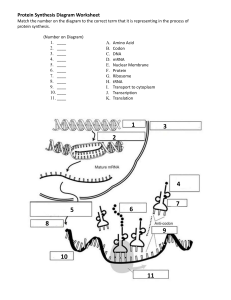
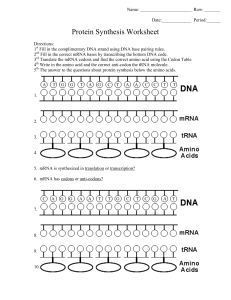
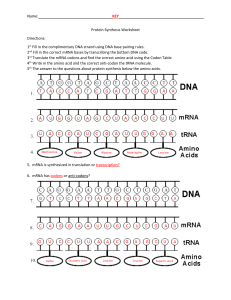
7.3 Translation Essential Idea 2.7: Genetic information in DNA can be accurately copied and can be translated to make the proteins needed by the cell Essential Idea 7.3: Informational transferred from DNA to mRNA is translated into an amino acid sequence What is Translation? Translation is the ___________ of ___________________(polymer of amino acid) _______________________ Translation happens in the cytoplasm / ER on a structure called ribosomes Ribosomes are made up __________ (ribosomal RNA) molecules and __________ Transcription Translation 1. After genes are ____________ into mRNA and ____________ into _______ mRNA, 2. These mRNA will exit the nucleus and bind to the ________ __________ of the ribosomes 3. Ribosomes has a large subunit and a small subunit which helps to bind the _________, _________ and amino acids together and ____________ the formation of _________ between the ___________________ Ribosomes 7.3.S3 The use of molecular visualisation software to analyse the structure of eukaryotic ribosomes and tRNA molecules. Recall: ribosomes in prokaryotes (70s) and in Eukaryotes (80s) Ribosomes are made up of a. protein – for ________________ b. ribosomal RNA – for ________ ____________ They have 2 subunits a. __________ subunit (30s) binds to the ___________ b. ______ subunit (50s) have 3 ________ _________ sites c. ___ (exit) site,___ (peptidyl) site, ___ (aminoacyl) site Structure of tRNA Have sections that become ______ ___________ by ________ __________ _______ base – ______________ (3 bases – attached to ________) which is part of the bottom loop Two other loops - associate with ______________ (T arm) & tRNA _____________ _______ (D arm) Base sequence __________ at the ________ end which forms site for _____________ to __________ _____________ – called the ____________ loop/stem tRNA activating enzyme 7.3.A1 tRNA-activating enzyme illustrate enzyme-substrate specificity and the role of phosphorylation tRNA molecule binds to a __________________ amino acid by _________________________________________________ 20 different types of tRNA activating enzyme – the enzyme’s _________ __________ is ___________ to each 20 _________ ____________ and specific to the correct __________ _________ (may recognise multiple tRNA due to _________________) This process uses __________ for energy, main function of ATP is to ___________ the _______ ____________ _______ to tRNA as ________ from this __________ is later used to _________ amino acid (____________ bond) during formation of the _______________ chain Step-by-step process 1. Specific ________ ________ & ____________ attached to active site of enzyme 2. ATP _____________ (break bond and release two phosphate) and ___________ amino acid 3. By forming a ________ _________ ____________ between _______________ and _________ (adenosine monophosphate – originally from ATP) 4. The high energy bond is transferred to ___________ ____________ the activated amino acid to tRNA & AMP is released 5. tRNA is ‘_______________’ and ready to use The GENETIC CODE – CODONS 2.7.U6 The amino acid sequence of polypeptides is determine by mRNA according to the genetic code 2.7.U7 Codons on three bases on mRNA correspond to one amino acid in polypeptide There are four different bases and 20 different type of amino acids So for the bases to code for the amino acids, we use a t_______ b_____ code – called c________ Three combination of bases – codes for a certain type of _________ ____________ The ______ of these codons on the __________ determines the amino acid ____________ of the polypeptides Note* there is also a STOP & START codon KEY Features of the Genetic Code The __________ __________ is _____________ - All living organisms uses the ________ triplet base ___________ (meaning this has evolved at the origin of life) In very limited organisms, some codons have different meaning, which raised some hypothesis that genetic code might not be universal, or that codons have occurred after the origin of life The code is ‘__________________’ – meaning that there are m_______ than o_____ c________ that codes for an amino acid o The degeneracy usually (not always) happens in the t_______ b________ p_________ o This also allows for ‘s__________ m______________ – meaning a mutation of the third base will not affect the production of protein Translation from mRNA codon • In order to translate an mRNA sequence into a polypeptide chain, it is important to establish the correct r____________ f____________ • An open reading frame starts with __________ (_____________ codon) and will continue in triplets to a _____________ codon (UAA, UAG, UGA) Codons on mRNA; Anti-codons on tRNA 2.7.U8 Translation depends on complementary base pairing between codons on mRNA and anticodons on tRNA • • __________________ (transfer RNA) has an ______________ that binds c_______________ to the ___________ of the mRNA (through base pairing) and each tRNA carries the ___________ _________ the corresponding triplet base codon on mRNA So as the ribosome ‘reads’ the mRNA from ________ to ____________direction, the tRNA with the complementary anti-codon to the mRNA will bind to the ribosome in sequence Initiation of Translation 7.3.U1 Initiation of translation involves assembly of components that carry out the process 1. ___________ binds to _________ ribosomal subunit 2. tRNA molecule carrying ___________ binds to ’start’ codon ____________ at the ______ site 3. ____________ ribosomal unit then bind to the ____________ ribosomal unit 4. The next codon _________ another ________ to bind to the __________ site Elongation of Translation 7.3.U2 Synthesis of the polypeptide involves a repeated cycle of events 5. _____________ _______ (condensation reaction) form between ________ _______ in ____ & _____ site 6. The first amino acid form P site is now _________________ attached to the amino acid from A site and the first tRNA is ________________ (no amino acid) 7. As the ribosome moves along the mRNA (5’ 3’), the deacylated tRNA moves from P to ____________ site and is _______________. 8. While the tRNA carrying the growing _________ _______ __________moves from _____ to _______ site 9. The next tRNA molecule that binds to the complementary base in the empty _______ site 10. This process is _________________ until a STOP codon is reached Termination of Translation 7.3.U3 Disassembly of components follows termination of translation • • When stop codon is reached, a r____________ f_________ binds to the _____ site that signals translation to stop (no tRNA for STOP codon) _____________ & _________ is released and ribosome d_____________ into two independent subunits Free Ribosomes 7.3.U4 Free ribosomes synthesize proteins for use primarily within the cell • • • • In Eukaryotes, ribosomes are separated from DNA by the nucleus Transcription happens in the __________ but _____________ happens in the _________ (fluid in cytoplasm) or the ______________ _________ After transcription, mRNA is transported from the nucleus and this transport requires m___________________ of mRNA (eg: ____ ___________ & addition of ________ t____ at 3’) Proteins destined for use in the ____________, _______________ and _______________ are synthesized by ________ ______________ in the _________________ ER Bound ribosomes 7.3.U5 Bound ribosomes synthesize proteins primarily for secretion or for use in lysosomes • Proteins that are used in the _______, _________ ___________, ____________, _________ _____________ or _____________ outside the ___________ are synthesized by ribosomes _________ to the __________ • Whether the ribosome is bound to the ER or free in the cytoplasm depends on the _________ _________________ on the _____________ being translated • The _________ _____________ is in the _________ part of the polypeptide and it can bind to a s___________ r______________ p______________ that stops _____________ until it binds to a ________________ on the ________surface • Once this happen, translation then continue and the polypeptide chain _____________ into the ER as it is being translated Prokaryotes – transcription & translation 7.3.U6 Translation occur immediately after transcription in prokaryotes due to absence of nuclear membrane • • • • In prokaryotes, ribosomes can start ________________ can _________ ____________ even while the rest of the mRNA is being transcribed from the DNA template This is because there is ______ ___________ __________________ ______________ it And possible because both _______________ and _____________ occur in the ____ to ____ direction No modification of mRNA in prokaryotes Polysomes (many ribosomes) 7.3.S1 Identification of polysomes in electron micrographs of prokaryotes and eukaryotes • • • • Polysomes = _____________ ______________ attached to a ________ __________ strand Visible under the electron microscope In prokaryotes, polysomes can be seen associated with ___________ strand In eukaryotes, polysomes occur in ____________ and next to _____________ In this eukaryotic polysome, the 5’ end is on the left as it has shorter polypeptide chain where the ribosomes have just started translating while the 3’ end is on the right where long chains of polypeptides have formed. Primary structure – chain of amino acid 7.3.U7 The sequence and number of amino acid in the polypeptide is the primary structure • Polypeptide – _________ of __________ with __________ _____________ and _______________ • With 20 types of amino acid, combined in any sequence and in any numbers brings a huge diversity of polypeptide • Primary structure controls all subsequent levels of protein organisation because it determines the nature of the interactions between R groups of different amino acids Secondary Structure 7.3.U8 The secondary structure is their formation of alpha helices and beta pleated sheets stabilised by hydrogen bonding • • • • • Chain of amino acid have ___________ covalent bonds in its ________________ due to _________ from _________ grp & ________ from ________________ grp of __________________________ amino acids Hydrogen bonds can form between Hydrogen of amine group of one amino acid to oxygen of carboxyl group of another amino acid along the chain • Form either ________________ or ____________________ Polypeptide coil / fold and hydrogen bonds provide ____________ ________________ Most fibrous proteins have secondary structure (eg: collagen & keratin) Tertiary Structure 7.3.U9 The tertiary structure is the further folding of the polypeptide stabilised by interaction between R groups • Further bonds formed between the _____________ side chains & surrounding water molecule ___________ the proteins into a complex ______________ • Tertiary ____________ maybe important for ______________ of protein (eg: specific active site shape for enzymes) • Eg: ________________, _________________, ___________________, _________________ • This structure is common in _____________ proteins Quaternary structure 7.3.U10 The quaternary structure exists in proteins with more than one polypeptide chain • Forms when _________________ ________________ interact together (haemoglobin is composed of four polypeptide chains) • Or interact with ___________________ prosthetic group (eg: Fe – haem group in haemoglobin) • Can be fibrous or globular proteins Shape Role Solubility Reason for insolubility/ solubility Sequence Stability Examples Fibrous Protein Globular protein Hydrophobic R groups are exposed as it form long strands Hydrophobic R groups can be folded into core of molecule and away from surrounding water molecules 7.3 Translation Essential Idea 2.7: Genetic information in DNA can be accurately copied and can be translated to make the proteins needed by the cell Essential Idea 7.3: Informational transferred from DNA to mRNA is translated into an amino acid sequence What is Translation? Translation is the synthesis of polypeptide (polymer of amino acid) on ribosomes Translation happens in the cytoplasm / ER on a structure called ribosomes Ribosomes are made up rRNA (ribosomal RNA) molecules and proteins Transcription Translation 4. After genes are transcribed into mRNA and modified into mature mRNA, 5. These mRNA will exit the nucleus and bind to the small subunit of the ribosomes 6. Ribosomes has a large subunit and a small subunit which helps to bind the mRNA, tRNA and amino acids together and catalyse the formation of bond between the amino acids Ribosomes 7.3.S3 The use of molecular visualisation software to analyse the structure of eukaryotic ribosomes and tRNA molecules. Recall: ribosomes in prokaryotes (70s) and in Eukaryotes (80s) Ribosomes are made up of c. protein – for stability d. ribosomal RNA – for catalytic activity They have 2 subunits d. Small subunit (30s) binds to the mRNA e. Large subunit (50s) have 3 tRNA binding sites f. E (exit) site, P (peptidyl) site, A (aminoacyl) site on large subunit Structure of tRNA Have sections that become double stranded by base pairing Triplet base – anticodon (3 bases – attached to mRNA) which is part of the bottom loop Two other loops - associate with ribosomes (T arm) & tRNA activating enzyme (D arm) Base sequence CCA at the 3’ end which forms site for attaching to amino acid – called the acceptor loop/stem tRNA activating enzyme 7.3.A1 tRNA-activating enzyme illustrate enzyme-substrate specificity and the role of phosphorylation tRNA molecule binds to a specific amino acid by tRNA-activating enzyme 20 different types of tRNA activating enzyme – the enzyme active site is specific to each 20 amino acids and specific to the correct tRNA molecule (may recognise multiple tRNA due to degeneracy) This process uses ATP for energy, main function of ATP is to transfer the high energy bond to tRNA as energy from this bond is later used to linked amino acid (peptide bond) during formation of the polypeptide chain Step-by-step process 6. Specific amino acid & ATP attached to active site of enzyme 7. ATP hydrolyse (break bond and release two phosphate) and activate amino acid 8. By forming a high energy bond between amino acid – AMP (adenosine monophosphate – originally from ATP) 9. The activated amino acid is then covalently bonded to tRNA & AMP is released 10. tRNA is ‘charged’ and ready to use The GENETIC CODE – CODONS 2.7.U6 The amino acid sequence of polypeptides is determine by mRNA according to the genetic code 2.7.U7 Codons on three bases on mRNA correspond to one amino acid in polypeptide There are four different bases and 20 different type of amino acids So for the bases to code for the amino acids, we use a triplet base code – called codons Three combination of bases – codes for a certain type of amino acid The order of these codons on the mRNA determines the amino acid sequence of the polypeptides Note* there is also a STOP & START codon KEY Features of the Genetic Code The genetic code is universal - All living organisms uses the same code (meaning this has evolved at the origin of life) In very limited organisms, some codons have different meaning, which raised some hypothesis that genetic code might not be universal, or that codons have occurred after the origin of life The code is ‘degenerate’ – meaning that there are more than one codon that codes for an amino acid o The degeneracy usually (not always) happens in the third base position o This also allows for ‘silent mutation’ – meaning a mutation of the third base will not affect the production of protein Translation from mRNA codon • In order to translate an mRNA sequence into a polypeptide chain, it is important to establish the correct reading frame • An open reading frame starts with AUG (start codon) and will continue in triplets to a termination codon (UAA, UAG, UGA) Codons on mRNA; Anti-codons on tRNA 2.7.U8 Translation depends on complementary base pairing between codons on mRNA and anticodons on tRNA • • tRNA (transfer RNA) has an anti-codon that binds complementarily to the codon of the mRNA and each carries the amino acid the corresponding amino acid So as the ribosome ‘reads’ the mRNA from 5’ to 3’ direction, the tRNA with the complementary anti-codon to the mRNA will bind to the ribosome in sequence Initiation of Translation 7.3.U1 Initiation of translation involves assembly of components that carry out the process 5. mRNA binds to small ribosomal subunit 6. tRNA molecule carrying methionine binds to ’start’ codon AUG at the P site 7. Large ribosomal unit then bind to the small ribosomal unit 8. The next codon signals another tRNA to bind to the A site Elongation of Translation 7.3.U2 Synthesis of the polypeptide involves a repeated cycle of events 5. Peptide bond (condensation reaction) form between amino acid in A & P site 6. The first amino acid form P site is now covalently attached to the amino acid from A site and the first tRNA is deacylated (no amino acid) 7. As the ribosome moves along the mRNA (5’ 3’), the deacylated tRNA moves from P to E site and is released. 8. While the tRNA carrying the amino acid chain moves from A to P site 9. The next tRNA molecule that binds to the complementary base in the empty A site 10. This process is repeated until a STOP codon is reached Termination of Translation 7.3.U3 Disassembly of components follows termination of translation • • When stop codon is reached, a release factor binds to the A site that signals translation to stop (no tRNA for STOP codon) Polypeptide & mRNA is released and ribosome disassembled into two independent subunits Free Ribosomes 7.3.U4 Free ribosomes synthesize proteins for use primarily within the cell • • • • In Eukaryotes, ribosomes are separated from DNA by the nucleus Transcription happens in the nucleus but translation happens in the cytosol (fluid in cytoplasm) or the endoplasmic reticulum After transcription, mRNA is transported from the nucleus and this transport requires modification of mRNA (eg: 5’ capping & addition of poly-A tail at 3’) Proteins destined for use in the cytoplasm, mitochondria and chloroplasts are synthesized by free ribosomes in the cytoplasm ER Bound ribosomes 7.3.U5 Bound ribosomes synthesize proteins primarily for secretion or for use in lysosomes • • • • Proteins that are used in the ER, Golgi apparatus, lysosomes, plasma membrane or secreted outside the cell are synthesized by ribosomes bound to the ER Whether the ribosome is bound to the ER or free in the cytoplasm depends on the signal sequence on the polypeptide being translated The signal sequence is in the initial part of the polypeptide and it can bind to a signal recognition proteins that stops translation until it binds to a receptor on the ER surface Once this happen, translation then continue and the polypeptide chain grows into the ER as it is being translated Prokaryotes – transcription & translation 7.3.U6 Translation occur immediately after transcription in prokaryotes due to absence of nuclear membrane • • • • In prokaryotes, ribosomes can start translation can occur immediately even while the rest of the mRNA is being transcribed from the DNA template This is because there is no nuclear membrane separating it And possible because both translation and transcription occur in the 5’ to 3’ direction No modification of mRNA in prokaryotes Polysomes (many ribosomes) 7.3.S1 Identification of polysomes in electron micrographs of prokaryotes and eukaryotes • • • • Polysomes = multiple ribosomes attached to a single mRNA molecules Visible under the electron microscope In prokaryotes, polysomes can be seen associated with DNA strand In eukaryotes, polysomes occur in cytoplasm and next to ER In this eukaryotic polysome, the 5’ end is on the left as it has shorter polypeptide chain where the ribosomes have just started translating while the 3’ end is on the right where long chains of polypeptides have formed. Primary structure – chain of amino acid 7.3.U7 The sequence and number of amino acid in the polypeptide is the primary structure • Polypeptide – chain of amino acid with specific sequence and length • With 20 types of amino acid, combined in any sequence and in any numbers brings a huge diversity of polypeptide • Primary structure controls all subsequent levels of protein organisation because it determines the nature of the interactions between R groups of different amino acids Secondary Structure 7.3.U8 The secondary structure is their formation of alpha helices and beta pleated sheets stabilised by hydrogen bonding • • • • • Chain of amino acid have polar covalent bonds in its back bone due to N--H from amine grp & C=O from carboxyl grp of non-adjacent amino acids Hydrogen bonds can form between amine group of one amino acid to carboxyl group of another amino acid along the chain • Form either ⍺ - helix or β – pleated sheet Polypeptide turn / fold and hydrogen bonds provide structural stability Most fibrous proteins have secondary structure (eg: collagen & keratin) Tertiary Structure 7.3.U9 The tertiary structure is the further folding of the polypeptide stabilised by interaction between R groups • Further bonds formed between the R group side chains & surrounding water molecule folds the proteins into a complex 3D structure • Tertiary structure maybe important for function of protein (eg: specific active site shape for enzymes) • Eg: H-bonds, disulphide bonds, ionic bonds and hydrophobic interactions • This structure is common in globular proteins Quaternary structure 7.3.U10 The quaternary structure exists in proteins with more than one polypeptide chain • Forms when multiple polypeptides interact together (haemoglobin is composed of four polypeptide chains) • Or interact with inorganic prosthetic group (eg: Fe – haem group in haemoglobin) • Can be fibrous or globular proteins