RANDOM FOREST
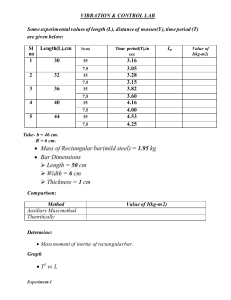
Importing Libraries
In [ ]: import
import
import
import
numpy as np
pandas as pd
matplotlib.pyplot as plt
seaborn as sns
Importing Dataset
In [ ]: df=pd.read_csv('drug200.csv')
df.head()
Age Sex
Out[ ]:
BP
Cholesterol
Na_to_K
Drug
0
23
F
HIGH
HIGH
25.355
drugY
1
47
M
LOW
HIGH
13.093
drugC
2
47
M
LOW
HIGH
10.114
drugC
3
28
F NORMAL
HIGH
7.798
drugX
4
61
F
HIGH
18.043
drugY
BP
Cholesterol
Na_to_K
LOW
In [ ]: x=df.iloc[:,:-1]
y=df.iloc[:,-1]
In [ ]: x.head()
Age Sex
Out[ ]:
0
23
F
HIGH
HIGH
25.355
1
47
M
LOW
HIGH
13.093
2
47
M
LOW
HIGH
10.114
3
28
F NORMAL
HIGH
7.798
4
61
F
HIGH
18.043
LOW
In [ ]: y.head()
Out[ ]:
0
drugY
1
drugC
2
drugC
3
drugX
4
drugY
Name: Drug, dtype: object
In [ ]: y.value_counts()
Out[ ]:
drugY
91
drugX
54
drugA
23
drugC
16
drugB
16
Name: Drug, dtype: int64
In [ ]: print("Shape of x:",x.shape)
print("Shape of y:",y.shape)
Shape of x: (200, 5)
Shape of y: (200,)
Data Preprocessing
Handeling The Null Values
In [ ]: x.isnull().sum()
Out[ ]:
Age
Sex
BP
Cholesterol
Na_to_K
dtype: int64
0
0
0
0
0
In [ ]: y.isnull().sum()
Out[ ]:
0
In [ ]: from sklearn.preprocessing import LabelEncoder
le=LabelEncoder()
x.iloc[:,1]=le.fit_transform(x.iloc[:,1])
x.iloc[:,2]=le.fit_transform(x.iloc[:,2])
x.iloc[:,3]=le.fit_transform(x.iloc[:,3])
<ipython-input-14-6bc19a4866e3>:3: DeprecationWarning: In a future version, `df.il
oc[:, i] = newvals` will attempt to set the values inplace instead of always setti
ng a new array. To retain the old behavior, use either `df[df.columns[i]] = newval
s` or, if columns are non-unique, `df.isetitem(i, newvals)`
x.iloc[:,1]=le.fit_transform(x.iloc[:,1])
<ipython-input-14-6bc19a4866e3>:4: DeprecationWarning: In a future version, `df.il
oc[:, i] = newvals` will attempt to set the values inplace instead of always setti
ng a new array. To retain the old behavior, use either `df[df.columns[i]] = newval
s` or, if columns are non-unique, `df.isetitem(i, newvals)`
x.iloc[:,2]=le.fit_transform(x.iloc[:,2])
<ipython-input-14-6bc19a4866e3>:5: DeprecationWarning: In a future version, `df.il
oc[:, i] = newvals` will attempt to set the values inplace instead of always setti
ng a new array. To retain the old behavior, use either `df[df.columns[i]] = newval
s` or, if columns are non-unique, `df.isetitem(i, newvals)`
x.iloc[:,3]=le.fit_transform(x.iloc[:,3])
In [ ]: x.describe()
Age
Sex
BP
Cholesterol
Na_to_K
count
200.000000
200.000000
200.000000
200.000000
200.000000
mean
44.315000
0.520000
0.910000
0.485000
16.084485
std
16.544315
0.500854
0.821752
0.501029
7.223956
min
15.000000
0.000000
0.000000
0.000000
6.269000
25%
31.000000
0.000000
0.000000
0.000000
10.445500
50%
45.000000
1.000000
1.000000
0.000000
13.936500
75%
58.000000
1.000000
2.000000
1.000000
19.380000
max
74.000000
1.000000
2.000000
1.000000
38.247000
Out[ ]:
Splitting the dataset into the Training set and Test
set
In [ ]: from sklearn.model_selection import train_test_split
x_train,x_test,y_train,y_test=train_test_split(x,y, test_size=0.2, random_state=29
print('The Shape of X_train:',x_train.shape)
print('The Shape of X_test:',x_test.shape)
print('The Shape of y_train:',y_train.shape)
print('The Shape of y_test:',y_test.shape)
The
The
The
The
Shape
Shape
Shape
Shape
of
of
of
of
X_train: (160, 5)
X_test: (40, 5)
y_train: (160,)
y_test: (40,)
Training the Random Forest model on the Training
set
In [ ]: from sklearn.ensemble import RandomForestClassifier
classification = RandomForestClassifier()
classification.fit(x_train,y_train)
Out[ ]: ▾ RandomForestClassifier
RandomForestClassifier()
Predicting the Test set results
In [ ]: y_pred = classification.predict(x_test)
output=pd.DataFrame()
output['Actual Value']=y_test
output['predicted Value']=y_pred
output.head()
Actual Value
predicted Value
145
drugX
drugX
197
drugX
drugX
75
drugY
drugY
119
drugY
drugY
141
drugY
drugY
Out[ ]:
Confusion Matrix And Accuracy
In [ ]: from sklearn.metrics import accuracy_score,confusion_matrix,ConfusionMatrixDisplay
import matplotlib.pyplot as plt
print("The accuracy of the above classification model is:")
print(accuracy_score(y_test,y_pred)*100," %")
print("\nConfusion Matrix\n")
matrix = ConfusionMatrixDisplay(confusion_matrix(y_test,y_pred),display_labels=clas
matrix.plot()
plt.show()
The accuracy of the above classification model is:
97.5 %
Confusion Matrix
In [ ]: from sklearn.tree import export_graphviz
from IPython.display import Image
import graphviz
for i in range(3):
tree = classification.estimators_[i]
dot_data = export_graphviz(tree,
feature_names=x_train.columns,
filled=True,
max_depth=1,
impurity=False,
proportion=True)
graph = graphviz.Source(dot_data)
display(graph)
Na_to_K <= 14.615
samples = 100.0%
value = [0.131, 0.062, 0.094, 0.262, 0.45]
True
False
Age <= 51.5
samples = 56.0%
value = [0.239, 0.114, 0.17, 0.477, 0.0]
(...)
samples = 44.0%
value = [0.0, 0.0, 0.0, 0.0, 1.0]
(...)
Age <= 31.5
samples = 100.0%
value = [0.094, 0.075, 0.088, 0.338, 0.406]
True
BP <= 0.5
samples = 25.7%
value = [0.256, 0.0, 0.116, 0.233, 0.395]
(...)
False
Na_to_K <= 14.829
samples = 74.3%
value = [0.034, 0.103, 0.077, 0.376, 0.41]
(...)
(...)
(...)
Na_to_K <= 14.829
samples = 100.0%
value = [0.1, 0.088, 0.088, 0.3, 0.425]
True
BP <= 0.5
samples = 55.7%
value = [0.174, 0.152, 0.152, 0.522, 0.0]
(...)
(...)
False
samples = 44.3%
value = [0.0, 0.0, 0.0, 0.0, 1.0]