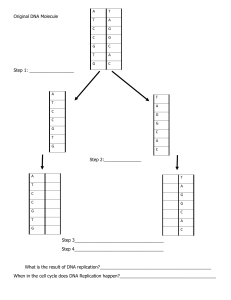
CHAPTER 14 DNA REPLICATION –MOLECULAR ASPECTS DNA REPAIR AND TELOMERES SUMMARY: DNA REPLICATION • The two strands of the DNA molecule unwind • DNA polymerase adds nucleotides to an existing chain • Direction of new synthesis is in the 5′→3′ direction, which is antiparallel to the template strand • Nucleotides enter a newly synthesized chain according to the A-T and G-C complementary base-pairing rules PROKARYOTIC REPLICATION • E. coli model • Single circular molecule of DNA • Replication begins at one origin of replication • Proceeds in both directions around the chromosome • Replicon – DNA controlled by an origin EUKARYOTIC DNA REPLICATION • Eukaryotes usually have multiple, large chromosomes. • multiple origins of replication 6 EUKARYOTIC REPLICATION • Complicated by • Larger amount of DNA in multiple chromosomes • Linear structure • Basic enzymology is similar • Requires new enzymatic activity for dealing with ends only MULTIPLE ORIGINS OF REPLICATION Origin DNA double helix Replication forks Replication direction REPLISOME • Enzymes involved in DNA replication form a macromolecular assembly • 2 main components • Primosome • Primase, helicase, SSB, Topoisomerase • Complex of 2 DNA pol III • One for each strand ENZYMES IN DNA REPLICATION • Many enzymes coordinate to replicate DNA. • DNA helicase unwinds DNA. • Primase initiates all new strands. • DNA polymerase III is the main polymerase. • DNA polymerase I forms the lagging strand. • DNA ligase binds Okazaki fragments together. • E. coli has 3 DNA polymerases • DNA polymerase I (pol I) • Acts on lagging strand to remove primers and replace them with DNA • DNA polymerase II (pol II) • Involved in DNA repair processes • DNA polymerase III (pol III) • Main replication enzyme • All 3 have 3′-to-5′ exonuclease activity – proofreading • DNA pol I has 5′-to-3′ exonuclase activity MOLECULAR MODEL OF DNA REPLICATION MOLECULAR MODEL OF DNA REPLICATION MOLECULAR MODEL OF DNA REPLICATION 19 TELOMERES • Specialized structures found on the ends of eukaryotic chromosomes • Protect ends of chromosomes from nucleases and maintain the integrity of linear chromosomes • Gradual shortening of chromosomes with each round of cell division • Unable to replicate last section of lagging strand TELOMERES • The RNA primer in DNA replication causes a problem for the replicating linear chromosomes of eukaryotes. • When the primer is removed, it leaves a gap at the 5′ end of the new DNA strand that DNA polymerase can’t fill, – causing the chromosome to shorten with each replication. • The ends of most eukaryotic chromosomes are protected by a buffer of noncoding DNA (the telomere), consisting of short, repeating sequences (the telomere repeat). 21 TELOMERASE • With each replication, some telomere repeats are lost, but the genes are unaffected. • Buffering fails when the entire telomere is lost. • Telomerase stops the shortening of telomeres by adding telomere repeats to chromosome ends. • An RNA section binds to DNA and is the template for addition of telomere repeats. • Active only in rapidly dividing embryonic cells, in germ cells, and in cancerous somatic cells. TELOMERE REPEAT 24 • Telomeres composed of short repeated sequences of DNA • Telomerase – enzyme makes telomere of lagging strand using and internal RNA template (not the DNA itself) • Leading strand can be replicated to the end • Telomerase developmentally regulated • Relationship between senescence and telomere length • Cancer cells generally show activation of telomerase REVIEW 1. What is the importance of complementary base pairing to DNA replication? 2. Why is a primer needed for DNA replication? How is the primer made? 3. DNA polymerase III and DNA polymerase I are used in DNA replication in E. coli. What are their roles? 4. Why are telomeres important? 26 DNA REPAIR • Mutagens – any agent that increases the number of mutations above background level • Radiation and chemicals • Importance of DNA repair is indicated by the multiplicity of repair systems that have been discovered REPAIR OF ERRORS IN DNA • Errors made during replication (base-pair mismatches) are corrected in a proofreading mechanism by DNA polymerases • After replication is complete, remaining base-pair mismatches are corrected by a DNA repair mechanism PROOFREADING MECHANISM • The proofreading mechanism allows DNA polymerases to back up and remove mispaired nucleotides • If a newly added nucleotide is mismatched, DNA polymerase reverses, using 3′→5′ exonuclease activity to remove the newly added incorrect nucleotide • DNA polymerase then resumes forward synthesis and inserts the correct nucleotide Template strand PROOFREADING DNA polymerase Polymerization activity of DNA polymerase adds DNA nucleotides to the new chain in the 5' 3' direction using complementary base pairing rules. New strand Rarely, DNA polymerase adds a mispaired nucleotide. New strand DNA polymerase recognizes the mismatched base pair.The enzyme reverses, using its 3' to 5' exonuclease to remove the mispaired nucleotide from the strand. DNA polymerase resumes its polymerization activity in the forward direction, extending the new chain in the 5' ⟶ 3' direction. DNA REPAIR MECHANISMS • DNA repair mechanisms correct base-pair mismatches that remain after proofreading (mismatch repair) • Distortions in DNA structure caused by mispaired bases provide recognition sites for mismatch repair enzymes • Repair enzymes cut the new DNA strand on each side of the mismatch, and remove a portion of the chain • DNA polymerase fills the gap with new DNA, and DNA ligase seals the nucleotide chain MISMATCH REPAIR Base-pair mismatch Template strand Mismatch repair proteins scan DNA for a mispaired base that was missed in proofreading and cleave the backbone of the new strand on each side of the mismatch. New strand The repair proteins remove several to many bases, including the mismatched base, leaving a gap in the DNA. A repair DNA polymerase fills in the gap with its 5’ to 3' polymerizing activity, using the template strand as a guide. Nick left after filled in gap DNA ligase seals the nick left after gap filling to complete the repair. EXCISION REPAIR • DNA damage occurs constantly in cells. • Base-excision repair mechanisms repair nonbulky damage by removing the erroneous base and replacing it with the correct one based on complementary pairing rules. • Nucleotide-excision repair repairs bulky distortions in DNA (such as in thymine dimers) by removing an entire segment of DNA. THYMINE DIMER MUTATION AND EVOLUTIONARY PROCESSES • Errors that remain in DNA after proofreading and DNA repair are a primary source of mutations (changes in DNA sequence that are passed on in replicated copies). • Mutation in a gene can alter the protein encoded by the gene, which may alter how the organism functions – mutations are the ultimate source of variability acted on by natural selection. DRAW A GRADIENT AT THE END OF SIX ROUNDS OF REPLICATION IN SEMI-CONSERVATIVE MODE • Bacteria start out in N14 and then get switched to N 15. Draw the gradient at the end of six rounds of replication in N15.