificity to cleave proteins into shorter peptides without completely digesting them to AA level. - Presence of acidic chyme into duodenum triggers the secretion of pancreatic enzymes into SI as result of CCK, which include: o Trypsin – inactive precursor trypsinogen, which is cleaved by enteropeptidase into active form, trypsin. Cleaves peptide bonds adjacent to lysine or arginine residue to activate suite of proteases. o Enzymatic regulation is critical to avoid premature activation of protease If activated early, can digest and damage pancreas, resulting in pancreatitis. - AA absorbed by intestinal epithelial cells via secondary active transport, so via cotransport with sodium ions, following concentration gradient by Na-K pump o AA exit cells via facilitated diffusion into circulatory system. NOTE - Carbs and proteins are similar as monomeric sugars are hydrophilic, which can readily dissolve in aqueous environment of intestinal lumen. - Lipids are hydrophobic, not dissolvable in aqueous environments. LIPIDS - Fat absorption aided by bile before released into SI. - Bile: mixture of bile salts, pigments and cholesterol that help emulsify or dissolve fat in aqueous lumen of SI; also amphipathic so hydrophobic regions can interact with triglycerides and cholesterol o Hydrophilic regions associate with water, forming small spherical micelles immersed in intestinal lumen. - By preventing lipids from aggregating into large clumps, increases SA accessible to pancreatic lipase for digestion. o Due to hydrophobic components, micelles easily diffuse through plasma membrane into intestinal epithelial cells at brush border. o In cytoplasm, fatty acids combine with monoglycerides to reform triglycerides which are them packaged into chylomicrons, which are fat droplets containing lipids and proteins which diffuse into lacteals. o Lacteals: small lymphatic vessels that drain into larger lymphatic vessels before emptying into venous circulation. VITAMINS AND MINERALS - Vitamins: organic substance required in small amounts o Can act as coenzymes (ex. vitamin K and blood clotting) - Lipid Soluble Vitamins: accumulate in adipose/fat tissue. o Vitamins A,D,E,K o Too many lipid soluble vitamins may cause unwanted consequences. (ex. excessive vitamin A = orange skin tint) - Water Soluble Vitamins: circulate in blood, and easily excreted in urine. o Vitamins C,B o Some B vitamins are stored in liver. FAT SOLUBLE VITAMINS - Vitamin A – essential for vision as it interacts with opsin to form protein, rhodopsin (present in rods of retina) and used for low light vision) o E.g. Retinol and Retinal - Vitamin D – acts as a hormone that regulates calcium and phosphate concentrations in blood stream. Also increases absorption of calcium, phosphate and other minerals. o Can be synthesized in skin from exposure to UV radiation. o 2 major forms are converted to biologically active form – calcitriol. - Vitamin K – synthesized by bacteria in large intestine and essential for blood coagulation. WATER SOLUBLE VITAMINS - Deficiency in Vitamin C o Common with sailor who spent time away from land and no access to fresh food. o Symptoms of scurvy – weakness, gum disease, excess bleeding, death. - Vitamin C – required for collagen synthesis. MINERALS - Minerals: inorganic substances required but not synthesized by the body and must be obtained in the diet. o Play role as cofactors. - Macrominerals – principal ions (Ca, Na,K) which are required in significant amounts - Trace Minerals – only needed in small/trace amounts (Iron, Copper, Zinc) GENE EXPRESSION GENE EXPRESSION AND CELL DIFFERENTIATION - Skin, liver, and muscle cells all possess the same genome. - What cells do with genes give diversity is gene expression. - During development, complex signaling patterns induce cell differentiation: where stem cells differentiate into different tissues and specialized cell types in body. STEM CELLS - Often undifferentiated cells that are capable of differentiating into more specialized cell types. Ex. embryonic stem cells and adult stem cells (somatic stem cells) o Somatic stem cells include: hematopoietic (generate blood cells), intestinal stem cells, and mesenchymal (generate fat, bone, and liver cells. STEM CELL POTENCY - Stem Cell Potency: limits on cell differentiation - Totipotent: can differentiate into any type of cell o In humans, only applies to early embryos, up to morula or 16-cell stage. - Pluripotent: can differentiate into any cells of ectoderm, mesoderm, or endoderm. o Cannot differentiate into cells of placenta. - Multipotent: can differentiate into a limited subset of cell types within a germ layer. APOPTOSIS - Apoptosis: programmed cell death. - Plays critical role in embryonic development to help find the boundaries of organs and tissues. o Ex. fingers and toes have webbed appearance, only through coordinated apoptosis that webbed structures disappear during embryogenesis. - Also important checkpoint for organelle dysfunction or DNA mutations. PROKARYOTIC GENE EXPRESSION - Operons: genetic system used by prokaryotes to regulate expression of specific genes. - Positively controlled genes: genes expressed when activator present; respond to activator molecule which binds and stimulates transcription. - Negatively controlled genes: genes expressed unless repressor present; respond to repressor molecules which bind a sequence, operator, within operon to turn off transcription. TYPES OF OPERONS - Lac operon: negative inducible (expression induced by removal of repressor, allowing transcription to occur) o Default = off position - Trp operon: negative repressible (expression repressed by binding of repressor) o Default = on position - Both are under negative control – repressor can turn off gene expression. LAC OPERON - E.Coli will use glucose as source of carbon but capable of metabolizing other sugars. - Lac operon contains genes for enzymes that can metabolize lactose (but prefer glucose) o Used when glucose stores are low and there is lactose in surplus. - - LacZ, LacY, and LacA encode for enzymes responsible for breaking down lactose to harness it’s energy and includes upstream promoter where RNA polymerase binds to initiate transcription. o Also contains CAP binding site and Promoter. Repressor is present, bound to operator and blocking transcription from RNA polymerase when lactose isn’t present o When lactose is present, can turn on these genes. o Allolactose (isomer) binds repressor and prevents it from interacting with operator so transcription can occur. CAP (CATABOLITE ACTIVATOR PROTEIN) - When glucose levels are low, cyclic AMPA levels are elevated within cell - cAMP binds to CAP which induces CAP to bind to CAP binding sequence to induce transcription. example of positive control o Positive Control: genes expressed when activator is present. TRP OPERON - Trp Operon contains genes for synthesis of AA tryptophan. o large amount of tryptophan represses the expression of these genes. o Under negative repressible control and exerts negative feedback on own synthesis. When tryptophan is absent, transcription is giving the go ahead, allowing tryptophan synthesis to occur without a repressor. EUKARYOTIC GENE EXPRESSION PROMOTER - Promoter: is short sequence upstream of target gene that serves as initial binding site for RNA polymerase to initiate transcription. - Many promoters will contain TATA box (30 BP upstream of coding sequence) o GC and CAAT Boxes are 10-150 BP upstream of TATA box o each has specific short sequence that’s recognized by proteins that recruit RNA polymerase to initiate transcription of downstream gene. o Ex. TATA box is bound to TATA binding protein which associates with other proteins to act as transcription factor for RNA polymerase. TRANSCRIPTION FACTORS - Transcription factors are proteins that bind to specific DNA sequences to regulate gene expression and recruiting other regulatory proteins that might affect gene transcription or that make chemical modifications to DNA - Specific transcription factors that cells express or that are activated in response to chemical signals affect the suite of genes that are active in the genes. ENHANCERS AND SILENCERS - Enhancers: sequences that promote enhanced expression of genes in response to appropriate stimuli. May be located upstream or downstream of target genes. o Bind transcription factors called activators which help DNA twist into hairpin loop to bring the enhancer region closer to gene sequence to facilitate initiation of transcription. o Ex. estrogen binding it’s nuclear estrogen receptor, which binds enhancers called estrogen response elements to enhance transcription of specific genes. - Silencers: sequences that when bound by repressor, silence or repress expression of specific genes. DNA MODIFICATION - In nucleus, DNA found around histones and condensed further into heterochromatin (or looser chromatin) - Acetylation or Deacetylation: adding or removal of acetyl groups to modify histones. Affects how tightly DNA is packaged. - Histone Acetyltransferases: adds acetyl groups to histones, reducing histone-DNA interaction which promotes DNA transcription. - Histone Deacetylases: removes acetyl groups from histones and promotes histone-DNA interactions, reducing DNA transcription. - Methylation: addition of methyl groups directly to cytosine or adenine nucleotides in DNA. o Often silences or deactivates gene sequences, preventing expression. - Epigenetics: heritable changes that affect gene expression without directly changing the genetic sequence itself. o Ex. fathers with prediabetes were more likely to have children with diabetes, due to changes in methylation in gametes. o Ex. people with mothers who were pregnant during Dutch famine (WWII) have higher rates of obesity and other health conditions, due to methylation state of specific genes. o some epigenetic changes can be transmitted through multiple generations. RNA INTERFERNCE (RNAi) - non coding RNA can interfere with gene expression by degrading mRNA prior to translation siRNA and microRNA microRNA is single stranded, siRNA is double stranded. For RNAi to occur, miRNA or microRNA is incorporated into RNA induced silencing complex (RISC), and then the silencing RNA strand pairs with complementary target mRNA transcript, inducing cleavage of mRNA transcript. TRANSCRIPTION AND TRANSLATION - RNA World Hypothesis: RNA stores genetic information and catalyzed reactions before DNA came. - SIMILARITIES BETWEEN DNA REPLICATION AND TRANSCRIPTION o Involve copying DNA into complementary nucleic acid o Occur in nucleus using same factors and coenzymes. o Ex. to access DNA for transcription, double helix needs to be unzipped by helicase enzyme, kept open by single stranded binding proteins. Tension created upstream relieved by topoisomerase enzyme. No need to worry about lagging/leaking strand or replicating ends of chromosomes. - RNA polymerase: enzyme that catalyzes transcription the polymerization of RNA. Locates specific sequences of genes that it is interested in. - Every gene has Upstream Promoter (in euks, TATA box) where it latches on with the help of transcription factors and begins to synthesize RNA, forming complementary base pairs with DNA sequence o TATA box: A-T rich sequence that’s recognized by RNA polymerase. o RNA uses uracil, instead of thymine. o Synthesizing precursor to messenger RNA (mRNA) - RNA polymerase reads the DNA strand in 3’ to 5’ direction, synthesizing the new RNA 5’ to 3’, until reaching transcription terminator sequence. - RNA polymerase has proofreading ability but not as accurate as DNA polymerase. o Error rate is higher for RNA polymerase - RNA can continuously synthesize RNA copy of any gene on either stand of DNA o Template Strand (antisense) Strand being read by RNA polymerase, sequence is complementary to sequence of newly synthesized RNA molecule. o Coding Strand (sense): DNA strand in double helix that is not being read; sequence is identical to new RNA molecule, but in RNA, uses uracil. o Coding and RNA sequences are nearly identical RNA POLYMERASE POST TRANSCRIPTIONAL MODIFICATIONS - Polyadenylation: addition of A nucleotides to RNA’s 3’ ends is catalyzed by polynucleotide adenyltransferase. - 3’ Poly(A) Trail o Facilitates nuclear export o Protects mRNA against degradation. - 5’ Cap o One nucleotide modification, addition of 7 -methylguanylate triphosphate cap that is attached in reverse directionality of mRNA transcript. o Also protects mRNA against degradation o Facilitates nuclear export and protects against attacks from 5’ exonucleases - RNA transcripts contain alternating segments of intron and exon sequences. o Introns: intervening, non-coding sequences that are excised or spliced out during RNA splicing. can contain regulatory sequences that modulate gene expression in the cell. o Exons: expressed, coding sequences that remain, that are ligated together and expressed in mature mRNA product. - Splicing is catalyzed by spliceosome, enzyme complex that is made of proteins and small nuclear RNA that, combined with form small nuclear ribonucleoproteins (snURPS) o Capable of enzymatic activity and considered ribozymes, RNA enzymes. - snRUPS recognize marker sequences at splice sites at the end of introns, pulling these ends together to form lariat loop structure. o Intron loop is spliced out as neighbouring exons are ligated together to form continuous mRNA product. - Given mRNA precursor can undergo splicing to produce more than one protein, based on how they are ligated together. o Ex. RNA sequence with exons 1-6 might splice all 6 exons to produce one protein or maybe 1,3,5,6 o Increases proteomic diversity. TRANSLATION - Central Dogma= DNA RNA Protein - Transcription: synthesis of RNA to DNA - DNA sequence is copied in complementary form but thymine placed with uracil. - mRNA nucleotides arranged in triplets, called codons, that each code for specific AA o info is not directly copied from RNA to protein, translational machinery deciphers RNA code and translates it into different sequences of AA. PROKARYOTES - no nucleus, just nucleoid region. - Translation and transcription occur simultaneously both temporally, at the same time, and geographically, on same mRNA molecule. EUKARYOTES - Transcription and post transcription modifications occur in nucleus - Once mRNA is decorated with 5’ and 3’ caps, protected against degradation and can be exported to cytoplasm for translation. - mRNA transcripts are read by ribosomes, molecular machines that translate mRNA into proteins. - Secreted Proteins: Proteins destined for cellular membrane or for secretion are synthesized on ribosomes are studded on membrane of rough ER and fed into ER for processing. o Then transferred to Golgi Apparatus and secretory vesicles until they reach their destination. - Cytoplasmic Proteins: read by free floating ribosomes in cytoplasm - Ribosomal RNA (rRNA): ribosomal component to ribosomes; rRNA and protein complexes that translate mRNA into protein. - Ribosomes are composed of ribosomal small subunit, that reads mRNA sequences and a large subunit which catalyzes formation of peptide bonds. - Refer ribosomes by sedimentation coefficients in Svedberg units, which tell us how fast they sediment in centrifuge, based on mass and size. o Prokaryotic ribosome = small 30S subunit and large 50s subunit, forming 70s subunit overall. o Eukaryotic ribosome = small 40s subunit and large 60s subunit, forming 80s subunit overall. TRANSFER RNA (tRNA) N - Cloverleaf, hairpin secondary structure and recognizes specific triplet codon on mRNA and attach corresponding AA into polypeptide sequence on ribosome. - Each tRNA has unique anticodon that’s complementary to specific mRNA codon. - Based on codon it recognizes, tRNA becomes charged when bound to corresponding AA. o One tRNA molecule can recognize many codons. - Degeneracy: multiple codons can correspond to 1 amino acid. - To charge a tRNA molecule with corresponding AA, aminoacyl-tRNA synthetase enzyme links C-terminus of particular AA to 3’ prime end of tRNA molecule o requires investment of ATP. o Energy from ATP is harnessed to power formation of peptide bond. STEPS OF TRANSLATION INITIATION - Translation is initiated, the small ribosomal subunits recognizes and binds specific sequence on mRNA o Prokaryotes = Shine-Dalgarno sequence in 5’ prime untranslated region. (region upstream of start codon that helps regulate translation) o Eukaryotes = 5’ prime cap. - Initiator tRNA binds start codon AUG on mRNA molecule; this tRNA is charged with methionine in eukaryotes (first AA in protein sequence) - Initiation factors facilitate the binding of large subunits to the small subunits, forming initiation complex. ELONGATION - Ribosome, and elongation factors reach mRNA sequence in 5’ – 3’ direction, going along mRNA transcript reading one triplet codon at a time between start and stop codons - Ribosome synthesizes polypeptide sequence from n-terminus to c-terminus, N-C polarity of all AA. - Ribosomal assembly line has 3 binding sites for tRNA o A (acceptor) site: accepts aminoacyl-tRNA complex o P (peptidyl) site: tRNA forms peptide bond between new, incoming AA and growing peptide chain o E (exit) site: uncharged tRNA molecule leaves ribosome o Each time ribosome reads next mRNA codon, tRNA molecule in each binding site move over one position, exposing A site for next tRNA site to bind. TERMINATION - When ribosome encounters one of three stop codons: UAA, UGA, UAG o AUG is the start codon (methionine in eukaryotes) o Stop codons do not encode for an AA, and recognized by release factors at p site that force ribosome to evict protein sequence as it moves onto new mRNA sequence to read. POST-TRANSLATIONAL MODIFICATION - post translational modifications occur in rough ER or in Golgi (addition of carbohydrates and protein cleavage) - proteins can be covalently linked to variety of functional groups. (ex. phosphorylation, addition of high energy phosphate group by kinase enzyme) o ex. glycosylation, addition of carbohydrate moieties (catalyzed in Golgi) relevant for ABO blood typing, based on presence or absence of glycoproteins on RBC. o Ex. ubiquitination, addition of ubiquinone protein which can be used to designate proteins for degradation in cells. - After synthesis, proteins must be folded into 3D conformation, which is facilitated by chaperone proteins in cytoplasm or ER. - Disulfide bonds between cysteine residues also form, linking 2 parts of the protein and maintaining folded tertiary structure. - In addition to folding, protein subunits may also be assembled together into protein complex after translation, forming quaternary structure of multi-subunit proteins. PROTEOLYTIC PROCESSING - May be cleaved at specific sites, creating 2 separate peptides or proteins with unique functions but also, cleavage of precursor protein or zymogen activates it. o Allows cells to regulate and where certain enzymes can be active. - Ex. processing of peptide hormones. Original peptide is preprohormone, which is cleaved to prohormone, and finally, to active form of hormone. (to prevent hormones from being prematurely activated.)