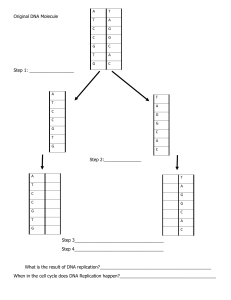
Histone acetylation can disrupt the interaction between adjacent nucleosomes A) because acetylated nucleosomes are targeted for degradation B) because it causes nucleosomes to release the DNA. C) because it prevents histone H1 from forming a bridge between histone H1 in two adjacent nucleosomes. D) because it prevents the N-terminal tail of histone H4 interacting with an acidic pocket in the H2A-H2B dimer in the next nucleosome. When histone H1 is incorporated into a nucleosome A) the nucleosome is about to be moved by a remodeling enzyme. B) it means that replication of the DNA has just occured. C) it increases the probability that condensation of the chromatin will occur.* D) it means that the underlying DNA is probably a core promoter. Histone acetylation can disrupt the interaction between adjacent nucleosomes A) because acetylated nucleosomes are targeted for degradation B) because it causes nucleosomes to release the DNA. C) because it prevents histone H1 from forming a bridge between histone H1 in two adjacent nucleosomes. D) because it prevents the N-terminal tail of histone H4 interacting with an acidic pocket in the H2A-H2B dimer in the next nucleosome. * Where would you most likely find chromatin in the beads on a string conformation? A) Heterochromatin B) Actively transcribed chromatin * C) Silenced chromatin D) Deacetylated chromatin E) 30 nm fiber chromatin There are five major types of histones in eukaryotic cells. One of these is not part of the structure of nucleosomes and is thought to participate in forming the 30-nm condensed chromatin fiber. Which histone is this? A. H1 B. H2A and B C. H3 D. H4 How is the structure of chromatin modified in cells to change transcriptional activity? .Draw template and nascent strands at a replication fork; label the 5' and 3' ends, and the leading and lagging strands. .Draw template and nascent strands at a replication fork; label the 5' and 3' ends, and the leading and lagging strands. Outline the general structure of the nucleosome. What is the evidence for this structure? [Structure: 146 bp of duplex DNA wrapped around a histone octamer (two copies of H3, H4, H2A, H2B) in 1.65 turns of a flat, left-handed superhelix. Evidence: 2 copies of each core histone (and ~1 of H1) per ~200 bp DNA, the 'beads-on-a-string' structure seen by electron microscopy, nuclease digestion of linker DNA to give a nucleosomal ladder (repeat 180-260 bp) following by trimming of the linker to give the core particle with 146 bp DNA, the structure of the core particle determined by X-ray crystallography.] Which of the following plays a substantial role in linking together sister chromatids immediately after replication? a) Cohesins b) Condensins c) Histones d) Topoisomerases Which is a characteristic of E. coli DNA polymerases? a) Pol I functions as a multimeric protein that participates in DNA repair. b) Pol I functions as a core enzyme that clamps around the DNA. c) Pol III functions as a holoenzyme that polymerizes DNA with high processivity. d) Pol III functions as a single polypeptide chain that can form phosphodiester bonds. 16.____ Which property is shared by E. coli DNA polymerase I and DNA polymerase III? a) Both enzymes require a template that can be either DNA or RNA. b) Both enzymes require a primer that can be either DNA or RNA. c) Both enzymes can use pyrophosphate as a substrate. d) Both enzymes can make and break N-glycosidic bonds. 17._____ Which describes the role of primase during replication? a) It catalyzes the formation of phosphodiester bonds using NTPs as substrates. b) It coordinates synthesis of the leading strand and the lagging strand. c) It functions as a holoenzyme that polymerizes in the 3’→ 5’direction. d) It uses an exonuclease activity to remove incorrect nucleotides. 18._____ Which function can be carried out by DNA replication proteins? a) Topoisomerases wind the DNA into a double-helix. b) DNA ligase can initiate new DNA chains c) SSB converts double-stranded DNA into single-stranded DNA. d) Helicases break hydrogen bonds in the DNA. In assembling a nucleosome, normally the …(1) histone dimers first combine to form a tetramer, which then further combines with two … (2) histone dimers to form the octamer. A. 1: H1–H3; 2: H2A–H2B B. 1: H3–H4; 2: H2A–H2B C. 1: H2A–H2B; 2: H1–H3 D. 1: H2A–H2B; 2: H3–H4 E. 1: H1–H2; 2: H3–H4 Proteins that contain SANT domains interact preferentially with which kind of histone tail? A. B. C. D. an acetylated histone tail an acetylated and methylated histone tail a methylated histone tail an unmodified histone tail The chromatin remodeling complexes play an important role in chromatin regulation in the nucleus. They … A. can slide nucleosomes on DNA. B. have ATPase activity. C. interact with histone chaperones. D. can remove or exchange core histone subunits. E. All of the above. Which of the following answer choices represents the correct evolutionary relationship between complexity of organisms and gene density (genes/Mb) A. B. C. D. Most organisms have a similar gene density. There is no relationship between organismal complexity and gene density. Less complex organisms have decreased gene density. Less complex organisms have increased gene density. At what point during mitosis does proteolytic cleavage of cohesin proteins occur? A. B. C. D. anaphase metaphase prophase telophase Which of the following is true regarding heterochromatin in a typical mammalian cell? A. B. C. D. E. About 1% of the nuclear genome is packaged in heterochromatin. The DNA in heterochromatin contains all of the inactive genes in a cell. Genes that are packaged in heterochromatin are permanently turned off. The different types of heterochromatin share an especially high degree of compaction. Heterochromatin is highly concentrated in the centromeres but not the telomeres. A nucleosome forms hydrogen bonds with what part of the DNA? A. B. C. D. with the phosphodiester backbone and with bases via the minor groove with the phosphodiester backbone and with bases via the major groove only with bases via the major groove only with bases via the minor groove In human cells, the alpha satellite DNA repeats … A. have a specific sequence indispensable for the seeding event that leads to chromatin formation. B. can be seen to be packaged into alternating blocks of chromatin, one of which contains the histone H3 variant CENP-A. C. are sufficient to direct centromere formation. D. are necessary for centromere formation. E. All of the above. The telomerase enzyme in human cells … A. B. C. D. E. has an RNA component. extends the telomeres by its RNA polymerase activity. polymerizes the telomeric DNA sequences without using any template. removes telomeric DNA from the ends of the chromosomes. creates the “end-replication” problem. What are the products of deamination of cytosine and 5-methyl cytosine, respectively? A. B. C. D. E. 1. Thymine and uracil Thymine in both cases Uracil and thymine Uracil in both cases Xanthine and hypoxanthine Which of the following repair pathways can accurately repair a double-strand break? A. B. C. D. Base excision repair Nucleotide excision repair Direct chemical reversal Homologous recombination 3. Nonhomologous end joining DNA glycosylases constitute an enzyme family found in all three domains of life. They can … A. B. C. D. E. add sugar moieties to DNA. remove sugar moieties from DNA. add a purine or pyrimidine base to DNA. remove a purine or pyrimidine base from DNA. remove a nucleotide from DNA. Methylation and subsequent deamination of cytosine produces what type of mutation after one round of DNA replication? A. B. C. D. G-to-A transition C-to-A transversion C-to-T transversion C-to-T transition Upon heavy damage to the cell’s DNA, the normal replicative DNA polymerases may stall when encountering damaged DNA, triggering the use of backup translesion polymerases. These backup polymerases … A. B. C. D. lack 3′-to-5′ exonucleolytic proofreading activity. are replaced by the replicative polymerases after adding only a few nucleotides. can create mutations even on undamaged DNA. may recognize specific DNA damage and add the appropriate nucleotide to restore the original sequence. E. All of the above. True or false Tens of thousands of replication origins are used each time a cell in our body replicates its DNA. ( ) Different cells in our body use different sets of replication origins. ( ) Both replication forks in a replication bubble are normally active in replication. ( ) Gene expression and chromatin structure can affect the choice of the origins to use as well as the order in which they are activated. Indicate true (T) and false (F) descriptions below regarding the nucleotide excision repair pathway. ( ) It involves recognition of distortions in the DNA double helix rather than specific base changes. ( ) It involves endonucleolytic cleavage and helicase-mediated strand removal. ( ) It involves cleavage by the AP endonuclease. ( ) It is coupled to the DNA transcription machinery of the cell. TTFT Indicate whether each of the following descriptions better applies to a centromere (C), a telomere (T), or an origin of replication (O 1. 2. 3. 4. ( ) It contains repeated sequences at the ends of the chromosomes. ( ) It is NOT generally longer in higher organisms compared to yeast. ( ) Each eukaryotic chromosome has many such sequences. ( ) There are normally two such sequences in each eukaryotic chromosomal DNA molecule. 5. ( ) There is normally one such sequence per eukaryotic chromosomal DNA molecule. 6. ( ) It is where DNA duplication starts in S phase. 7. ( ) It attaches the chromosome to the mitotic spindle via the kinetochore structure. Essay Explain why each chromosome in a eukaryotic cell contains multiple origins of replication but includes one and only one centromere. Describe the process by which the histone tail modifications that are present at a given genomic region are maintained during DNA replication. How does the distribution of histones during replication compare to the semi-conservative replication of DNA itself? What are the "tails" of the histone proteins, and how are they distinguished from the rest of the protein? What are the "tails" of the histone proteins, and how are they distinguished from the rest of the protein?