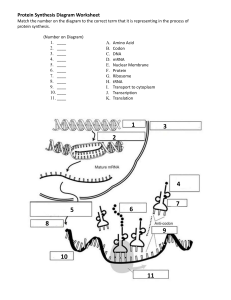
Module 13 The genetic code o Codons: 3 base sequence of DNA that codes for an amino acid (triplet) Expressed as mRNAs in the 5’ to 3’ orientation start/initiation codon: AUG = methionine Stop codons: UAA, UAG, UGA DONT code for amino acids o Reading frame: linear sequence of codons in a nucleic acid defined by a start codon and ending with a stop codon Start with AUG and from there every 3 letters are a codon that codes for one amino acid Characteristics of the genetic code o The genetic code is unambiguous. Each of the 61 triplets that code for amino acids, will only code for 1 amino acid. o The genetic code is degenerate Most amino acids are coded by more than one codon o The genetic code is universal Most living organisms use the exact same code, but some exceptions exist o Commaless: there are no breaks between the codons in a reading frame. All of the bases of the translated sequences are part of codons. No letters left in between o Non-overlapping: the triplets in a reading frame are in a tandem sequence and do not overlap o Translation on ribosomes will result in a polypeptide Mutations o o Substitution mutation can occur because of unrepaired errors in DNA replication but can also occur spontaneously. Spontaneous cytosine deamination Cytosine deaminated results in a uracil in DNA Consequences: o Deaminated cytosine becomes a uracil o C can pair with G, but uracil cannot o Results in a mismatch o The cell has mechanisms to fix this, but sometimes those mechanisms can fail o The strand with uracil will become template for an adenine o Results in a permanent substitution mutation Substitution mutations in protein coding genes o When a base is replaced by a different base in DNA, the result is a permanent single codon change o Mutations occur in DNA and are copied to mRNAs o 3 types of substitution mutations Silent: the resulting new codons code for the same amino acid as the original codon Missense: the new codon codes for a different amino acid Nonsense: the new codon is a stop codon Resulting in a shorter polypeptide o AGU= Ser If the U is replaced with a C, then it will produce a different codon However, it just so happens that AGC also code for Ser so it will stay the same This is a silent mutation There is no consequence in the protein that’ll be synthesized o AGU= Ser If the G is replaced with an A AAU codes for a different amino acid, ASN This is a missense mutation A different amino acids is replaced in the protein o AGA= Arg If the A is replaced with a U UGA is a stop codon Nonsense mutation The protein synthesis is terminated prematurely Consequences of these mutation o May or may not have a big effect depending on the situation o A single amino acid substitution in the hemoglobin protein results in sickle cell anemia Insertion mutations o Frameshift: reading frame has been altered Results in a completely different polypeptide Very dire consequences Trinucleotide repeat expansions o Top strand is the template The mRNA that is translated from the templates will have the same sequence as the nontemplate (the bottom strand) o The bottom strand is CAG repeats CAG= glutamine o Because of complementarity between C’s and G’s a hairpin forms on the newly synthesized strand o Causing part of the template strand to be replicated twice and increasing the number of repeats on the newly synthesized strand Additional 5 repeats of the trinucleotide has been added o When the new strand becomes template for the next round of DNA replication, the new gene now contains 13 repeats of the trinucleotide instead of the original 8 Trinucleotide repeats expansion o This example happens in Huntington’s disease An autosomal dominant disease caused by expansions of CAG trinucleotide repeats in the huntington gene 11-35 CAG repeats= normal phenotype 36-39 CAG repeats= borderline phenotype You may or may not develop the disease 40+ CAG repeats= disease phenotype Translation in bacteria o Basic requirements: mRNA Charged transfer RNA (tRNAs) (the molecules that actually read the genetic code in the mRNA Ribosome Other requirements: initiation factors, elongation factors, and energy sources No primers needed o Transfer RNA (tRNA) By itself, tRNAs are no use unless they have an amino acid attached to it 3’ end is the amino acids attachment site Anticodon arm reads the genetic code Rare bases (red dots) Unusual bases found in tRNA, not found in other nucleic acids RNA have A,C,G, and U Inosine: modified/deaminated adenine o Enzymatically modified base from adenine to inosine o Sometimes shows up in the anticodon loop in tRNA molecules o o Because of degeneracy, the codons that code for the same amino acid, have the same sequence except the 3rd letter is changed. This allows the cell to employ a trick The wobble hypothesis Proposed by Crick He proposed the interaction between the codon in mRNA and the first letter of the anticodon in the tRNA. The anticodon is the 3 letter word that base pairs with the codon. UCC= Ser Pairing at the 3rd codon is relaxed AGG can recognize UCC or UCU One tRNA can read more than one codon in the mRNA, so the cell does not have to make too many tRNAs Essential amino acids for humans o The R or variable group distinguishes an amino acid from the rest o Top portion is the same in all of them o The carboxyl group (COOH) is the portion of the amino acid that will be attached to the 3’ end of the tRNA Charging of a tRNA o This is a charged tRNA that can now participate in protein synthesis o Charging is catalyzed by 20 different Aminoacyl tRNA synthases Requires ATP The correct amino acids need to be charged to the correct tRNA, the one that has the anticodon, that can read the codon of the mRNA that codes for the amino acid that its carrying Recognition of tRNAs by Aminoacyl tRNA synthases o It can recognize a tRNA as the correct tRNA to charge with the correct amino acid is because of certain positions within the tRA that include bases that identify that tRNA as the correct one Charging of a tRNA o Needs energy because you are creating a new covalent bond between an amino acid and tRNA o Step 1: Amino Acid Activation: ATP is hydrolyzed, loses 2 phosphates and the resulting AMP is attached to the carboxyl group in the amino acid Results in aminoacyl adenylic acid (AA-AMP) Cell expends energy o Step 2: Charging Exchange the bond between the amino acid and AMP with the bond between the amino acids and the 3’ end of the correct tRNA AMP is detached from the amino acid, and the amino acids is attached the 3’ end of the tRNA Results in Aminoacyl tRNA The prokaryotic ribosome o Ribosome: large particle of rRNA and proteins where translation occurs o 2 subunits: small and large o The 2 subunits dont get together unless they have an mRNA associated with it. o The Svedberg unit (S) is not a measure of molecular weight, but a measure of the rate at which particles sediment in a centrifugal field. Depends on weight, size, and shape o Svedberg values Small subunit: 30s Large subunit: 50s Complete ribosome: 70s o Sites in the ribosome Peptydyl (P): where the AUG will sit initially Aminoacyl (A): where the 2nd codon will be Exit (E): occupied briefly by tRNA molecules that have already donated the amino acid they are carrying to a growing chain of polypeptide o Steps in initiation of translation: The mRNA binds to the small ribosomal subunit, with the AUG codon positioned on the P site f-Met-tRNA binds the AUG codon The large ribosomal subunit joins the complex This process requires GTP (energy) plus a series of initiation factors (IFs) Elongation of translation o EF-tu (elongation factor) and GTP facilitate the binding of the 2nd tRNA to the 2nd codon at the A site o o o o o The amino acid on the 1st tRNA is transferred and forms a peptide bond with the amino acid on the 2nd tRNA A dipeptide forms The 1st tRNA moves to the E site The mRNA is shifted to place the 2nd codon in the P site and bring the 3rd codon into the A site The tRNA carrying the 3rd amino acids binds the 3rd codon on the A site The dipeptide on the 2nd tRNA is transferred to form a tripeptide attached to the 3rd tRNA The 2nd tRNA moves to the E site with the help of EF-G and GTP 3rd codon moves to the P site Summary o Initiation is complete when the 1st tRNA carrying the 1st amino acid binds to the codon on the mRNA which is in the P site. o At any point, the tRNA in the P site has a chain of amino acids o A new tRNA carrying the amino acids, binds to the aminoacyl site o The last amino acids that came in, along with the chain that already formed, will detach from the tRNA in the P site and form a peptide bond with the amino acids attached to the tRNA on the aminoacyl site. o The position of the mRNA will shift so the uncharged tNRA will move to the exit site and will be eliminated o The original AUG is now empty and available, now another ribosome can assemble there Explains Simultaneous Transcription and Translation in bacteria As soon as AUG appears it can be recognized by a ribosome This CANNOT happen in eukaryotes It can happen in bacteria because there is no physical separation between where the DNA is and the ribosomes are. Termination of translation o A stop codon moves to the A site o No tRNA binds to the stop codon o Release factor 1 (RF1) binds to the stop codon o The GTP-dependent Release factor 3 release the polypeptide chain from the last tRNA and the translation complex dissociates The diphtheria toxin o Inhibits the last step of the polypeptide elongation cycle o Produced by Corynebacterium diphtheriae o Inhibits the eukaryotic elongation factor 2 (EF2) Initiation of translation can happened, but not elongation Cell is unable to make new proteins Cell will die, will result in major organ failure Module 14 Levels of gene regulation o The mechanisms of gene regulation, or genetic control, determine where, when, and how much a gene is expressed o Occurs at many levels o Level of transcription Dna that is condensed cannot be transcribes Important for eukaryotes Results in pre mRNA The amount of processing that occurs can also be subject to regulation o Ex: addition of a poly a tail The process DNA is translated into a polypeptide by ribosome and tRNA o Likely inactive polypeptide o Requires posttranslational modification before it can become an active protein Regulation of transcription o Every gene is an RNA-coding region (a transcribed region) o Nearby regulatory regions are NOT transcribes o DNA binding proteins recognize specific sequences in the regulatory regions near the gene and either activate or repress the transcription of the coding region Gene regulation in prokaryotes: The Operon o Operon: a cluster of structural genes with related functions under the control of a common regulatory system that can respond to changes in the environmental condition. o Operons are common in prokaryotes but rare in eukaryotes o Genes A, B, and C are structural genes The products are involved in a related function o o o Promoter and operator overlap and come before the structural genes The regulator protein turns on or off the expression of the structural genes Operons are rare in eukaryotes o Transcription of the polycistronic mRNA from the lac operon Structural genes: lacZ lacY lacA When they are transcribed, it is transcribed into a single Polycistronic mRNA Translation will result in 3 separate proteins Lac operon o The products of the structural genes of the lac operon in E. coli are involved in the utilization of lactose for energy o These genes will only be expressed if lactose is available, but also only if the cell needs to use lactose for energy o Glucose is the preferred source of energy for the cell, therefore: The genes of the lac operon are expressed only if the glucose is absent and lactose is available The products of the structural genes of the lac operon o lacZ gene product: ꞵ -galactosidase Breaks down lactose into galactose and glucose, which can enter glycolysis the harvest energy for the cell Can also isomerize lactose allolactose o lacY gene product: permease A membrane transporter of lactose Facilitates the entry of lactose into the bacterial cell o lacA gene product: transacetylase Function in lactose metabolism unknown Might be involved in the removal of by-products of lactose digestion from the cell Chromosomal components of the lac operon o Structural genes under operon control: lacZ, lacY, and lacA 3 of them are controlled by a common regulatory system which includes a promoter (Plac) and an operator (lacO) The promoter and operator overlap o lacI (i gene): codes for the repressor protein o Control regions: lacP (promoter or Plac): binding site for RNA polymerase lacO (lac operator): binding site for the repressor protein Pi: promoter for lacI The state of the lac operon in the absence of lactose o In the absence of lactose, the regulator protein (a repressor) binds to the operator, blocking RNA polymerase from binding to the promoter o No expression of the structural genes (repression of gene expression) o This system is leaky Very small amounts of ꞵ -galactosidase and permase are still made. A little bit of transcription and production of the products will still happen. The state of the lac operon in the presence of lactose o The little bit of permase made by the cell will allow some of the lactose to come in o Ꞵ -galactosidase that was made from the leaky system, will turn lactose into allolactose Allolactose will be the inducer of the operon, because it will bind the repressor protein and inactivate it. The repressor protein cannot bind the operator RNA polymerase can bind the promoter region Transcription and translation of structural genes will happen o The operon inducer is allolactose (a lactose metabolite that inactivates the repressor protein). It activates the expression of the genes needed for the use of lactose as an energy source o This will only happen if glucose is not available Catabolite repression of the lac operon o If glucose is available, the lac structural genes are OFF, even if lactose is present. The binding of RNA polymerase to the promoter occurs only if glucose is absent. o Effect of glucose levels on CAP activity The activity of the enzyme adenylate cyclase (which catalyzes the hydrolysis of ATP into cAMP + PP) is induced when glucose is absent Cytoplasmic levels of cAMP (the “catabolite”) increase cAMP activates the catabolite activator protein (CAP) Activated CAP facilitates the binding of RNA polymerase to the lac promoter The catabolite activator protein (CAP) is necessary for the stable binding of RNA polymerase to the promoter. It is active only when glucose is absent In the presence of glucose, cAMP levels decrease, and CAP remains inactive o RNA polymerase does not bind the promoter efficiently o The operon is OFF States of the lac operon o Glucose (+) Lactose (+) = Operon (OFF) Glucose and lactose are available Glucose present means adenylate cyclase will not be activated, so cyclic amp will not be made, so CAP will not be able to assist the binding of RNA polymerase to the promoter Lactose present means allolactose will inactive the repressor protein so the operator is not occupied, so RNA polymerase could bind the promoter CAP is not there to help, so RNA will not bind the promoter efficiently so the operon will remain off o Glucose (+) Lactose (-) = Operon (OFF) Glucose present means CAP will not be able to assist Lactose absent means the repressor protein is active and binding to the operator Operon is off o Glucose (-) Lactose (-) = Operon (OFF) No glucose means adenylate cyclase is activated, cyclic AMP is made, CAP can assist RNA polymerase to bind the promoter efficiently No lactose means the repressor protein is active and will bind to the operator o Glucose (-) Lactose (+) = Operon (ON) Transcription will happen The trp operon o In prokaryotes o One of the 20 amino acids used for making proteins o The 5 structural genes of the trp operon code for proteins that are necessary for the biosynthesis (making of trp) of trp in the cell. trpE, trpD, trpC, trpB, and trpA Are transcribed as a polycistronic mRNA beginning with the 5’ untranslated region (5’ UTR) o If trp is absent: the structural genes are expressed o If trip is present: the structural genes are turned off by trp itself o Promoter (trpP); binding site of RNA polymerase o Operator (trpO): binding site of the repressor protein The operator overlaps the promoter Both are just before the beginning of the structural genes o Repressor gene (trpR): codes for the repressor protein o Repressor gene promoter (Pr) o The expression of the trpR results in production of the regulate protein It is inactive in the absence of trp It will not be able to bind the operator RNA polymerase will be able to bind the promoter and transcribe the structural genes Resulting in the enzymes necessary for the biosynthesis of the amino acid trp o Trp present: trp binds the repressor protein and activates it. The binding of the trpactivated repressor to the operator prevents the binding of RNA polymerase to the promoter. How do we know that these specific protein-DNA interactions happen? o Electrophoresis Fragments of DNA migrate in a gel in an electric field toward the plus end. The speed of migration depends on the size of the fragment of DNA. Smaller= faster migration What if the fragment of DNA has a protein bound to it? (the operator of the trp operon) The electrophoretic mobility shift assay (EMSA) o EMSA is a method used for detecting specific DNA-protein interactions, such as the specific binding of transcription factors and other regulatory proteins to regulatory regions of the chromosome o Ex: establishing the specific bingsing of the trp repressor to the trp operator in the presence of tryptophan o Mix in a test tube: DNA fragment: trpO E. coli cell extract: contains all cellular proteins Trp (also run a control experiment with no trp) Specific antibodies against the trp repressor and other cellular pretins, such as the lac repressor Antibodies recognize specific antigens 5 experiments (test tubes) Conduct electrophoresis on each of the test tubes 1. DNA alone o Migrates fast and far 2. DNA + cell extract (no trp) o Cannot activate the repressor protein and will not affect the mobility of the DNA 3. DNA + cell extract + trp o Trp will bind and activate the repressor protein. The repressor protein will bind the DNA. o DNA now has a protein so the migration will be slower (mobility shift) 4. DNA + cell extract + trp + anti-lac repressor antibody o The antibody finds the lac repressor protein in the cell extract and binds to it. It will not affect the binding of the activated trp repressor protein to the DNA o Result is similar to the 3rd experiment 5. DNA + cell extract + trp + anti-trp repressor antibody o Add antibody against the repressor proteins o The repressor protein is active so it will bind the DNA. the antibody binds the repressor protein o Results in the 2nd mobility shift o Slows the migration of the DNA even more The first 3 experiments tells us that in the presence of trp, something in the cell extract becomes activated and binds the operator of the trp operon Last 2 experiments identify the protein binding to the DNA. The protein that binds the DNA in the presence of trp is the trp operon repressor protein. Gene regulation in eukaryotes o Classification of eukaryotic genes according to the RNA polymerases that transcribe theme: RNA polymerase 1: large rRNA genes (5.8s 18s, and 28s) RNA polymerase 2: all mRNA-coding genes (all protein-coding genes) RNA polymerase 3: small rRNA (5s) and all tRNA genes. The consequence of histone acetylation o Transcriptional regulation is about opening of DNA by partially disassociating it from histones to allow RNA polymerase to bind o Nuclear DNA in eukaryotes is always in the presence of histones o Lysine (K) is an amino acids we see a lot in histone proteins Has a positive charge amino group Can interact with NDA tightly because DNA has negative charge in the phosphate groups o Histone acetyltransferase can add an acetyl group to lysine, eliminating the positive charge The histone acetylated will not be able to interact with DNA very tightly o Histone acetylation weakens the interaction between basic histones and the acidic DNA molecule, causing chromatin decondensation. o Histone deacetylase removes the acetyl group from the lysine and you go back to the tight interaction between histone and DNA Covalent modification of histones o Transcription activators recruit histone acetyl transferases (HATs), causing chromatin decondensation. o HATs act as coativators, but they do not bind to DNA o Transcription repressors recruit histone deacetylases (HDACs), causing chromatin condensation. o HDACs act as corepressors, but they do not bind to DNA Control of flowering Arabidopsis thaliana o Flowering locus C (FLC) codes for a transcription factor that represses flowering, but is only expressed if the histones on the locus are acetylated. o Mutations in FLC result in premature flowering o The gene product of Flowering locus D (FLD) is a histone deacetylase that inactivates the FLC locus Components of a transcription regulation system in eukaryotes o o o The term transcription factor refers to any transcription regulator protein that binds to a specific DNA sequence. The transcription regulator protein can be positive (activates transcription) or negative (repress transcription). Cis-acting elements are DNA sequences that are necessary for the control of transcription(the regulatory regions of the chromosome): promoters, enhancers, and silencers. Trans-acting factors are proteins that are necessary for the control of transcription (the transcription factors that bind to the cis-acting elements) Whether transcription happens or not Regulatory regions o Promoter Is always right before the +1 (transcription initiation site) and in very close proximity Recognized by the transcription initiation complex (basal transcription apparatus) Sufficient only for basal (unstimulated) transcription o Conserved elements within the promoter region of eukaryotic egens transcribed by RNA polymerase 2 TATA box: located 25-30 bases upstream from the +1, and present in the promoters of all mRNA-coding genes Enhancers Required for stimulated transcription (up-regulation) Sequences vary widely and are recognized by a large variety of transcription activators Therefore: the activation of a gene in any particular cell will depend on whether the cell has the right activators to bind the gene enhancers Enhancers determine where, when, and how much transcription occurs Tissue specific enhancer (recognized by an activator present only in βpancreatic cells Ensures only one type of cell in your body expresses the gene Role of regulatory regions o Promoter alone Only the basal transcription factors may bind to DNA Only the basal transcription initiation complex may form Only very low or undetectable transcription can occur So low, that it’s biologically irrelevant o With the right enhancers Transcriptional activators bind to enhancers Stimulated transcription (biologically significant) o Enhancers can be distant upstream, downstream or in an inverted position (always on the same chromosome), and they can still be effective in stimulating transcription But you can't touch the promoter Trans-acting factors: DNA-binding proteins o Transcription factors are transcription regulator protein that bind to specific DNA sequences o Transcription factors have two domains: A DNA-binding domain Assembly of the RNA polymerase 2 transcription initiation complex on the promoters of eukaryotic protein-coding genes o RNA polymerase 2 transcribes all protein-coding genes in eukaryotes o The purpose of this complex is to direct RNA polymerase 2 to the correct place on the promoter behind the gene transcription site (+1), because the polymerase does not recognize specific DNA sequences This complex is not sufficient for biologically significant levels gene expression o Transcriptional activators, which bind the upstream, (and downstream in some cases) sequence elements (enhancers) are necessary for stimulated (biologically significant) levels of transcription o The main function of the mediator complexes is to transmit signals from the transcription factors to the polymerase o A trans-activating or trans-repressing domain The RNA polymerase 2 transcription initiation complex (or basal transcription apparatus) contains TFIID, other TFIIs, and RNA polymerase 2. DNA loops to allow transcriptional activators, bound to enhancer, to interact with the initiation complex Inducible regulation of the yeast GAL structural genes o The induction of the GAL structural genes: o The products of GAL structural genes in yeast are proteins that metabolize the sugar galactose o o o o The transcription of these genes requires the enhancer UASg (upstream activating sequence of GAL genes) which is permanently occupied by the transcription activator GAL4 In the absence of galactose, the GAL80 protein blocks GAL4 and prevents it from activating the transcription of the GAL structural genes When galactose is available, it activates the GAL3 protein, which interacts with GAL80 to displace it and expose the GAL4 trans-activating domain A mediator interacts with both GAL4 and the transcription initiation complex Translational regulation o The translation of an mRNA can be regulated by the extent of a cell’s requirement for the protein product o Translational regulation of Ferritin and the Transferrin receptor: soluble iron atoms are necessary for the function of many enzymes, but an excess is toxic. o Iron is transported in the blood by the protein Transferrin o Transferrin receptors, on the surface of cells, interact with the Transferrin/iron complex and transport it into the cytoplasm, where iron is then released o if levels of free cytoplasmic iron increase too much, the cell synthesizes Ferritin, a protein that sequesters iron atoms o in the absence of iron, the Iron regulatory protein binds the iron response element (IRE), a stem-loop structure present in both the Ferritin and the Transferrin receptor mRNAs o o o In the absence of iron, the Iron regulatory protein binds the iron response element of the Transferrin receptor mRNA, stabilizing it and promoting translation free iron binds the Iron regulatory protein and prevents it from binding IRE, causing the destabilization of theTransferrin receptor mRNA and disrupting protein synthesis (this prevents the transport of excess of iron) Translational regulation of Ferritin: in the absence of iron, the Iron regulatory protein binds the iron response element of the Ferritin mRNA, inhibiting translation free iron can bind the Iron regulatory protein to allow Ferritin synthesis Post translational regulation: proteolytic cleavage o Just because a polypeptide is made, doesnt mean its a functioning protein o Insulin is translated in preproinsulin Signal sequence Connecting polypeptide A and B functional domains o Signal sequence is removed first by proteolytic cleavage Results in proinsulin o A and B are joined by disulfide bonds o Connecting polypeptide is removed Results in functional insulin hormone Post translational regulation: protein phosphorylation o Requires the hydrolysis of an ATP molecule to place a phosphate group on an amino acid Serine in this example o Consequence Activation of a protein Deactivation Destruction o Protein kinase catalyzes phosphorylation o You need protein phosphatase to reverse phosphorylation Removes phosphate group Phosphoserine hydrolysis in this example Module 16 Recombinant DNA Technology o making recombinant DNA molecules o using expression vectors o applications of recombinant DNA technology Genome Editing Technology o CRISPR-Cas9 targeted gene editing Forensic DNA Profiling o restriction fragment length polymorphisms o short tandem repeats/PCR loci o DNA fingerprints Creating recombinant DNA molecules o Putting together restriction fragments of DNA from different origins The pUC18 polylinker region inside the lacZ gene o Plasmid used as vectors o o o o o Ampicillin resistance gene Any bacterium that picks up this plasmid will be able to grow in medium that has the antibiotic Bacterium without it will be killed Selection for cells carrying the plasm lacZ Polylinker region Collection of restriction sites o Ex: HindIII If you isolated plasmid and incubated it with HindIII restriction in the nuclease, the entire plasmid would be linearized because there is only 1 HindIII site You would be disrupting the lacZ gene Creating recombinant DNA molecules with a cloning vector o To use a plasmid such as pUC18 as a cloning vector: the plasmid is cut with (for example) the restriction enzyme EcoR1 (as a result, the plasmid is linearized) the foreign DNA containing a gene of interest is also treated with EcoR1 so that it will have the same “sticky ends” as the linearized plasmid when mixed, the result will be a recombinant plasmid the recombinant plasmid is introduced into bacteria (transformation: the bacteria will pick up the DNA) as the bacteria multiply, they also multiply the recombinant plasmid with them (cloning) o o o o o o o Antibiotic selection If the nutrient growth medium also contains an antibiotic, the only bacteria that will be able to multiply and form colonies will be those that carry the correct antibiotic resistance gene. Example: ampR (ampicillin resistance gene) in plasmids such as pUC18 and pUC19. The bacteria growing in colonies will be able to grow in the presence of ampicillin Treat the plasmid with a restriction enzyme Digest the foreign DNA with the same restriction enzyme so it will have the same overhangs at the end as the linearized plasmid Put them together and you get the recombinant DNA molecule When that happens the lacZ gene is disrupted by the forgein DNA Remember the restriction site is in the middle of the gene This is what we want lacZ+ gene disrupted by foregin DNA: the plasmid is now lacZThere is a possibility that the foregin DNA will instead close again without any foreign DNA because it has those complementary overhangs The lacZ gene will not be disrupted Plasmid will remain lacZ+ Expression vectors o Use of the pUC19 expression vector ampR is the ampicillin resistance gene the polylinker region inside the lacZ+ gene is a cluster of restriction sites; any of which may be chosen to insert a gene of interest (and this would disrupt the lacZ+ gene) lacZ+, or any gene inserted in the polylinker region, will be under the control of the T7 phage viral promoter (which is the binding site for the T7 phage RNA polymerase),and the lac operator (lacO, the binding site for the lac operon repressor protein) the host bacterium for this plasmid is a lacZ-strain that has been engineered to carry the viral T7 phage RNApolymerasegene under the control of the lac operon promoter and operator in its chromosome o lacZ+, or any gene inserted in the polylinker region, will be under the control of the T7 phage viral promoter (which is the binding site for the T7 phage RNA polymerase), and the lac operator (lacO, the binding site for the lac operon repressor protein). Use of the pUC19 expression vector o IPTG (Isopropyl β-D-1-thiogalactopyranoside) is allolactose analog, and can therefore induce the expression of both the T7 phage RNA polymerase gene in the engineered host bacterium chromosome, and of any gene in front of the T7 phage virus promoter in the plasmid (by binding to the lac operon repressor and inactivating it). Unlike allolactose, IPTG can not be digested by b-galactosidase, so its concentration will remain constant during the experiment. o If the lacZ+ gene in the plasmid is intact, those cells will turn blue in an X-gal assay. o If the plasmid’s lacZ gene has been disrupted, those cells will not turn blue in an X-gal assay because they are lacZ-(they have no lacZ+ gene in their own cellular genome)and therefore can not make their own b-galactosidase. The ampicillin/X-gal blue-white double-selection assay to identify E. coli colonies that contain cloned DNA fragments o To screen for cells that have picked up a plasmid with an inserted DNA fragment following transfection, lacZ-cells are grown on agar medium containing the antibiotic ampicillin, IPTG, and the lactose analog X-gal. o any bacterium that takes up a plasmid will become resistant to ampicillin (the antibiotic) and will grow to form a colony of cells o plasmid vectors that did not integrate any foreign DNAfragment in their polylinker regions will have an intact lacZ gene and will express b-galactosidase o recombinant plasmids (those with a foreign DNA fragment inserted in their polylinker regions) will have a disrupted lacZ gene and will not express bgalactosidase