1.1.
BIOTECHBOLOGY
ENGINEERING
&
BIOPROCESS
✓ We have come a long way with genetic
manipulation. For thousands of years,
genetic engineering has been practiced at
the level of breeding and selection (trial
and error procedure to generate best
offspring)
✓ Now, genetic engineering is done in a
purposeful, predetermined manner with
the advent of techniques to manipulate
DNA at the molecular level.
➢ We can determine which genes are
problematic, and we can attempt to
correct those defective genes.
➢ Identify stem cells for progenitors
of all differentiated tissue; this
allowed us to make artificial organs
in bioreactors starting with these
stem cells.
➢ Use of biological molecules to make
up biological sensors the tests for
diabetes tests for prostate cancer
that's for liver damage.
▪ microchips have these
biological
molecules
implanted on them; they
make up of what is called as
biosensor
✓ Biological systems are very complex and
beautifully constructed, but they obey the
rules of chemistry and physics and are
thus susceptible to engineering analysis.
✓ Living cells are, to a degree, predictable
processes to use them can be rationally
designed and constructed on commercial
(large) scales.
✓ This is the job of the bioprocess engineer.
Biotechnology
✓ In the figure, the plant is modified by
inserting genetic material using a vector.
✓ With this revolution, new visions and
hopes have emerged:
➢ Large-scale culture of bacteria that
can degrade plastics.
➢ Genetically modified plants that are
safe to eat and resistant to
pestilence or even changes in
weather
conditions
(natural
pesticides)
➢ Develop new means of treating
people with rare genetic diseases
(Called gene therapy)
✓ Biotechnology implies the use or
development of methods or direct genetic
manipulation for a socially desirable goal.
➢ The “desirable” goals:
▪ Production of a particular
chemical
▪ Production of better plants
or seeds
▪ Gene therapy
▪ Use of specially designed
organisms
to
degrade
wastes
✓ Biotechnology is correlated to the use of
sophisticated techniques outside the cell
for genetic manipulation.
✓ the use of living systems and organisms to
develop or make product
✓ Biotechnology is also interpreted in a
broader sense and is equated with applied
biology.
➢ This might include engineering as a
subcomponent of biotechnology.
Bioengineering
✓ Bioengineering is a broad title and
includes
agricultural,
electrical,
mechanical, industrial, environmental,
chemical engineers, etc.
Bioengineers or Biological Engineering
✓ Emphasizes on the
animals and plants.
applications
to
Biochemical Engineering
✓ It is usually meant the extension of
chemical engineering principles to
systems using a biological catalyst to bring
about desired chemical transformations.
✓ It is focused on fermentation engineering,
application of engineering principles to
microscopic biological systems that are
used to create new products by synthesis,
including the production of protein from
suitable raw materials.
✓ It is usually divided into two:
➢ Bioreaction engineering
➢ Bioseparations
Biomedical Engineering
✓ Considered to be completely separate
from biochemical engineering.
✓ Mostly focused on biomedical devices or
implants
✓ The boundary between biomedical and
biochemical engineering is increasingly
vague specifically in:
➢ The areas of cell surface receptors
➢ Animal cell culture
Biomolecular Engineering
✓ Defined by the National Institutes of
Health as “…research at the interface of
biology and chemical engineering and is
focused at the molecular level.”
✓ Application of engineering principles and
deign concepts to medicine and biology
✓ Under its umbrella:
➢ Tissue engineering
➢ Genetic engineering
➢ Neural engineering
➢ Pharmaceutical engineering
➢ Clinical engineering
➢ Bioinformatics
➢ Biomechanics
Biological Systems Engineering
✓ develops technology to monitor the
conditions of where the process of making
pharmaceuticals takes place
✓ Ex: bioprocess design, biocatalysis,
bioseparation, bioinformatics, bioenergy
Environmental Health Engineering
✓ application of engineering principles to
the control of the environment for the
health, comfort, and safety of human
beings. It includes the field of life-support
systems for the exploration of outer space
and the ocean
Biomimetics
✓ the imitation of models, systems, and
elements of nature for the purpose of
solving complex human problems.
✓ Ex: velcro, designed after George de
Mestral noticed how easily burs stuck to a
dog's hair
Bionics
✓ an integration of Biomedical, focused more
on the robotics and assisted technologies.
✓ Ex: prosthetics
Bioprinting
✓ utilizing biomaterials to print organs and
new tissues
Systems Biology
✓ The study of biological systems.
Other Related Fields:
✓ Bioelectrical Engineering
✓ Biomechanical Engineering
✓ Biorobotics
1.2.
BIOLOGISTS & ENGINEERS DIFFER IN
THEIR APPROACH TO RESEARCH
✓ Trainings of biologists and engineers are
distinctly different.
Human Factors and Ergonomics Engineering
✓ application of engineering, physiology,
and psychology to the optimization of the
human–machine relationship.
✓ Ex: physical ergonomics, cognitive
ergonomics, human–computer interaction
Difference between Bioprocess Engineering
and Biochemical Engineering
✓ In addition to chemical engineering,
bioprocess engineering, include the work
of:
➢ Mechanical;
➢ Electrical;
➢ And industrial engineers
to apply the principles of their disciplines
to processes based on using living cells or
subcomponents of such cells.
✓ Problems of detailed equipment design,
sensor development, control algorithms,
and manufacturing strategies can be
solved through the mentioned disciplines.
✓ Meanwhile, biochemical engineering is
➢ More limited in the sense that it
primarily draws from chemical
engineering principles
➢ Yet more broader in the sense that
it is not restricted to well-defined
artificially constructed processes;
can be applied to natural systems
✓
✓
✓
✓
On biology:
In life sciences, mathematical theories and
quantitative methods (except statistics)
play secondary roles
Most progress in biology are due to
experimental tools
➢ Results are qualitative and
descriptive models are formulated
and tested
Biologists
often
have
incomplete
backgrounds in mathematics but are very
strong in terms of laboratory tools and on
the interpretation of laboratory data from
complex systems.
Biologists are better at the formation of
testable hypotheses, experimental design,
and data interpretation from complex
systems.
On Engineering:
✓ Engineers usually have a background in
physical and mathematical sciences.
✓ Often, theory leads to mathematical
formulations.
➢ The validity of the theory is tested
by comparing predicted response
to those in experiments.
➢ Quantitative
models
and
approaches, even to complex
systems, are strengths.
✓ Engineers are typically unfamiliar with
experimental techniques and strategies
used by life scientists.
Complementary
✓ The skills of engineers and life scientists
are complementary.
✓ To convert the promises of molecular
biology into new processes to make new
products require the integration of these
skills.
✓ Typical route for the development of a
bioprocess:
✓ STEPS 1 TO 12: Handled by life scientist
✓ A broad range of disciplines is involved in
bioprocessing.
✓ STEPS 12 TO 13: Handled by bioprocess
engineer; the one responsible for the
design of a bioreactor; determine if batch
or semi-batch; on step 12, one has to know
the maximum possible anticipated agitate
or speeed
✓ STEPS 14 ONWARD: No life scientist
involved, only bioprocess engineer
✓ Scientists working in this area are
constantly confronted with biological,
chemical, physical, engineering, and
sometimes medical questions.
✓ In bioprocessing, we have involvement of
a broad range of disciplines.
➢
➢
➢
➢
➢
➢
➢
➢
1.3.
PENCILLIN HISTORY
✓ In September 1928, Alexander Fleming is at
St. Mary’s Hospital in London
➢ He is trying to isolate the
bacterium, Staphylococcus aureus,
which causes boils
➢ The technique in use was to grow
the bacterium on the surface of a
nutrient solution.
➢ One
of
the
dishes
was
contaminated inadvertently with a
foreign particle; he noticed that no
bacteria grew near the invading
substance.
He recognized that the cell killing
must be due to antibacterial agent.
He recovered the foreign particle
and found that it was a common
mold of the Penicillium genus (later
identified as Penicillium notatum)
Fleming nurtured the mod to grow
though crude extraction methods.
He obtained a tiny quantity of
secreted material.
▪ He discovered that this
material had powerful
antimicrobial
properties
and named it penicillin.
He preserved the culture, but its
discover lay dormant for over a
decade.
In World War II, the discovery of
penicillin was resurrected due to
the need of an antibiotic with
minima side effects and broader
compatibility that sulfa drugs.
Howard Florey and Ernst Chain of
Oxford
build
on
Fleming’s
observations.
Norman
Heatley
produced
sufficient material for Chain and
Florey to test penicillin’s effectivity.
▪ Though Heatley is a
biochemist, he performed as
a bioprocess engineer.
▪ He developed an assay to
monitor
the
penicillin
amount made as so to
determine the kinetics of
fermentation.
▪ He developed a culture
technique that could be
easily implemented.
▪ He devised a novel backextraction
process
to
recover
very
delicate
product.
After
months,
they
produced enough penicillin
to treat laboratory animals.
18 months after the project, they
treated a London bobby for a blood
infection; he recovered.
Penicillin supply was exhausted,
and the man relapsed and died.
Many companies, assisted by
universities aided the development
penicillin including Merck, Pfizer,
Squibb, and the USDA Northern
Regional Research Laboratory in
Peoria Illinois.
Early clinical successes were so
dramatic that in 1943, the War
Production Board appointed A.L
Elder to coordinate the activities of
producers to increase penicillin
supply.
▪ The fermentation route was
chosen.
Typical problems in most new
fermentation processes:
▪ A valuable product made at
very low levels.
▪ Low rate of production per
unit volume necessities very
large
and
inefficient
reactors.
▪ Also, the low concentration
(titer)
made
product
recovery and purification
difficult.
In 1939, final concentration of
penicillin fermentation broth was
one part per million (ca. 0.001 g/l);
gold is more plentiful in sea water.
Penicillin is fragile and unstable.
▪ This means that there are
significant constrains on the
approaches
used
for
recovery and purification.
Life scientists at the Northern
Regional Research Laboratory
made major contribution to the
▪
➢
➢
➢
➢
➢
➢
➢
➢
penicillin program like the
following:
▪ The development of a corn
steep liquor-lactose based
medium; this increased the
productivity about tenfold.
▪ Its worldwide search for
better producer strains of
Penicillium led to the
isolation of a Penicilium
chrysogenum strain.
• This strain is isolated
from
a
moldy
cantaloupe at
a
Peoria fruit market.
• It is proven to have
superior to hundreds
of other isolates
tested.
• Its progeny have
been used in almost
all
commercial
fermentations.
➢ Another problem in penicillin
production is on deciding on the
manufacturing process:
▪ One method involved the
growth of the mold on the
surface of moist bran.
• This is discarded
because
of
difficulties
in
temperature control,
sterilization,
and
equipment size.
▪ The
surface
method
involved growth of the mold
on top of a quiescent
medium.
• This used a variety of
containers, including
milk bottles.
• “bottle plant” is used
to
indicate
this
manufacturing
technique.
•
▪
This method gave
relatively high yields
but had a long
growing cycle and
was labor intensive.
• The
first
manufacturing
plants were bottle
plants because the
method worked and
can be implemented
quickly.
• Surface
method
would not meet the
full
need
for
penicillin.
The
submerged
tank
process was favored by
engineers.
• It
presented
challenges in terms
of
both
mold
physiology and in
tank design and
operation.
• Large volume of
absolutely clean oil
and dirt-free sterile
air were required.
• Large agitators are
required to prevent
organism entry.
✓ Problems in recovery and purification also
exist.
➢ A combination of pH shits and rapid
liquid-liquid extraction proved
useful.
✓ Soon, tanks of about 10,000 gal were built.
✓ Pfizer completed in less than six months
the first penicillin plant for commercial
production.
➢ It has 14 tanks (with 7000-gal
capacity)
✓ In World War II, there is enough penicillin
for almost 100,000 per year.
✓ From 1939 to now, the yield of penicillin
gone from 0.001 g/l to over 50 g/l of
fermentation broth.
✓ Schematic of penicillin production
process:
1.3 .
BIOPROCESSES:
CONSTRAINTS
REGULATORY
✓ The U.S Food and Drug Administration
(FDA) and its equivalents in other
countries must ensure the safety and
efficacy of medicines.
✓ There have been tragic examples where a
small process change has allowed a toxic
trace compound to form or become
incorporated in the final product, resulting
in severe side effects, including death.
✓ Thus, even slight process changes may
require new clinical trials to test the safety
of the resulting product. Since clinical
trials are very lengthy and expensive,
process improvements are made under a
limited set of circumstances.
✓ Bioprocess
engineers
in
the
pharmaceutical
or
biotechnology
industry’s primary concern is not the
reduction of manufacturing cost, but the
production of consistently high-quality
amounts to satisfy the medical needs of the
population.
✓ A typical drug undergoes 6.5 years of
development from the discovery stage
through preclinical testing in animals.
✓ Human clinical trials are conducted in
three phases:
➢ Phase 1 clinical trials (about 1 year)
are used to test safety; 30 to 80
volunteers involved.
➢ Phase 2 clinical trials (about 2
years) use 100 to 300 patients and
the emphasis is on efficacy (if it
helps the patient) and the
determination of side effects (or if
it exist)
➢ Phase 3 clinical trials (about 3
years) are those with promising
results with about 1000 to 3000
patients.
✓ Clinical trials are presented in the FDA for
review (about 18 months).
✓ The drug is then approved if it is well
design, has statistically significant
improvements, and has acceptable side
effects.
✓ Continued monitoring will be held for
possible adverse effects.
✓ The whole drug discovery-throughapproval process takes an average of 15
years and costs about 400 million
dollars (in 1996).
✓ Only 1 in 10 drugs that enter human
clinical trials receives approval.
✓ Recent FDA reforms decreased the time to
obtain drug approval for life-saving drugs
in treatment of diseases like cancer and
AIDS.
✓ Drugs sold on the marker or used in
clinical trials must come from facilities
that are certified as GMP (Good
Manufacturing Practice)
➢ It
concerns
the
actual
manufacturing facility, design,
layout, equipment and procedures,
training of production personal,
control of process inputs (raw
materials and cultures), and
product handling
➢ The design layout must prevent
contamination of the product and
dictates the flow of material,
personnel, and air.
➢ Equipment and procedures must
be validated.
▪ Procedures also include the
cleaning and sterilization
and not only the operation.
➢ Computer software used to
monitor and control the processes
must be validated.
➢ Offline assays done in the
laboratory must satisfy GLB (Good
Laboratory Practices)
➢ Procedures should be documented
by SOPs (Standard Operating
Procedures)
➢ GMP Guidelines stress the need
for documented procedures to
validate performance.
➢ “Process
Validation
is
establishing documented evidence
which provides a high degree of
assurance that a specific process
will consistently produce a product
meeting
its
predetermined
specifications
and
quality
characteristics” and that “There
shall be written procedures for
production and process-control to
assure that products have identity,
strength, quality, and purity they
purport or are presented to
possess.”
➢ Key concepts in process validation:
▪ Written documentation
▪ Consistency of procedures
▪ Consistency of products
▪ Demonstrable measures of
product
quality
(particularly purity and
safety)
✓ The key point is that process changes
cannot be made without considering their
considerable regulatory impact.
✓ Research is part of the timeline champion
amalgam of discovery only on drug or
quantum product, then there has to be
research and this the length of time.
1.1.
CELL DIVERSITY
✓ Our focus in microbial diversity or small
cells.
✓ Living things are not confined to the
familiar temperate realm of land, water,
and sunlight inhabited by plants and planteating animals.
✓ They can be found in the darkest depths of
the ocean, in hot volcanic mud, in pools
beneath the frozen surface of the
Antarctic, and buried kilometers deep in
the Earth’s crust.
➢ The aforementioned are some of
the most unique microorganisms.
➢ They are inaccessible.
➢ Very microscopic in size.
✓ Some cells grow at -20oC (in brine), while
other can grow at 120oC (under high
pressures).
➢ This can be achieved if there is high
concentration of salts; this allows
freezing point depression
➢ With high pressures, then you can
still have liquid water at this high
temperature
➢ Some cells can grow at very
extreme condition which otherwise
would be very deadly to plants and
animals.
✓ In terms of optimal temperature for
growth, cells can be classified as:
➢ Psychrophiles: cells that grow
best at low temperatures (below
20oC)
➢ Mesophiles: cells that grow best in
the range of 20oC to 50oC
▪ Plant cells and animal cells
are considered mesophiles
➢ Thermophiles: cells that grow
best at temperatures greater than
50oC
▪ for thermophiles, they have
enzymes that are mostly
heat
resistant,
heat
sensitive; hence, they are
thoroughly stable
▪ Some organisms like this are
used in biotechnology: In
PCR, we have this special
enzyme polymerase that
can still function at high
temperatures and that
enzyme has been isolated
from a specific thermal
fighter.
✓ Many organisms also have pH optima far
from neutrality, with some growing best
in highly acidic conditions, and some in
highly basic conditions.
➢ Although most organisms grow
only where water activity is high,
some organisms can grow on
barely moist solid surfaces or in
highly saline solutions.
✓ Cells also differ in oxygen requirements:
➢ Aerobic: require oxygen for
growth and metabolism
▪ like human beings
➢ Anaerobic: growth inhibited by
the presence of oxygen
▪ tetanus
➢ Facultative: growth is possible
either aerobically or anaerobically
▪ it can shift to metabolism
that makes use of oxygen if
there is no oxygen; then it
shifts to another kind of
metabolism, that does not
require oxygen
✓ Energy & Carbon Source:
✓ Coccis (pl. cocci): spherical cells
✓ Bacillus (pl. bacilli): rod-shaped cells
✓ The smallest cells: hard to identify the
shape; has a separate category
✓ Spirillum (pl. spirilla): spiral cells
✓ The shape is important because one has to
know that morphology, its requirements
(anaerobic or aerobic), pH for growth,
temperature, etc.
Primary Cell Types
✓ Despite the diversity of cells, all cells are
classified as one of two primary cell types:
➢ eukaryotic and prokaryotic.
➢ Living organisms obtain their free energy
and carbon in different ways.
➢ On Energy Sources:
Phototrophs: when energy
is from sunlight; they feed
on light as energy source
▪ Chemotrophs:
when
energy is derived from the
consumption of chemical
compounds.
✓ Not only do microorganisms occupy a
wide range of habitats and have different
growth requirements, but they also come
in a wide range of sizes and shapes.
✓ Some cells may even change shape in
response to changes in their local
environment.
▪
✓ Main differences between eukaryotic
and prokaryotic cells:
1. Compartmentalization of DNA (DNA in
organelles):
In prokaryotes, DNA is exposed and
floating in the cytoplasm; called “naked
DNA”
2. Number of DNA molecules:
Prokaryotes usually have only one circular
DNA while eukaryotes have many.
3. Membrane bound organelles:
There are many membrane bound
organelles present in eukaryotes and not
in prokaryotes such as the mitochondria,
ER, golgi apparatus, etc.
Prokaryotes
✓ Above is the bacterium Vibrio cholerae
and its simple organization:
➢ Flagellum: a helical appendage at
one end; rotates as a propeller to
drive the cell forward; it can infect
human small intestine to cause
cholera
➢ DNA is floating inside the
cytoplasm and there are a lot of
ribosomes scattered around.
➢ Plasma membrane: surrounding
your microorganism.
✓ Bacteria Gram Strains:
✓ Bacteria gram strains differentiate
bacteria in two classes: gram positive
cells and gram negative cells.
✓ Procedure:
➢ We use crystal violet and initially,
all material will be stained with
crystal violet, and we add iodine as
a fixating agent, then we wash it
with alcohol. Upon washing it with
alcohol, some cells will retain that
crystal violet stain; some will be
clear again.
➢ We will then add the second dye.
✓ Gram positive microorganisms: have a
thick peptidoglycan layer; peptidoglycan
means you have macromolecules that are
combinations of carbohydrates and
proteins.
➢ Underneath this, there is just one
plasma membrane.
➢ E.coli is one of the most famous
examples of this type of cell.
✓ Gram
negative
microorganisms:
peptidoglycan layer is thinner, although
there is an additional outer membrane.
➢ there is an additional outer
membrane, hence, sandwiching the
peptidoglycan layer.
✓ Genome analyses have made it clear that
prokaryotes comprise two distinct groups:
bacteria (or eubacteria) and the
archaea (or archaebacteria).
✓ Tree of Life:
✓ The green areas are called the “missing
links”
✓ Eukaryotes came from archaea, and not
bacteria
✓ Three main domains of life:
➢ Bacteria (prokaryotes)
➢ Acraea (prokaryotes)
➢ Eukaryotes
(where
humans
evolved from)
✓ Archaea
➢ Archaea are often found inhabiting
environments that humans avoid,
such as bogs, sewage treatment
plants, ocean abysses, salt brines,
and hot acid springs—most of
theme are extremophiles (those
who
inhabit
extreme
environments)
Eukaryotes
✓ Fungi, algae, protozoa, and animal and
plant cells constitute the eukaryotes.
✓ Eukaryotes are 5-10 times larger than
prokaryotes in diameter and have a true
nucleus, in addition to a number of cellular
organelles inside the cytoplasm.
✓ In cell wall and cell membrane structure,
eukaryotes are similar to prokaryotes.
✓ Plants have cell walls, but animal cells
don’t—for this treason, animal cells are
very shear-sensitive and fragile. This
factor significantly complicates the design
of large-scale bioreactors for animal cells.
✓ Mixing does not kill the cells: plant cells
can withstand mixing, but it is a limited
case for animal cells.
✓ This is an animal cell because it does not
have a cell wall.
✓ Part of the cytoskeleton (gives
mechanical support, strength and
structure to the cell):
➢ Microtubule
➢ Intermediate filaments
➢ Actin filaments
✓ Main parts:
➢ Nucleus
➢ Nuclear membrane or nuclear
envelope: it has force because
information has to come out of this
set of this component
➢ Nucleolus: this is the site for
ribosome synthesis
➢ Ribosomes: scattered all around
the cytoplasm (the black dots), but
they can be attached to the ER.
▪ The ribosomes are the
“protein factories”
➢ Rough endoplasmic reticulum: ER
with attached ribosome; involved
in
protein
synthesis
and
medication (attachment of sugars
to
proteins
to
produce
glycoproteins; also include the
attachment of lipids to proteins to
produce lipoproteins).
➢ Smooth endoplasmic reticulum:
does not have attached ribosomes;
involved in lipid synthesis
➢ Centrosomes: involved in the
reproduction
of
the
cell;
attachment of microtubules; also
involved in locating the center of
the cell (it does that by extending
the microtubules until it reaches
the periphery of the cells;
important in cell division)
➢ Golgi apparatus or bodies: involved
in packaging and release of
materials that has to be secreted by
the cell; packaging of protein that
has to be sent outside the cell; also
involved in modification.
➢ Plasma membrane: serves as a
semipermeable barrier.
➢ Mitochondrion: powerhouse of the
cell;
provide
energy
by
synthesizing ATP (energy currency
of the cell)
➢ Lysosome: responsible for the
degradation of whatever needs to
be degraded; degradation sites
✓ The most common eukaryotic cells used in
bioprocesses are:
1. Saccharomyces cerevisiae: used in both
winemaking or simply alcohol production
from as well as in baking for yeast
- One strain produced ethanol
in an anaerobic process
- Other strain produced CO2
(for baking) in aerobic
process
2. Aspergillus niger: used in the
production of citric acid.
3. Penicillium chrysogenum: production of
penicillin
4. Algae: used in wastewater treatment as
well as in simultaneous production of
single cell protein (commonly used as a
feed for livestock or food for humans;
considered as “food of the future”); also
used in producing biofuels and biooils
1.2.
CELL COMPOSITION
Chemical Components
✓ Living organisms are made of only a small
selection of the 92 naturally occurring
elements, four of which—carbon (C),
nitrogen (N), hydrogen (H), and oxygen
(O)—make up 96.5% of an organism’s
weight.
✓ By weight, macromolecules are the most
abundant carbon-containing molecules in
a living cell.
✓ The four elements in red constitute 99%
of the total number of atoms present in the
human body.
✓ An additional of seven elements,
highlighted in blue, together represent
about 0.9% of the total.
✓ The elements in green are required in
trace amounts by humans; needed in small
amounts; if they exceed in this amount,
they become toxic.
✓ The elements in yellow are those unclear
if they are essential in humans.
✓ The small organic molecules of the cell are
carbon-based compounds that have
molecular weights in the range of 1001000 and can contain up to 30 or so carbon
atoms.
✓ Some of these are used as monomers to
construct
giant
polymeric
biomacromolecules. Others act as energy
sources to be broken down via metabolic
pathways.
✓ Broadly speaking, cells contain four major
families of small organic molecules:
sugars, fatty acids, nucleotides, and
amino acids. These four families of small
organic molecules, together with their
polymers, account for a large fraction of
the cell mass.
✓ We have 3 volumetric macromolecules:
o Only lipids are not polymeric while
polysaccharide,
protein,
and
nucleic acids are.
Proteins
✓ Proteins are the most abundant organic
molecules in living cells. They are built
from amino acid monomers, and have
diverse biological functions:
➢ structural
(glycoproteins,
collagen, keratin, microtubules,
etc.)
➢ catalytic (enzymes)
➢ transport (hemoglobin, serum
albumin, membrane transport
proteins, etc.)
➢ regulatory (hormones)
➢ protective (antibodies, thrombin,
etc.)
✓ We have 20 amino acids (make up proteins
in living cells) and some of these have
hydrophobic residues, charged residues,
and can either be acidic or basic, some
have aromatic groups.
✓ The general structure (R is the residue):
✓ Amino acids require nitrogen.
✓ Some of the residues also contain sulfur.
✓ The sequence of proteins when attached
together make up your primary structure;
that primary structure dictates how the
protein will look like in 3D space, so that
will give rise to your secondary structure.
✓ Combinations of different secondary
structures will make up the final tertiary
structure.
✓ When different units combine to form a
much larger structure, it is called the
quaternary structure.
✓ In terms of structure, cellulose is a
carbohydrate and cellulose is responsible
for the rigidity of plant cells.
✓ Storage in plants – starch
✓ Storage in animals or humans – glycogen
✓ Most common carbohydrates:
✓ Cellulose has an upward pointing
connection
✓ Starch and glycogen have downward
pointed connections
✓ We cannot degrade cellulose, but we
can degrade starch; this is due to their
connection differences
✓ Ex. Hemoglobin, a transport protein found
in blood, is responsible for transporting
oxygen and carbon dioxide; it is consisting
of 4 subunits. Hence, it is a quaternary
structure, and those subunits are tertiary
structures.
Carbohydrates
✓ Carbohydrates are the most abundant
organic molecules in nature. These
molecules play key roles as structural and
storage compounds in cells, as well as in
some aspects of chemical signaling in
animals and plants.
✓ Carbohydrates are represented by the
general “hydrated carbon” formula,
(𝐶𝐻2 𝑂)𝑛 , 𝑛 ≥ 3.
✓ Another function of carbohydrates is that
when they are clasped with a protein, they
can shield that protein from recognition by
the immune system.
Lipids
✓ Lipids are hydrophobic biological
compounds that are insoluble in water, but
soluble in nonpolar solvents like benzene,
chloroform, and ether. They are usually
present in plasma membranes.
✓ Cells can alter the mix of lipids in their
membranes to compensate, at least
partially, for changes in temperature or to
increase their tolerance to the presence of
chemical agents such as ethanol.
✓ Lipids are actually in general class of
molecules that can be extracted by
organics solvents; anything that can be
extracted from organic solvents are lipids.
✓ Cholesterol is a good structural support.
✓ Other molecules also serve as the OH
source for esterification; in this case, fatty
acids react with a phosphate group to form
a phospholipid.
✓ Lipids make up out cell membrane.
➢ Not all cell membranes are made up
of phospholipids, but others are
made up of special lipids like
sphingolipids.
➢ Lipids are important for protection
and cell membrane structure.
➢ Cell membrane is made up of lipids
and some protein (it can span
either the whole membrane of be
present in just one layer or only in
the surface); carbohydrates can
also be found on these proteins
➢ The small orange circles in the
figure are cholesterol; they assist in
making the cell membrane more
rigid;
prevent
freezing
of
membrane at low temperatures
✓ If your product is intracellular, it means
inside the cell, you must be able to destruct
the lipid membrane to be able to get the
product from the interior of the cell.
➢ To do that, we must know what the
membrane is composed of.
Nucleic Acids
✓ Nucleic acids play the central role in the
reproduction
of
living
cells.
Deoxyribonucleic acid (DNA) stores and
preserves the genetic information, while
ribonucleic acid (RNA) converts
segments of DNA into short “manuals” for
use by ribosomes in protein synthesis,
among other functions like regulation and
structure.
✓ There are also other classes aside from
DNA or RNA like SNRNA, MI RNA, etc.
✓ 4 nucleotides make up the nucleic acids
o These nucleotides have
nitrogenous base
o Contains sugar; this sugar is the
deoxyribose for ribose in RNA
o It also has at least one phosphate
groups
o To give rise nucleic acids,
phosphate and sugar groups must
have a reaction
✓ 4 base base pairs: A, T, G, C
➢ However, in RNA, thymine is
replaced with uracil
1.3. CELL NUTRIENTS
✓ Most of the products formed by organisms
are produced as a result of their response
to environmental conditions, such as
nutrients, growth hormones, and ions.
✓ The
qualitative
and
quantitative
nutritional requirements of cells need to
be determined experimentally to optimize
growth and product formation.
✓ Cell nutrients can be classified into two
categories:
CELL COMPOSITION
✓ A cell’s composition differs greatly from its
environment, and it must selectively
remove desirable compounds from its
extracellular environment and retain
other compounds within itself (not spill).
✓ Since the cell differs greatly in composition
from
its
environment,
active
concentration differences are present, and
the cell must expend energy to maintain
itself
away
from
thermodynamic
equilibrium, or practically, death.
✓ Allowing equilibrium in living systems
means death.
✓ The intracellular composition of cells
varies depending on the type and age of
the cells, and the composition of nutrient
media in which the cells are growing.
✓ Different
compositions
between
organisms:
➢ Macronutrients:
needed
in
concentrations larger than 10-4M;
carbon,
oxygen,
nitrogen,
hydrogen, sulfur, phosphorus,
Mg2+, and K+ are major
macronutrients
➢ Micronutrients:
needed
in
concentrations less than 10-4M;
trace elements such as Mo2+, Zn2+,
Cu2+, Mn2+, Ca2+, Na+, vitamins,
growth hormones, and metabolic
precursors are micronutrients
Macronutrients
✓ Carbon compounds are major sources of
cellular carbon and energy. The most
common carbon sources in industrial
fermentations are molasses (sucrose),
starch (glucose, dextrin), corn syrup, and
waste sulfite liquor (glucose).
➢ In lab fermentations, glucose, sucrose,
and fructose are the most common
carbon sources. Methanol, ethanol, and
methane also constitute cheap carbon
sources for some fermentations.
➢ In aerobic fermentations, about 50% of
substrate carbon is converted into cell
mass, and 50% is used as energy
source. In anaerobic fermentations,
more carbon is converted to products,
and a smaller fraction is converted to
cell mass (<30%).
➢ In anaerobic fermentations, there is a
higher yield in product compared to
aerobic fermentations.
✓ Nitrogen constitutes about 10-14% of cell
dry weight. The most widely used sources
are ammonia or the ammonium salts
(chloride, sulfate, or nitrate), proteins,
peptides, and amino acids.
➢ Urea may also be used as a nitrogen
source by some organisms. Organic
nitrogen sources such as yeast extract
and peptone are expensive compared
to ammonium salts but are
nevertheless
common
in
lab
fermentations.
Micronutrients
✓ Trace elements are essential to microbial
nutrition—lack of essential trace elements
increases the time lag from inoculation to
growth in culture and may decrease the
growth rate.
✓ Micronutrients are grouped into three:
1. those that are most widely needed
(Fe, Zn, Mn)
2. those that are needed under
specific growth conditions (Cu, Co,
Mo, Ca, Na, Cl, Ni, Se)
3. Those that are rarely required (B,
Al, Si, Cr, V, Sn, Be, F, Ti, Ga, Ge, Br,
Zr, W, Li, I)
✓ In addition, chelating agents such as
EDTA, citric acid, polyphosphates,
histidine, tyrosine, and cysteine, are
needed to form soluble compounds with
ions that are prone to precipitation in the
culture medium.
➢ We never use pure sources because it
is expensive.
Other macronutrients and their function:
✓ Growth factors may also be added to
stimulate the growth and synthesis of
some metabolites. Vitamins, hormones,
and amino acids are major growth factors.
Growth Media
✓ Two major types of growth media are
defined and complex media:
➢ Defined media are those whose
components are present at specific
amounts. Although more expensive,
defined media have the advantage of
reproducibility and better control over
fermentation. Product recovery and
purification are often easier and
cheaper.
➢ Complex media are those containing
natural compounds whose chemical
compositions are not exactly known,
such as yeast extracts, peptone,
molasses, and corn steep liquor.
Complex media are usually able to
provide the necessary growth factors,
vitamins, hormones, and trace
elements, resulting in higher cell
yields, compared to defined media.
Enzymes
✓ Enzymes are biological catalysts (of
nature) that mediate the transformation of
molecules or energy from one form into
another.
✓ The catalytic activity of enzymes
accelerates typical reactions to usually 103
to 1017 times faster than the uncatalyzed
rate.
✓ Although nearly all enzymes are proteins,
some enzymes are also RNA molecules—
called ribozymes.
✓ By utilizing the full repertoire of
intermolecular forces (e.g. hydrophobic
reactions, hydrostatic reactions, van der
Waals
reactions),
enzymes
bring
substrates together in an optimal
orientation, the requirement for a reaction
to occur.
✓ In the illustration, the coils are alpha
helices; sheets are beta sheets;
everything else are random coils.
✓ Growth media can either be solid or liquid
➢ In solid form, usually dissolved in
agarose; agarose is a powder but if you
melt it with hot water, it will start on
polymerizing and form the ager gel.
➢ Medias also have different colors;
yellow for agarose.
➢ Some microorganisms prefer to grow
on surfaces, some prefer to grow in
solutions.
✓ Rate enhancement
enzymes:
by
selected
✓ Like any catalyst, enzymes provide an
alternate pathway for the reaction to
occur. This pathway requires a lower
activation energy.
✓ Because enzymatic pathways have
lowered EA, enhancements in reaction
rates can be enormous.
✓ Activation energy and rate of reaction
are
inversely
related;
this
independence is exponential.
𝐸𝐴
✓ Arrhenius equation (𝑘 = 𝐴𝑒 −𝑅𝑇 ) is
exponential but also an inverse one.
✓ Enzymes lower the activation energy
by facilitating the formation of, and
stabilizing, transition states.
➢ proximity effects: bringing
substances closer together; for
a biomolecular reaction, a
collision of two substrates
should occur
➢ orientation effects: everything
must be oriented for there to
have a reaction
✓ An important property of enzymes is that
they are highly specific: one enzyme
usually catalyzes a single chemical
reaction or a set of closely related
reactions.
✓ For example:
➢ Proteases: only catalyze the
hydrolysis of peptide bonds
between specific amino acids in
proteins; e.g. papain (from
papaya and can hydrolize any
peptide bonds), trypsin (can
cleave peptide bonds but only
those involving amino acids like
arginine
or
lysine),
chymotrypsin
➢ Amylases: work only on
glycosidic bonds
between
glucose molecules in starch
➢ Lipases: only attack fats,
degrading them into fatty acids
and glycerol
✓ In addition, enzymes act on specific
substrates or they can only bind to a
specific
enantiomer—they
are
stereospecific.
✓ We can think of it as if the enzyme having
insertion sites or “active sites” that can
only bind complimentary substrates
➢ E.g. ibuprofen: we have R and S
ibuprofen enantiomers, and the
body can only metabolize one
✓ Enzymes usually work under mild
conditions: pH 4 to 9 and temperatures
between 20oC and 70oC.
✓ The Enzyme Commission number (EC
number) is a numerical classification
scheme for enzymes based on the chemical
reactions they catalyze.
✓ Every EC number is associated with a
recommended name for the respective
enzyme.
✓ The current (6th) edition of the
nomenclature scheme is published by the
International Union of Biochemistry
and Molecular Biology.
✓ Top-level EC Numbers:
Class
Reaction Catalyzed
Typical
Reaction
Enzyme
example(s)
with trivial
name
EC 1
Oxidoreductases
Oxidation/reduction
reactions; transfer
of H and O atoms or
electrons from one
substance
to
another
Transfer
of
a
functional
group
from one substance
to another. The
group
may
be
methyl-,
acyl-,
amino- or phosphate
group
Formation of two
products from a
substrate
by
hydrolysis
Non-hydrolytic
addition or removal
of groups from
substrates. C-C, C-N,
C-O or C-S bonds
may be cleaved
Intramolecule
rearrangement, i.e.
isomerization
changes within a
single molecule
Join together two
molecules
by
synthesis of new CO, C-S, C-N or C-C
bonds
with
simultaneous
breakdown of ATP
Catalyse
the
movement of ions or
molecules
across
membranes or their
separation within
membranes
AH + B → A
+
BH
(reduced)
A + O → AO
(oxidized)
Dehydrogenase,
oxidase
AB + C → A
+ BC
Transaminase,
kinase
AB + H2O
→ AOH +
BH
Lipase, amylase,
peptidase,
phosphatase
RCOCOOH
→ RCOH +
CO2 or [XA+B-Y] →
[A=B + XY]
ABC → BCA
Decarboxylase
X + Y + ATP
→ XY +
ADP + Pi
Synthetase
EC 2
Transferases
✓ Every enzyme code consists of the letters
“EC” followed by four numbers
separated by periods, representing
progressively finer classification of the
enzyme.
✓ For
example,
the
tripeptide
aminopeptidases are EC 3.4.11.4:
➢ EC 3 –hydrolase enzymes
➢ EC 3.4 –hydrolases acting on
peptide bonds
➢ EC 3.4.11 –hydrolases cleaving
off amino-terminal amino acids
from a polypeptide
➢ EC
3.4.11.4
–hydrolases
cleaving off amino-terminal
amino acids from a tripeptide
EC 3
Hydrolases
EC 4
Lyases
EC 5
Isomerases
EC 6
Ligases
EC 7
Translocases
Isomerase,
mutase
Transporter
✓ Expasy (database for enzymes)
✓ The structure above is the human carbonic
anhydrase II and its zinc site.
➢ (Left) Notice that the zinc ion is
bound to the imidazole rings of
three histidine residues as well
as to a water molecule.
➢ (Right) Notice the location of
the zinc site in a cleft near the
center of the enzyme.
✓ Many enzymes require small molecules
called cofactors for their catalytic activity.
This is where the micro or macronutrients
come in. Generally, these cofactors are able
to execute chemical reactions that cannot
be performed by the standard set of
twenty amino acids.
✓ An enzyme bound to its cofactor is called a
holoenzyme; without the cofactor, the
enzyme is known as an apoenzyme.
Apoenzyme + cofactor = holoenzyme
✓ Cofactors can be either metals or small
organic molecules called coenzymes.
These coenzymes are often derived from
vitamins.
✓ Tightly bound coenzymes (like covalently
attached) called prosthetic groups;
loosely associated coenzymes are
sometimes considered “cosubstrates.”
✓ Above is a quaternary structure of
deoxyhemoglobin.
➢ Hemoglobin, which is
composed of two 𝛼 chains and
two 𝛽 chains, functions as a
pair of 𝛼𝛽 dimers,
➢ (A) a ribbon diagram; (B) a
space-filling model
➢ For hemoglobin the cofactors
are both metal ion and organic
molecule.
a similar manner to the fitting of a key
into a lock.
➢ Here, both enzyme and substrate are
rigid. For a long time, this was the
preferred model for E·S complex
formation.
The Enzyme-Substrate Complex
✓ The catalyzed reaction pathway proceeds
through an active intermediate (a
“transition state”), called the enzymesubstrate complex (E·S complex).
✓ The substrate binds with a specific active site
of the enzyme to form the E·S complex.
✓ There are also sits called the allosteric sites.
✓ Much of the catalytic power is attributed to
the binding energy of the substrate to the
enzyme through multiple noncovalent bonds
with specific functional groups on the enzyme.
All noncovalent bonds:
➢ electrostatic
(ionic)
interactions
(represented by the + and – signs)
➢ hydrogen bonds
➢ van der Waals forces
➢ π-πstacking interactions
✓ There are two models for enzymesubstrate interactions:
➢ In the lock-and-key model, proposed
by Emil Fischer in 1890, the E·S
complex is pictured as being formed in
✓ In the induced fit model (actually the
more useful model), both the enzyme and
substrate are distorted.
➢ These changes in conformation distort
one or more of the substrate bonds,
making these bonds weaker and easier
to break.
➢ Thus, the mechanism of enzymatic
catalysis is dynamic, involving
structural changes with multiple
intermediates of both substrates and
the enzyme.
Mechanisms
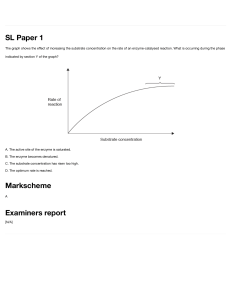
✓ For a constant amount of enzyme, the rate
of catalysis increases with substrate
concentration, but begins to level off and
approach a maximum at higher substrate
concentrations—a
phenomenon
recognized as saturation.
➢ This means that we have an upper limit
to the catalysis that can be done by
your enzymes.
✓ In 1913, Leonor Michaelis and Maud
Menten proposed a simple model to
account for these kinetic characteristics.
✓ The critical feature in their treatment is
that a specific ES complex is a necessary
intermediate in catalysis:
negligible, i.e. at the beginning of the
reaction).
✓ We want an expression (rate law) that
relates the rate of catalysis to the
concentrations of substrate and enzyme,
and the rates of the individual steps.
✓ The rate of product formation is:
𝑑 [𝑃 ]
−𝑟𝑠 = 𝑟𝑝 = 𝑣 = (
) = 𝑘2 [𝐸𝑆]
𝑑𝑡
Where
➢ 𝑟𝑝 = 𝑟𝑎𝑡𝑒 𝑜𝑓 𝑝𝑟𝑜𝑑𝑢𝑐𝑡𝑖𝑜𝑛 𝑓𝑜𝑟𝑚𝑎𝑡𝑖𝑜𝑛
➢ 𝑟𝑠 = 𝑟𝑎𝑡𝑒 𝑜𝑓 𝑠𝑢𝑏𝑠𝑡𝑟𝑎𝑡𝑒 𝑐𝑜𝑛𝑠𝑢𝑚𝑝𝑡𝑖𝑜𝑛
➢ 𝑣 = 𝑣𝑜𝑙𝑙𝑢𝑚𝑒𝑡𝑟𝑖𝑐 𝑟𝑒𝑎𝑐𝑡𝑖𝑜𝑛 𝑟𝑎𝑡𝑒
Notes
➢ ES is a transition state and cannot be
isolated. Hence, it cannot be measured or
quantified.
➢ We convert ES to actually measurable
quantities: 𝑘2 [𝐸𝑆(𝑆, 𝐸𝑜 )] or the initial
substrate concentration and initial
enzyme concentration
THE RATE OF PRODUCTION FORMATION:
➢ E and S formed ES complex in a
rapid equilibrium.
➢ Once the ES complex is formed, it
will follow one or two things:
1. Either the substrate dissociates
to reform E and S.
2. Or the substrate becomes
processed to form the product,
P.
Note: The formation of the product from the ES is
irreversible with rate constant k2.
Michaelis-Menten Kinetics: Derivation
✓ It is assumed that the ES complex is
established rather rapidly, and the rate of
the reverse reaction of the second step is
negligible (when product accumulation is
𝒅[𝑷]
∴ −𝒓𝒔 = 𝒓𝒑 = 𝒗 = (
) = 𝒌𝟐 [𝑬𝑺]
𝒅𝒕
𝒘𝒉𝒆𝒓𝒆 𝒌𝟐 [𝑬𝑺] → 𝒌𝟐 [𝑬𝑺(𝑺, 𝑬𝒐 )]
➢ The goal is to derive ES and let it have
measurable properties.
THE TIME VARIATION OF [ES]:
𝒅[𝑬𝑺]
= 𝒌𝟏 [𝑬][𝑺] − 𝒌−𝟏 [𝑬𝑺] − 𝒌𝟐 [𝑬𝑺]
𝒅𝒕
➢ The time variation allows us to relate ES to
S; E is the amount of enzyme that will not
be bound to any substrate (also
immeasurable).
ENZYME CONSERVATION EQUATION
[𝑬]𝟎 = [𝑬] + [𝑬𝑺]
➢ It means that whatever amount of enzyme
we add to the reaction will be split into
two.
➢ Parts of the enzyme added will be bound to
the substrate while the other will remain
unbonded.
➢ In the equations so far, the only known
parameters are [𝑺] and [𝑬]𝟎; we want to
replace this with ES in the final equation.
➢ Two ways to derive the equation from the
starting equations:
1. Rapid Equilibrium Assumption
2. Pseudo-Steady
State
Hypothesis
(PSSH)
1. RAPID EQUILIBRIUM ASSUMPTION
✓ Michaelis and Menten, along with
V.C.R. Henri, used the assumption that
equilibrium between the enzyme
and substrate is rapid, forming the
ES complex. An equilibrium constant
can therefore be used to express [ES] in
terms of [S].
✓ Because this is rapid, the 𝑘2 can be
neglected.
✓ The equilibrium constant, written in
the form of a dissociation constant
(reciprocal of 𝑲𝑪 ), is
′ (=
𝐾𝑚
[𝐸][𝑆]
1
𝑘−1
)=(
)=
[𝐸𝑆]
𝐾𝐶
𝑘1
Note:
For a reversible reaction 𝐸 + 𝑆 ⇌ 𝐸𝑆,
𝐾𝑐 =
[𝐸𝑆]
𝑘1
=
[𝐸][𝑆] 𝑘−1
Derivation:
′
✓ Write 𝐾𝑚
as the ratio of the rate constants,
𝑘−1 [𝐸][𝑆]
′
𝐾𝑚
=
=
→ (𝒆𝒒. 𝟏)
[𝐸𝑆]
𝑘1
✓ However, [E] cannot be quantified.
Instead, we want [E] in terms of the initial
amount of enzyme. Hence, using Enzyme
Conservation Equation:
[𝐸]0 = [𝐸] + [𝐸𝑆] → (𝒆𝒒. 𝟐)
✓ Rearranging (eq.1) we know that:
′ [
𝐾𝑚
𝐸𝑆]
[𝐸 ] =
→ (𝒆𝒒. 𝟑)
[𝑆]
✓ Plugging (eq.3) to (eq.2)
𝐾 ′ [𝐸𝑆]
[𝐸]0 = 𝑚
+ [𝐸𝑆] → (𝑬𝒒. 𝟒)
[𝑆 ]
✓ Factoring out [ES]
𝐾′
[𝐸]0 = [𝐸𝑆] ( 𝑚 + 1) → (𝒆𝒒. 𝟓)
[𝑆]
✓ Expressing [ES] in terms of [𝐸]0
[𝐸 ] 0
[𝐸𝑆] =
→ (𝒆𝒒. 𝟔)
′
𝐾𝑚
(
+ 1)
[𝑆 ]
✓ Simplifying by multiplying both the
numerator and the denomination with [S]
[𝐸]0
[𝑆 ]
[𝐸𝑆] =
(
)
𝐾′
[ ]
( 𝑚 + 1) 𝑆
[𝑆]
[𝐸]0 [𝑆]
∴ [𝐸𝑆] = ′
→ (𝒆𝒒. 𝟕)
𝐾𝑚 + [𝑆]
✓ Since we now have a measurable quantity
for [ES], we can use the rate of product
formation equation:
𝑑[𝑃]
𝑟𝑠 = 𝑟𝑝 = 𝑣 = (
) = 𝑘2 [𝐸𝑆]
𝑑𝑡
𝑑[𝑃]
𝑑 [𝑆]
𝑣=(
)=−
= 𝑘2 [𝐸𝑆]
𝑑𝑡
𝑑𝑡
✓ Simplifying:
RATE LAW FOR RAPID EQUILIBRIUM
𝒗 = (𝒌𝟐 )
[𝑬]𝟎 [𝑺]
𝑲′𝒎 + [𝑺]
2. PSEUDO-STEADY STATE HYPOTHESIS
(PSSH) or the QUASI-STEADY STATE
ASSUMPTION
✓ In many cases, the rapid equilibrium
assumption is not valid, although the
enzyme-substrate reaction will still show
saturation-type kinetics.
✓ Hence, this assumption is the most
acceptable one.
✓ The PSSH was first proposed by G.E. Briggs
and J.B.S. Haldane. In a closed system,
after a brief transient, and if [𝑺]𝟎 ≫
[𝑬]𝟎 ,
𝒅[𝑬𝑺]
𝒅𝒕
≈ 𝟎. (there will be an instance
wherein the [ES] complex will be constant)
✓ This is as long as the substrate remains at
a high concentration relative to th4
enzyme concentration.
✓ The PSSH assumes (eq. 1) to be 0.
𝑑 [𝐸𝑆]
= 𝑘1 [𝐸][𝑆] − 𝑘−1 [𝐸𝑆] − 𝑘2 [𝐸𝑆] = 0
𝑑𝑡
→ (𝒆𝒒. 𝟐)
✓ Factor-out the [ES]
𝑘1 [𝐸][𝑆] = (𝑘−1 + 𝑘2 )[𝐸𝑆] → (𝒆𝒒. 𝟑)
✓ Isolate [ES]
𝑘1
[𝐸][𝑆] → (𝒆𝒒. 𝟒)
𝑘−1 + 𝑘2
Recall the enzyme conservation equation:
[𝐸]0 = [𝐸] + [𝐸𝑆] → (𝒆𝒒. 𝟓)
Isolate [E]
[𝐸] = [𝐸]0 − [𝐸𝑆] → (𝒆𝒒. 𝟔)
Substitute (eq. 6) to (eq.4)
𝑘1
[𝐸𝑆] =
([𝐸]0 − [𝐸𝑆])[𝑆]
𝑘−1 + 𝑘2
→ (𝒆𝒒. 𝟕)
Factor the introduced [ES]
𝑘 [𝐸] [𝑆] 𝑘1 [𝐸𝑆][𝑆]
[𝐸𝑆] = 1 0
−
→ (𝒆𝒒. 𝟖)
𝑘−1 + 𝑘2
𝑘−1 + 𝑘2
Transfer the second term to the left
𝑘 [𝐸𝑆][𝑆] 𝑘1 [𝐸]0 [𝑆]
[𝐸𝑆] + 1
=
→ (𝒆𝒒. 𝟖)
𝑘−1 + 𝑘2
𝑘−1 + 𝑘2
𝑘1 [𝑆]
𝑘1 [𝐸]0 [𝑆]
∴ [𝐸𝑆] (1 +
)=
𝑘−1 + 𝑘2
𝑘−1 + 𝑘2
→ (𝒆𝒒. 𝟗)
Introduce a new 𝐾𝑚 (without the prime)
𝑘−1 + 𝑘2
𝐾𝑚 =
→ (𝒆𝒒. 𝟏𝟎)
𝑘1
Using (eq. 10) to (eq. 9) for simplification
[𝐸𝑆] =
✓
✓
✓
✓
✓
✓
✓
[𝐸𝑆] (1 +
✓ PSSH is at steady state since it is a batch
reaction:
𝑑[𝐸𝑆]
𝑃𝑆𝑆𝐻:
=0
𝑑𝑡
✓ Also known as the Briggs-Haldane
assumption.
Derivation:
✓ Using the time variation of [ES] equation:
𝑑[𝐸𝑆]
= 𝑘1 [𝐸][𝑆] − 𝑘−1 [𝐸𝑆] − 𝑘2 [𝐸𝑆]
𝑑𝑡
→ (𝒆𝒒. 𝟏)
[𝑆]
[𝐸]0 [𝑆]
)=
→ (𝒆𝒒. 𝟏𝟏)
𝐾𝑚
𝐾𝑚
✓ Simplify by multiplying entire equation
with 𝐾𝑚
{[𝐸𝑆] (1 +
[𝑆]
[𝐸]0 [𝑆]
} (𝐾𝑚 ) → (𝒆𝒒. 𝟏𝟐)
)=
𝐾𝑚
𝐾𝑚
∴ [𝐸𝑆](𝐾𝑚 + [𝑆]) = [𝐸]0 [𝑆] → (𝒆𝒒. 𝟏𝟑)
✓ Rearranging to isolate [ES]
[𝐸]0 [𝑆]
[𝐸𝑆] =
→ (𝒆𝒒. 𝟏𝟒)
(𝐾𝑚 + [𝑆])
✓ Accounting the rate of production
formation equation:
𝑑 [𝑃 ]
𝑣=(
) = 𝑘2 [𝐸𝑆] → (𝒆𝒒. 𝟏𝟓)
𝑑𝑡
✓ Substituting (eq.14) to (eq. 15)
RATE LAW FOR PSSH
𝒗 = (𝒌𝟐 )
APPLYING THE FINAL MICHAELIS-MENTEN
EQUATION
✓ For a given total enzyme concentration, a
sketch of the rate of disappearance of the
substrate (=rate of formation of product)
is shown:
✓ This is called a Michaelis-Menten plot:
[𝑬]𝟎 [𝑺]
(𝑲𝒎 + [𝑺])
✓ We will use PSSH because it is more
general; the same result as the rapid
equilibrium except the dissociation
constant. Hence, PSSH accounts the third
reaction or k2.
✓ 𝑘2 is also called 𝑘𝑐𝑎𝑡
Significant Parameters
✓ The parameter 𝒌𝒄𝒂𝒕 (unit 𝟏/𝒔) is also
called the turnover number and
represents the number of substrate
molecules converted to product in a given
time, on a single-enzyme molecule when
the enzyme is saturated with substrate.
✓ The blue line is the plot for MichaelisMenten equation for the given data points
✓ We are satisfying the saturation constraint
because there is an upper limit.
✓ We have two extremes and 1 condition:
✓ Ideal for enzymes: 𝒉𝒊𝒈𝒉 𝒌𝒄𝒂𝒕 , 𝒍𝒐𝒘 𝑲𝑴
✓ The constant 𝑲𝑴 is called the Michaelis
constant and, for simple systems, is a
measure of the attraction of the enzyme
for its substrate. It is also called the
affinity constant.
✓ Since [𝐸]0 will be constant for a given
reaction, we can group together 𝑘𝑐𝑎𝑡 and
[𝐸]0 into one parameter, which we call the
maximum velocity 𝒗𝒎𝒂𝒙:
MAXIMUM VELOCITY FORMULA
𝒗𝒎𝒂𝒙 = 𝒌𝒄𝒂𝒕 [𝑬]𝟎
MICHAELIS-MENTEN EQUATION
𝒗𝒎𝒂𝒙 [𝑺]
𝒗=
𝑲𝑴 + [𝑺]
1. At low substrate concentrations ([𝑺] ≪
𝑲𝑴)
𝑖𝑓 [𝑆] ≪ 𝐾𝑀 , [𝑆] + 𝐾𝑀 ≈ 𝐾𝑀
𝒗𝒎𝒂𝒙[𝑺]
𝑽𝒎𝒂𝒙
) [𝑺]
𝒗≈
≈(
𝑲𝑴
𝑲𝑴
Note: This is approximately 1𝑠𝑡 order,
when substrate concentration is very low.
2. At high substrate concentrations ([𝑺] ≫
𝑲𝑴)
𝑖𝑓 [𝑆] ≫ 𝐾𝑀 , [𝑆] + 𝐾𝑀 ≈ [𝑆]
𝒗𝒎𝒂𝒙[𝑺]
𝒗≈
≈ 𝒗𝒎𝒂𝒙
[𝑺]
Note: This is approximately 0𝑡ℎ order; at high
concnetration, it becomes constant until it is
asymptotic to vmax.
One way to compare the catalytic efficiencies of
two enzymes is to compare their ratios
𝒌𝒄𝒂𝒕
𝑲𝑴
.
✓ We can get 𝑘𝑐𝑎𝑡 once we know 𝑣𝑚𝑎𝑥
𝑣𝑚𝑎𝑥 = 𝑘𝑐𝑎𝑡 [𝐸]0
𝑣𝑚𝑎𝑥
𝑘𝑐𝑎𝑡 =
[𝐸]0
Protein Engineering
3. When [𝑺] = 𝑲𝑴,
✓ This implies that 𝐾𝑀 also has units
of concentration.
𝑤ℎ𝑒𝑛 [𝑆] = 𝐾𝑚
𝒗𝒎𝒂𝒙[𝑺]
𝒗𝒎𝒂𝒙 [𝑺] 𝒗𝒎𝒂𝒙[𝑺]
𝒗=
=
=
𝑲𝑴 + [𝑺] [𝑺] + [𝑺]
𝟐[𝑺]
𝒗𝒎𝒂𝒙
∴𝒗=
𝟐
✓ This means that when the susbtrate
concentration is equal to 𝐾𝑀 , the
reaction will be running at half the
maximum velocity.
✓ the Michaelis constant is equal to
the substrate concentration at
which the rate of reaction is equal
to one-half the maximum rate.
In summary
For reactions described by MichaelisMentenkinetics, two parameters are needed:
✓ 𝒗𝒎𝒂𝒙 (𝒌𝒄𝒂𝒕 [𝑬]𝟎 ) which is a function of the
total enzyme concentration, and
✓ 𝑲𝑴, which is not a function of total enzyme
concentration; it is only a function of rate
parameters.
Two enzymes may have the same values for
𝑘𝑐𝑎𝑡 but different affinities for the substrate
(represented by 𝐾𝑀 ).
✓ We start with a natural form of enzyme
with a goal of designing an ezyme that
starts from the same native enzyme but is
more efficient.
✓ For one to imprve enzymes, we must do
interventions immediately in DNA; they
sometimes use mutageneis.
✓ The overall goal of protein engineerring is
catalytic efficiency together with
catalytic stability.
Linearizing Michaelis-Menten Equations
✓ Given rate versus substrate concentration
data, the parameters 𝑣𝑚𝑎𝑥 and 𝐾𝑀 may be
obtained by curve-fitting or linear
regression.
✓ There are several ways of linearizing the
Michaelis-Mentenequation.
Exercise 1.4.1
Determine the Michaelis-Menten parameters
𝑣𝑚𝑎𝑥 and 𝐾𝑀 for the reaction:
The rate of reaction is given as a function of urea
concentration (𝐶𝑆 ):
✓ Lineweaver-Burk is also called the “double
reciprocal” method and the most common:
➢ We invert Michaelis-Menten on both sides:
𝑣𝑚𝑎𝑥 [𝑆]
𝑓𝑟𝑜𝑚 𝑣 =
𝐾𝑀 + [𝑆]
𝑡𝑜 𝑡ℎ𝑖𝑠
Solution (Exercise 1.4.1):
✓ Using the Lineweaver-Burk Equation
✓ Initial Assumptions:
1 𝐾𝑀 + [𝑆]
=
𝑣
𝑣𝑚𝑎𝑥 [𝑆]
𝑡ℎ𝑒 𝑛𝑢𝑚𝑒𝑟𝑎𝑡𝑜𝑟 𝑐𝑎𝑛 𝑏𝑒 𝑓𝑎𝑐𝑡𝑜𝑟𝑒𝑑 𝑡𝑜 2:
1 𝐾𝑀 + [𝑆]
=
𝑣
𝑣𝑚𝑎𝑥 [𝑆]
1
𝐾𝑀 1
1
=
∙
+
𝑣 𝑣𝑚𝑎𝑥 [𝑆] 𝑣𝑚𝑎𝑥
✓ The Michaelis-Menten Equation:
𝑣𝑚𝑎𝑥 [𝑆]
𝑣=
→ (𝒆𝒒. 𝟏)
𝐾𝑀 + [𝑆]
✓ Lineweaver-Burk equation:
1 𝐾𝑀 + [𝑆]
=
→ (𝒆𝒒. 𝟐)
𝑣
𝑣𝑚𝑎𝑥 [𝑆]
✓ Separating the numerators:
1
𝐾𝑀 1
1
=
∙
+
𝑣 𝑣𝑚𝑎𝑥 [𝑆] 𝑣𝑚𝑎𝑥
1
1
✓ Plot 𝑉 as y, [𝑆] as x
[𝑆]
1
✓ Therefore, a plot of 𝑡 ln ( [𝑆]0 ) 𝑣𝑠.
1
([𝑆]0 −[𝑆])
Results in a line of slope − 𝐾 and intercept
𝑀
𝑡
𝑣𝑚𝑎𝑥
𝐾𝑀
.
𝑲𝑴 𝒂𝒏𝒅 𝒗𝒎𝒂𝒙
✓ While 𝐾𝑀 is an intrinsic parameter,
𝑣𝑚𝑎𝑥 is not.
o 𝐾𝑀 is a function of rate parameters
and is expected to change with
temperature or pH.
o 𝑣𝑚𝑎𝑥 is a function of both the rate
parameter 𝑘𝑐𝑎𝑡 (𝑘2 ) and the initial
enzyme concentration.
✓ For highly purified enzyme preparations,
it may be possible to express [𝐸]0 in terms
of M or g/L.
✓ From the Excel plot,
𝑦 = 0.02073𝑥 + 0.75308
𝐾𝑀
Where 0.02073 is the 𝑣
𝑚𝑎𝑥
; 0.75308 is the 𝑣
1
𝑚𝑎𝑥
✓ Solving for 𝑣𝑚𝑎𝑥
𝑣𝑚𝑎𝑥 =
1
𝒌𝒎𝒐𝒍
= 𝟏. 𝟑𝟑 𝟑
0.75308
𝒎 ∙𝒔
✓ Solving for 𝐾𝑀
𝐾𝑀 = (𝑚)(𝑣𝑚𝑎𝑥 )
𝐾𝑀 = (0.02073)(1.33) = 𝟎. 𝟎𝟐𝟕𝟓
𝒌𝒎𝒐𝒍
𝒎𝟑
Use of Batch Reactor
✓ A batch reactor can also be used to study
time course variation of [S].
✓ Integration of
𝑣=−
✓ Yields
𝑑 [𝑆]
𝑣𝑚𝑎𝑥 [𝑆]
=
𝑑𝑡
𝐾𝑀 + [𝑆]
𝐾𝑀
ln[𝑆]0
[𝑆]0 − [𝑆]
𝑣𝑚𝑎𝑥 −
= 𝑡
[𝑆]
𝑡
✓ When the enzyme is part of a crude
preparation, its concentration is in terms
of units. One enzyme unit (U or IU) is
defined as the amount of enzyme that
catalyzes the conversion of one micromole
of substrate to product per minute, under
specific reaction
(usually
optimal)
conditions.
✓ The SI unit is the katal, which is defined as
the amount of enzyme that catalyzes the
conversion of one mole of substrate to
product per second.
1.1.
ALLOSTERY
Kinetics of Allosteric Binding
✓ Some enzymes have multiple binding sites,
and the binding of one substrate can
facilitate the binding of the subsequent
substrates. This is known as cooperative
binding or allostery.
o The affinity of the second substrate
increases because of the binding of
the first substrate; this is true for all
subsequent binding substrates.
✓ The activity of allosteric enzymes can be
altered by regulatory molecules binding
on allosteric sites; their properties can
thus be adjusted to meet the immediate
needs of a cell.
o Binding of substrates in allosteric
sites helps enzyme regulation;
there is cooperative binding, thus,
affinity will increase.
✓ Michaelis-Menten kinetics fails to describe
the kinetics of cooperative binding;
allosteric
enzymes
often
display
sigmoidal plots.
✓ When one oxygen molecule binds
(refer to the hemoglobin), the binding
of that first oxygen molecule causes a
structural change in the hemoglobin. It
enhances the binding of other oxygen
molecules.
✓ The rate expression in this case of
allosteric enzymes becomes a threeparameter rate law:
✓ The dark yellow color: one pair or 𝛼
and 𝛽 monomers.
RATE EXPRRESION (ALLOSTERIC
ENZYMES)
𝒅[𝑺]
𝒗𝒎𝒂𝒙 [𝑺]𝒏
𝒗=−
= ′′
𝒅𝒕
𝑲𝒎 + [𝑺]𝒏
where n is the Hill coefficient, and
𝑛 > 1 indicates cooperativity.
For n=1, the expression reduces to the
Michaelis-Menten expression.
✓ The Hill coefficient can be determined
by linear regression:
ln (
𝑣
𝑣𝑚𝑎𝑥 − 𝑣
✓ A plot of ln 𝑣
′′
) = 𝑛 ln[𝑆] − 𝑙𝑛𝐾𝑚
𝑣
𝑚𝑎𝑥 −𝑣
versus ln[𝑆] gives a
Enzyme Inhibition: Competitive
✓ In competitive inhibition, the enzyme
binds either substrate or inhibitor.
Competitive inhibitors are usually
substrate analogs and compete with
substrate for the active site of the enzyme.
straight line whose slope is n.
1.2.
ENZYME INHIBITION
✓ Enzyme inhibition is the opposite of
allostery because you are decreasing
the catalytic activity of the enzyme.
✓ It is a way of regulating enzyme activity
as well.
✓ Enzyme activity can be inhibited by the
binding of specific small molecules and
ions.
✓ The ions responsible are called
inhibitors.
✓ Like allostery, inhibition also serves as
a major control mechanism in
biological systems. In addition, many
drugs and toxic agents act by inhibiting
enzymes.
✓ Enzyme inhibition can be irreversible
or reversible:
1. Irreversible inhibitors dissociate
very slowly from its target enzyme;
they are either covalently or noncovalently bound to it. Examples are
heavy metals (Pb, Cd, Hg, etc.) (toxic).
2. Reversible
inhibitors
rapidly
dissociate from the enzyme-inhibitor
complex, and can be competitive,
uncompetitive, or noncompetitive.
In some cases, the substrate can also be
inhibitory if accumulated at high
concentrations.
✓ The effect of inhibitors is seen as a
decrease in 𝐾𝑀 .
✓ The inhibition scheme can be described as:
Enzyme Inhibition (Competitive) Derivation:
✓ Our goal is to replace [ES] with a
measurable quantity
𝑣 = 𝑘2 [𝐸𝑆] → (𝒆𝒒. 𝟏)
✓ Note that the only measurable quantities
are:
[𝑆], [𝐸]0 , [𝐼 ]
✓ Begin with the balance on the transition
states
𝑑 [𝐸𝑆]
= 𝑘1 [𝐸][𝑆] − (𝑘−1 + 𝑘2 )[𝐸𝑆]
𝑑𝑡
→ (𝒆𝒒. 𝟐)
✓ Using the PSSH approach, we can see to it
that,
𝑑 [𝐸𝑆]
= 𝑘1 [𝐸][𝑆] − (𝑘−1 + 𝑘2 )[𝐸𝑆] = 0
𝑑𝑡
→ (𝒆𝒒. 𝟑)
✓ Rewriting in terms of [E]
𝑘 + 𝑘2 [𝐸𝑆]
[𝐸] = −1
∙
→ (𝒆𝒒. 𝟒)
[𝑆]
𝑘1
✓ Recall that
𝑘−1 + 𝑘2
= 𝐾𝑀
𝑘1
∴ 𝑢𝑠𝑖𝑛𝑔 (𝒆𝒒. 𝟒),
[𝐸𝑆]
𝐾𝑀 ∙
→→ (𝒆𝒒. 𝟓)
[𝑆]
✓ Performing balance on the inhibitor’s side.
At steady state, which occurs in the middle
of the reaction,
𝑑 [𝐸𝐼 ]
= 𝑘3 [𝐸][𝐼 ] − 𝑘−3 [𝐸𝐼 ] = 0
𝑑𝑡
Note that:
✓ Isolating [ES] to have [ES] as a function of
all measurable properties
[𝐸]0
[𝐸𝑆] =
→ (𝒆𝒒. 𝟏𝟏)
𝐾𝑀 𝐾𝑀 [𝐼 ]
(1 +
)
[𝑆] + 𝑘𝐼 [𝑆]
✓ Now that [ES] is now a function of
measurable properties, we circle back to
(𝒆𝒒. 𝟏). then substitute (eq. 11)
𝑣 = 𝑘2 [𝐸𝑆] → (𝒆𝒒. 𝟏)
✓
✓
✓ Isolating [EI]
[𝐸][𝐼 ]
𝑘
[𝐸𝐼 ] = 3 [𝐸][𝐼 ] =
→ (𝒆𝒒. 𝟔)
𝑘−3
𝑘𝐼
✓ Substitute (eq.6) to (eq.5)
𝐾 [𝐸𝑆][𝐼 ]
[𝐸] = 𝑀
→ (𝒆𝒒. 𝟕)
𝑘𝐼 [𝑆]
✓ Using the enzyme conservation equation
and the third form of [EI]:
[𝐸]0 = [𝐸] + [𝐸𝑆] + [𝐸𝐼] → (𝒆𝒒. 𝟖)
✓ Write everything in terms of [ES] from
(eq.7)
𝐾
𝐾 [𝐼]
[𝐸]0 = 𝑀 [𝐸𝑆] + [𝐸𝑆] + 𝑀 [𝐸𝑆]
[𝑆 ]
𝑘𝐼 [𝑆]
→ (𝒆𝒒. 𝟗)
✓ Factor out [ES]
𝐾
𝐾 [𝐼 ]
[𝐸]0 = [𝐸𝑆] (1 + 𝑀 + 𝑀 ) → (𝒆𝒒. 𝟏𝟎)
[𝑆] 𝑘𝐼 [𝑆]
𝑘2 [𝐸]0
→ (𝒆𝒒. 𝟏𝟐)
𝐾𝑀 𝐾𝑀 [𝐼 ]
(1 +
)
[𝑆] + 𝑘𝐼 [𝑆]
Multiply the numerator and denominator
with [S]
[𝑆]
𝑘2 [𝐸]0
𝑣=
∙
𝐾
𝐾 [𝐼 ] [𝑆]
(1 + 𝑀 + 𝑀 )
[𝑆] 𝑘𝐼 [𝑆]
𝑘2 [𝐸]0 [𝑆]
∴
→ (𝒆𝒒. 𝟏𝟑)
𝐾𝑀 [𝐼 ]
([𝑆] + 𝐾𝑀 +
)
𝑘𝐼
Rearranging the denominator in the form
of 𝐾𝑀
𝑘2 [𝐸]0 [𝑆]
→ (𝒆𝒒. 𝟏𝟒)
[𝐼 ]
𝐾𝑀 (1 + 𝐾 ) + [𝑆]
𝐼
Recall that,
𝑣𝑚𝑎𝑥 = 𝑘2 [𝐸]0
And
[𝐼 ]
𝐾𝑀,𝑎𝑝𝑝 = 𝐾𝑀 (1 + )
𝐾𝐼
Therefore, using the recalled equations
and (eq. 14),
𝒗𝒎𝒂𝒙[𝑺]
𝒗=
→ (𝒆𝒒. 𝟏𝟓)
𝑲𝑴,𝒂𝒑𝒑 + [𝑺]
𝑣=
✓
✓
THE RATE LAW FOR COMPETETIVE
INHIBITION IS:
𝒗𝒎𝒂𝒙[𝑺]
𝒗𝒎𝒂𝒙 [𝑺]
=
[𝑰]
𝑲𝑴,𝒂𝒑𝒑 + [𝑺]
𝑲𝑴 (𝟏 + 𝑲 ) + [𝑺]
𝑰
where
𝒗=
𝑲𝑴,𝒂𝒑𝒑 = 𝑲𝑴 (𝟏 +
[𝑰]
)
𝑲𝑰
✓ The net effect of competitive inhibition is
an apparently higher value of𝐾𝑚 . Since
𝑣𝑚𝑎𝑥 is unaffected, high concentrations of
substrate can overcome the inhibition to
reach 𝑣𝑚𝑎𝑥.
✓ A competitive inhibitor reduces the
rate of catalysis by reducing the
proportion of enzyme molecules bound
to a substrate.
o If there is an inhibitor in the
enzyme, you can kick that out; only
when substrate concentration is
high.
✓ Only competitive inhibition can overcome
high substrate concentration.
Enzyme Inhibition: Noncompetitive
✓ In noncompetitive inhibition, the
inhibitors are not substrate analogs.
Inhibitors bind sites other than the active
site and reduce enzyme affinity to the
substrate. Noncompetitive inhibitors can
bind free enzyme or the ES complex.
✓ Assumes the dissociation constant for ESI
and EI are the same (not functional version
of the proteins because ES is the only
functional one)
✓ The inhibition scheme can be described as:
Derivation
THE RATE LAW FOR
NONCOMPETETIVE INHIBITION IS:
𝒗=
𝒗𝒎𝒂𝒙,𝒂𝒑𝒑 [𝑺]
𝒗𝒎𝒂𝒙[𝑺]
=
[𝑰]
𝑲𝑴 + [𝑺]
(𝟏 + ) (𝑲𝒎 + [𝑺])
𝑲𝑰
where
𝒗𝒎𝒂𝒙,𝒂𝒑𝒑 =
𝒗𝒎𝒂𝒙
[𝑰]
𝟏+𝑲
𝑰
✓ This means that 𝑣𝑚𝑎𝑥 decreases at higher
concentrations.
✓ The maximum possible rate decreases if
𝑣𝑚𝑎𝑥 is affected.
✓ The net effect of noncompetitive inhibition
is an apparently lower value of 𝑣𝑚𝑎𝑥.
Therefore,
substrate
concentrations
cannot
overcome
noncompetitive
inhibition; the initial 𝑣𝑚𝑎𝑥cannot be
restored.
✓ A noncompetitive inhibitor lowers the
concentration of functional enzyme.
The resulting solution behaves as a
more dilute solution of the enzyme
does (same amount of enzyme but k2
decreases).
Enzyme Inhibition: Uncompetitive
✓ In uncompetitive inhibition, the inhibitors
bind to the ES complex only; this implies
that substrate must bind first before
uncompetitive inhibitors can take effect.
✓ The inhibition scheme can be described as:
Comparing and Contrast Enzyme Inhibitions
✓ Measurements of the rates of catalysis at
different concentrations of substrate and
inhibitor can serve to distinguish the three
types of reversible inhibition.
✓ For Competitive Inhibition
THE RATE LAW FOR UNCOMPETETIVE
INHIBITION IS:
Where
✓ For Noncompetitive Inhibition
✓ The net effect of uncompetitive inhibition
is a decrease in both 𝐾𝑚 and 𝑣𝑚𝑎𝑥. The
rate is more sensitive to changes in 𝑣𝑚𝑎𝑥,
so the net result is a decrease in reaction
rate. As in uncompetitive inhibition, high
substrate
concentrations
cannot
overcome uncompetitive inhibition.
✓ Because some unproductive ESI
complex will always be present,
[𝑬]𝟎 will be lower and so will 𝒗𝒎𝒂𝒙. Also,
because ES is consumed to form ESI, the
equilibrium shifts to more binding of S,
lowering the apparent value of 𝑲𝒎.
✓ For Uncompetitive Inhibition
✓ The following scheme may be used to
describe the pH dependence of enzymatic
reaction rates for ionizing enzymes:
THE RATE LAW (ACCOUNTING
EFFECTS OF Ph)
✓ SUMMARY:
➢ 𝑣𝑚𝑎𝑥 𝑐ℎ𝑎𝑛𝑔𝑒𝑠: noncompetitive
➢ 𝐾𝑚 𝑐ℎ𝑎𝑛𝑔𝑒𝑠: competitive
➢ 𝑣𝑚𝑎𝑥 𝑎𝑛𝑑 𝐾𝑚 𝑐ℎ𝑎𝑛𝑔𝑒: uncompetitive
1.3.
EFFECTS OF pH AND TEMPERATURE
Effects of pH
✓ Recall that some amino acid residues have
side chains that are ionizable; certain
enzymes have these groups on their active
sites, and their extents of protonation
depend on the prevailing pH of the solution.
✓ Changes in solution pH result in changes in
enzyme activity due to different
ionizations, which may also result in
changes in the three-dimensional shape of
the enzyme. In some cases, the pH of the
medium can also affect the ionization state
of the substrate, and hence its affinity to
the enzyme.
✓ For these reasons, enzymes are only
active over a certain pH range.
✓ As a result of this behavior, the pH
optimum of the enzyme is between pK1
and pK2.
✓ Theoretical prediction of the pH optimum
of enzymes requires a knowledge of the
active site characteristics of enzymes,
which are very difficult to obtain. It is
usually determined experimentally.
substitution of the previous expression
gives:
Effects of Temperature
✓ The rate of enzyme-catalyzed reactions
increases with temperature up to a
certain limit only, above which, activity
decreases with temperature due to
denaturation (there will be no 3D
structure anymore).
✓ The initial increase in the activity with
temperature is called temperature
activation, and this follows the Arrhenius
equation:
𝑬𝒂
𝒌𝟐 = 𝑨𝒆−𝑹𝑻
✓ The descending part is called the
temperature deactivation or thermal
denaturation
stage.
Denaturation
kinetics follows a first-order rate law:
✓ The rate of enzyme-catalyzed reactions
increases with temperature up to a
certain limit only, above which, activity
decreases with temperature due to
denaturation.
✓ The denaturation constant also follows the
Arrhenius equation:
✓ Since 𝑣𝑚𝑎𝑥 = 𝑘2 [𝐸] (not [𝐸]0 if it is
changing) (exponentially decreasing),
Where
𝑬𝒂
𝒌𝟐 = 𝑨𝒆−𝑹𝑻
[𝑬] = [𝑬]𝟎 𝒆−𝒌𝒅 𝒕
𝑬𝒂
𝒌𝒅 𝜶 𝑨𝒅 𝒆−𝑹𝑻
✓ The activation energies of enzymecatalyzed reactions are within the 4-20
kcal/mol range, whereas deactivation
energies vary between 40-130 kcal/mol
range.
✓ Higher activation energy means it is more
sensitive to temperature changes and viceversa; denaturation is more dominant at
high temperatures.
✓ There is a point wherein two rates are
equal called the optimal point.
✓ As a result, enzyme denaturation is more
sensitive to temperature changes; a rise in
temperature from 30o to 40oC results in a
1.8-fold increase in enzyme activity, but a
41-fold increase in enzyme denaturation.
✓ Isolate [ES],
[𝐸𝑆] =
1. ENZYME INHIBITION: COMPETETIVE
DERIVATION (RAPID EQUILIBRIUIM
APPROACH)
✓ In the Rapid Equilibrium Approach, we
assume that 𝑘2 is extremely small.
Defining the constants,
[𝐸][𝑆]
𝐾𝑀′ =
→ (𝒆𝒒. 𝟏)
[𝐸𝑆]
[𝐸][𝐼 ]
𝐾𝐼 =
→ (𝒆𝒒. 𝟐)
[𝐸𝐼 ]
✓ We then invoke the enzyme conservation,
and remember that for competitive
inhibition, enzyme exists in three forms:
[𝐸]𝑂 = [𝐸] + [𝐸𝑆] + [𝐸𝐼 ] → (𝒆𝒒. 𝟑)
✓ The objective is to express [ES] as a
function of measurable properties:
[𝐸]𝑂 , [𝑆], [𝐼]
✓ Express (eq.3) as a function of [ES] from
(eq.1) and (eq.2)
′ [
[𝐸][𝐼 ]
𝐾𝑚
𝐸𝑆]
+ [𝐸𝑆] +
→ (𝒆𝒒. 𝟒)
[𝑆 ]
𝐾𝐼
✓ Note that,
𝐾 ′ [𝐸𝑆]
[𝐸] = 𝑚
[𝑆 ]
′ [
[𝐼 ] 𝐾𝑀′ [𝐸𝑆]
𝐾𝑚 𝐸𝑆]
[
]
[
]
∴ 𝐸𝑂=
+ 𝐸𝑆 +
∙(
)
[𝑆]
[𝑆]
𝐾𝐼
→ (𝒆𝒒. 𝟓)
✓ Factor [ES] out,
[𝐸]𝑂 =
[𝐸]𝑂 = [𝐸𝑆](
𝐾𝑀′
𝐾𝑀′ [𝐼 ]
+1+
∙
) → (𝒆𝒒. 𝟔)
[𝑆]
[𝑆] [𝐾𝐼 ]
[𝐸]𝑂
→ (𝒆𝒒. 𝟕)
𝐾𝑀′ [𝐼 ]
[𝑆] + 1 + [𝑆] ∙ 𝐾𝐼
𝐾𝑀′
✓ Multiply the numerator and denominator
with [S],
[𝐸]𝑂
[𝑆]
[𝐸𝑆] = ′
′ [ ]∙
𝐾𝑀
𝐾𝑀 𝐼 [𝑆]
[𝑆] + 1 + [𝑆] ∙ 𝐾𝐼
[𝐸]𝑂 [𝑆]
∴ [𝐸𝑆] =
→ (𝒆𝒒. 𝟖)
′
′ [𝐼 ]
𝐾𝑀 + [𝑆] + 𝐾𝑀 ∙ 𝐾
𝐼
✓ Factor 𝐾𝑀′
[𝐸]𝑂 [𝑆]
[𝐸𝑆] =
→ (𝒆𝒒. 𝟗)
[𝐼 ]
′
𝐾𝑀 + (1 + 𝐾 ) + [𝑆]
𝐼
✓ We can now express everything in terms of
𝑣
𝑑 [𝑃]
𝑑[𝑆]
𝑣=
=−
= 𝑘2 [𝐸𝑆] → (𝒆𝒒. 𝟏𝟎)
𝑑𝑡
𝑑𝑡
✓ Substituting (eq.9) to (eq.10)
[𝐸]𝑂 [𝑆]
𝑣=
[𝐼 ]
𝐾𝑀′ + (1 + 𝐾 ) + [𝑆]
𝐼
✓ Note that,
[𝐼 ]
𝐾𝑀′ ,𝑎𝑝𝑝 = 𝐾𝑀′ + (1 + ) → (𝒆𝒒. 𝟏𝟏)
𝐾𝐼
✓ Thus, the final equation is:
𝒗=
𝒗𝒎𝒂𝒙 [𝑺]
(𝑲𝒎,𝒂𝒑𝒑 + [𝑺])
2. ENZYME
INHIBITION:
COMPETETIVE DERIVATION
EQUILIBRIUIM APPROACH)
NON
(RAPID
[𝐸]𝑂
[𝑆]
∙
′ [ ]
[𝐼 ] [𝑆]
𝐾𝑀 𝐼
+
1
+
+
[𝑆 ]
𝐾𝐼 [𝑆] 𝐾𝐼
→ (𝒆𝒒. 𝟕)
[𝐸]𝑂 [𝑆]
∴ [𝐸𝑆] =
[𝐼 ]
𝐾 ′ [𝐼 ]
[𝑆 ]
𝐾𝑀′ + [𝑆] + 𝑀
+
𝐾𝐼
𝐾𝐼
(
)
→ 𝒆𝒒. 𝟖
✓ Factor 𝐾𝑀′
[𝐸𝑆] =
𝐾𝑀′
[𝐸]𝑂 [𝑆]
[𝐼 ]
[𝐼 ]
𝐾𝑀′ (1 + 𝐾 ) + [𝑆] (1 + 𝐾 )
𝐼
𝐼
→ (𝒆𝒒. 𝟗)
Group the denominator
[𝐸]𝑂 [𝑆]
[𝐸𝑆] =
→ (𝒆𝒒. 𝟏𝟎)
[𝐼 ]
′ + [𝑆])
(1 + ) (𝐾𝑚
𝐾𝐼
Expressing in terms of v
𝑣 = 𝑘2 [𝐸𝑆] → (𝒆𝒒. 𝟏𝟏)
Substituting (eq.10) to (eq.11)
[𝐸]𝑂 [𝑆]
𝑣 = 𝑘2
[𝐼 ]
′ + [𝑆 ])
(1 + ) (𝐾𝑚
𝐾𝐼
[𝐸]𝑂
[𝑆]
𝑣=
∙ ′
→ (𝒆𝒒. 𝟏𝟐)
[𝐼 ] (𝐾𝑚 + [𝑆])
(1 + )
𝐾𝐼
Note that
[𝐸 ]𝑂
𝑣𝑚𝑎𝑥,𝑎𝑝𝑝 =
→ (𝒆𝒒. 𝟏𝟑)
[𝐼 ]
(1 + )
𝐾𝐼
Thus, the final equation is,
[𝐸𝑆] =
✓ Defining the constants,
[𝐸][𝑆] [𝐸𝐼 ][𝑆]
𝐾𝑀′ =
=
→ (𝒆𝒒. 𝟏)
[𝐸𝑆]
[𝐸𝑆𝐼 ]
✓ Note that in this model, we assume that
Km for the formation of the ES complex is
the same as the formation of the ESI
complex from EI + S. Same goes for I.
[𝐸][𝐼 ] [𝐸𝑆][𝐼 ]
𝐾𝐼 =
=
→ (𝒆𝒒. 𝟐)
[𝐸𝐼 ]
[𝐸𝑆𝐼 ]
✓ For the total enzyme balance,
[𝐸]𝑂 = [𝐸] + [𝐸𝑆] + [𝐸𝐼 ] + [𝐸𝑆𝐼 ]
→ (𝒆𝒒. 𝟑)
✓ The objective is to express [ES] as a
function of measurable properties:
[𝐸]𝑂 , [𝑆], [𝐼]
✓ Express (eq.3) as a function of [ES] from
(eq.1) and (eq.2)
[𝐸][𝐼 ] [𝐸𝑆][𝐼 ]
𝐾𝑀′
[𝐸]𝑂 =
[𝐸𝑆] + [𝐸𝑆] +
+
[𝑆 ]
𝐾𝐼
𝐾𝐼
→ (𝒆𝒒. 𝟒)
✓ Note that:
𝐾′
[𝐸] = 𝑀 [𝐸𝑆]
[𝑆]
′
𝐾𝑀
𝐾 ′ [𝐼 ]
[𝐸𝑆] + [𝐸𝑆] + 𝑀 [𝐸𝑆]
∴ [𝐸]𝑂 =
[𝑆]
𝐾𝐼 [𝑆]
[𝐼 ]
+ [𝐸𝑆] → (𝒆𝒒. 𝟓)
𝐾𝐼
✓ Factor out the [ES]
𝐾′
𝐾 ′ [𝐼 ] [𝐼 ]
[𝐸]𝑂 = [𝐸𝑆] ( 𝑀 + 1 + 𝑀 + )
[𝑆 ]
𝐾𝐼 [𝑆] 𝐾𝐼
→ (𝒆𝒒. 𝟔)
✓ Isolate [ES] and multiply the numerator
and denominator with [S]
✓
✓
✓
✓
✓
𝒗=
𝒗𝒎𝒂𝒙,𝒂𝒑𝒑 [𝑺]
(𝑲′𝒎 + [𝑺])
3. ENZYME INHIBITION: UNCOMPETETIVE
DERIVATION (RAPID EQUILIBRIUIM
APPROACH)
✓ Factor out [S]
[𝐸]𝑂 [𝑆]
[𝐸𝑆] =
→ (𝒆𝒒. 𝟖)
[𝐼 ]
+ [𝑆] (1 + 𝐾 )
𝐼
✓ Multiply numerator and denominator by
𝐾𝑀′
[𝐼]
(1 + 𝐾 )
𝐼
[𝐸]𝑂 [𝑆]
[𝐼 ]
(1 + )
𝐾𝐼
[𝐸𝑆] =
→ (𝒆𝒒. 𝟗)
𝐾𝑀′
+
[𝑆]
[𝐼 ]
(1 + )
𝐾𝐼
✓ we can now express the equation in terms
of v,
𝑣 = 𝑘2 [𝐸𝑆] → (𝒆𝒒. 𝟏𝟎)
✓ Defining the constants,
[𝐸][𝑆]
𝐾𝑀′ =
→ (𝒆𝒒. 𝟏)
[𝐸𝑆]
✓ Note that in this model, we assume that
Km for the formation of the ES complex is
the same as the formation of the ESI
complex from EI + S. Same goes for I.
[𝐸𝑆][𝐼 ]
𝐾𝐼 =
→ (𝒆𝒒. 𝟐)
[𝐸𝑆𝐼 ]
✓ For the total enzyme balance,
[𝐸]𝑂 = [𝐸] + [𝐸𝑆] + [𝐸𝑆𝐼 ] → (𝒆𝒒. 𝟑)
✓ The objective is to express [ES] as a
function of measurable properties:
[𝐸]𝑂 , [𝑆], [𝐼]
✓ Express (eq.3) as a function of [ES] from
(eq.1) and (eq.2)
[𝐼 ]
𝐾𝑀′
[𝐸𝑆] + [𝐸𝑆] + [𝐸𝑆]
[𝑆 ]
𝐾𝐼
→ (𝒆𝒒. 𝟒)
✓ Factor out the [ES]
[𝐼 ]
𝐾′
[𝐸]𝑂 = [𝐸𝑆] ( 𝑀 + 1 + + ) → (𝒆𝒒. 𝟓)
[𝑆 ]
𝐾𝐼
✓ Isolate [ES] and multiply the numerator
and denominator with [S]
[𝐸]𝑂
[𝑆]
[𝐸𝑆] = ′
∙
→ (𝒆𝒒. 𝟔)
[𝐼 ] [𝑆]
𝐾𝑀
+
1
+
[𝑆]
𝐾𝐼
[𝐸]𝑂 [𝑆]
∴ [𝐸𝑆] =
→ (𝒆𝒒. 𝟕)
[𝐼 ]
′
[
]
[
]
𝐾𝑀 + 𝑆 + 𝐾 𝑆
𝐼
[𝐸]𝑂 =
✓ Substituting (eq.9) to (eq.10)
𝑘2 [𝐸]𝑂
[𝐼 ] ∙ [𝑆]
(1 + )
𝐾𝐼
𝑣=
→ (𝒆𝒒. 𝟏𝟏)
𝐾𝑀′
[𝑆]
+
[𝐼 ]
(1 + )
𝐾𝐼
✓ Take note that,
𝑣𝑚𝑎𝑥 = 𝑘2 [𝐸]𝑂 → (𝒆𝒒. 𝟏𝟐)
𝑣𝑚𝑎𝑥,𝑎𝑝𝑝 =
𝐾𝑚,𝑎𝑝𝑝 =
𝑘2 [𝐸]𝑂
→ (𝒆𝒒. 𝟏𝟑)
[𝐼 ]
(1 + )
𝐾𝐼
𝐾𝑀′
→ (𝒆𝒒. 𝟏𝟒)
[𝐼 ]
(1 + )
𝐾𝐼
✓ Hence, the final equation is,
𝒗=
𝒗𝒎𝒂𝒙,𝒂𝒑𝒑 [𝑺]
𝑲𝑴,𝒂𝒑𝒑[𝑺]
Exercise 1.5.1
The hydrolysis of urea by urease shows
inhibition. Hydrolysis data are given:
✓ Solve for x-intercept:
𝑥 − 𝑖𝑛𝑡𝑒𝑟𝑐𝑒𝑝𝑡:
1
1 𝐾𝑚
1
𝑦 − 𝑖𝑛𝑡 = = 0 =
∙
+
[𝑆] 𝑣𝑚𝑎𝑥 𝑣𝑚𝑎𝑥
𝑣
✓ Rearranging,
1
1
1
1
=
𝐾𝑚 + 1 →
=−
[𝑆]
𝑣 [𝑆 ]
𝐾𝑚
✓ Plot in excel only the chosen points
What type of inhibition do the data show?
Based on the type of inhibition, determine Km and
KI.
Solution to Exercise 1.5.1
✓ We have two substrate concentrations,
plotting those data:
✓ The resulting graph is:
✓ We are going to select specific points:
o We need inhibitions points that
would be very close to each other
or exactly the same like as follows,
✓ Recall Lineweaver-Burk equation:
1
1 𝐾𝑚
1
=
∙
+
𝑣 [𝑆] 𝑣𝑚𝑎𝑥 𝑣𝑚𝑎𝑥
✓ Calculate Km represented by each line
𝑠𝑙𝑜𝑝𝑒
𝐾𝑚 =
𝑦 − 𝑖𝑛𝑡𝑒𝑟𝑐𝑒𝑝𝑡
a. [𝐼 ] = 0
0.0102
𝐾𝑀𝐼 =
= 𝟎. 𝟎𝟔𝟎𝟒
0.1689
b. [𝐼 ] = 0.0012
0.0153
𝐾𝑀2 =
= 𝟎. 𝟎𝟔𝟎𝟒
0.2533
c. [𝐼 ] = 0.0044
0.0436
𝐾𝑀3 =
= 𝟎. 𝟎𝟖𝟎𝟒
0.5422
d. [𝐼 ] = 0060
0.0573
𝐾𝑀4 =
= 𝟎. 𝟎𝟗𝟔𝟔
0.5433
From the accumulated data, the inhibitor changes
vmax only since Km is not statistically different from
one another. Henceforth, the inhibitor is
NONCOMPETETIVE.
✓ Solve for 𝐾𝑚 average (obtained data)
𝑲𝒎,𝒂𝒗𝒈 = 𝟎. 𝟕𝟒𝟒𝟓 𝑴
✓ Solve for 𝐾𝐼
✓ Recall that for noncompetitive inhibition,
𝑣=
[𝑆 ]
𝑣𝑚𝑎𝑥
∙
[𝐼 ] 𝐾𝑚 + [𝑆]
1+ 𝐾
𝐼
𝑣𝑚𝑎𝑥,𝑎𝑝𝑝 =
𝑣𝑚𝑎𝑥
[𝐼 ]
1+
𝑘2
✓ We can use any point, for our case, we used
0.33
→ 𝑓𝑟𝑜𝑚 𝑡ℎ𝑒 𝑝𝑙𝑜𝑡
𝑣𝑚𝑎𝑥 =
1
= 𝟑. 𝟗𝟓
0.2533
✓ Choose inhibitor point where 1/v=0.33
and [I]=0.0012
∴ 𝑣 = 0.33; 𝑣𝑚𝑎𝑥 = 3.95; [𝐼 ] = 0.0012;
[𝑆] = 0.2; 𝐾𝑚 𝑎𝑣𝑔 = 0.7445
3.95
0.2
0.33 =
∙
0.0012 0.7445
1+ 𝐾
𝐼
✓ Solve for 𝐾𝐼
𝑲𝑰 = 𝟔𝒙𝟏𝟎−𝟑 𝑴