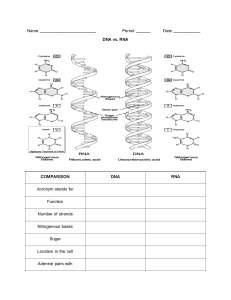
Spring 2022 Microbiology II course Virology- Week 4 (Viral Genetics) By Assist. Prof. Dr. Dana Khdr Sabir 2021- 2022 Learning outcome 1- Basic revision of molecular genetics 2- Viral classification 3- Viral variations 2 PART 1: What is Genetics? • Genetics is a branch of biology concerned with the study of genes, genetic variation, and heredity in organisms The central dogma of molecular biology explains the flow of genetic information, from DNA to RNA, to make a functional product, a protein. Central Dogma of Molecular Biology Comparison Full Name Function Structure Length Sugar DNA RNA Deoxyribonucleic Acid Ribonucleic Acid DNA replicates and stores genetic information. It is a blueprint for all genetic information contained within an organism. RNA converts the genetic information contained within DNA to a format used to build proteins, and then moves it to ribosomal protein factories. DNA consists of two strands, arranged in a double helix. These strands are made up of RNA only has one strand, but like DNA, is subunits called nucleotides. Each nucleotide made up of nucleotides. RNA strands are contains a phosphate, a 5-carbon sugar shorter than DNA strands. molecule and a nitrogenous base. DNA is a much longer polymer than RNA. The sugar in DNA is deoxyribose, which contains one less hydroxyl group than RNA’s ribose. RNA molecules are variable in length, but much shorter than long DNA polymers. RNA contains ribose sugar molecules, without the hydroxyl modifications of deoxyribose. 4 Comparison DNA RNA The bases in DNA are Adenine (‘A’), Thymine (‘T’), Guanine (‘G’) and Cytosine (‘C’). RNA shares Adenine (‘A’), Guanine (‘G’) and Cytosine (‘C’) with DNA, but contains Uracil (‘U’) rather than Thymine (T). Adenine and Thymine pair (A-T) Cytosine and Guanine pair (C-G) Adenine and Uracil pair (A-U) Cytosine and Guanine pair (C-G) Location DNA is found in the nucleus, with a small amount of DNA also present in mitochondria. RNA forms in the nucleolus, and then moves to specialised regions of the cytoplasm depending on the type of RNA formed. Reactivity Due to its deoxyribose sugar, which contains one less oxygen-containing hydroxyl group, DNA is a more stable molecule than RNA, which is useful for a molecule which has the task of keeping genetic information safe. RNA, containing a ribose sugar, is more reactive than DNA and is not stable in alkaline conditions. RNA’s larger helical grooves mean it is more easily subject to attack by enzymes. Ultraviolet (UV) Sensitivity DNA is vulnerable to damage by ultraviolet RNA is more resistant to damage light. from UV light than DNA. 5 Bases Base Pairs Main differences between DNA, RNA 6 The Genetic Code: Codons • The genetic code is read in three- letter grouping called codons. • A codon is a group of three nucleotide bases in messenger RNA that specifies a particular amino acids. 7 8 PART 2: Viral Classification • Taxonomy is the science of categorizing and assigning names (nomenclature) to organisms based on similar characteristics. • It is important to note that viruses, since they are not alive, belong to a completely separate system that does not fall under the tree of life (Phylogenetic Tree). 9 Virus Classification and Taxonomy Why classification? 1. It allows scientists to contrast viruses and to reveal information on newly discovered viruses by comparing them to similar viruses. 2. It also allows scientists to study the origin of viruses and how they have evolved over time. The classification of viruses is not simple, however—there are currently over 2800 different viral species with very different properties! 10 Viral Classification • Since viruses lack ribosomes (and thus rRNA), they cannot be classified within the Three Domain Classification scheme with cellular organisms. • Dr. David Baltimore derived a viral classification scheme, one that focuses on the relationship between a viral genome to how it produces its mRNA. • The Baltimore Scheme recognizes seven classes of viruses. 11 Notes: • Viruses are only classified using order, family, genus, and species • • • • Order: Ends in virales suffix (such as picornavirales) Family: Ends in- viridae suffix, subfamilies are indicated with virinae suffix Such as Picornaviridae. Genus: Ends in –virus suffix. Such as Enterovirus Species: Generally the common names of the virus. 12 Virus Classification and Taxonomy The Baltimore classification system: • This system categorizes viruses based on the type of nucleic acid genome and replication strategy of the virus. The system also breaks down single-stranded RNA viruses into those that are positive strand (+) and negative strand (−). • Positive-strand (also positive-sense or plus-strand) RNA is able to be immediately translated into proteins; as such, messenger RNA (mRNA) in the cell is positive strand. 13 Virus Classification and Taxonomy • Negative-strand (also negative-sense or minusstrand) RNA is not translatable into proteins; it first has to be transcribed into positive-strand RNA. • Baltimore also took into account viruses that are able to reverse transcribe, or create DNA from an RNA template, which is something that cells are not capable of doing. 14 Seven viral classes: • Class I: dsDNA viruses • Class II: ssDNA viruses • Class III: dsRNA viruses • Class IV: positive-sense (+) ssRNA viruses • Class V: negative-sense (-) ssRNA viruses • Class VI: RNA viruses that reverse transcribe • Class VII: DNA viruses that reverse transcribe 15 16 DNA viruses (Class I, II, and VII) Class I: dsDNA • DNA viruses with a dsDNA genome have a genome exactly the same as the host cell that they are infecting. Therefore, many host enzymes can be utilized for replication and/or protein production. • The flow of information follows a conventional pathway: dsDNA → m R N A → protein. • DNA-dependent RNA-polymerase producing the mRNA and the host ribosome producing the protein. • The genome replication, dsDNA → dsDNA, requires a DNA-dependent DNApolymerase from either the virus or the host cell. Example like: bacteriophages T4 and lambda 17 18 DNA viruses (Class I, II, and VII) • Class II: ssDNA • ssDNA viruses can either have the same base sequence as the mRNA (plus-strand DNA) or be complementary to the mRNA (minus-strand DNA). • For the plus-strand DNA viruses, the complementary to the viral genome must be manufactured first, forming a double-stranded replicative form (RF). This can be used to both manufacture viral proteins and as a template for viral genome copies. • For the minus-strand DNA viruses, the genome can be used directly to produce mRNA but a complementary copy will still need to be made, to serve as a template for viral genome copies. Example: Parvoviruses 19 Class II 20 DNA viruses (Class I, II, and VII) • Class VII: DNA viruses that use reverse transcriptase • The viruses of this class has a DNA genome that is partially double-stranded, but contains a single-stranded region. • After gaining entrance into the cell’s nucleus, host cell enzymes are used to fill in the gap with complementary bases to form a dsDNA closed loop • Gene transcription yields a plus-strand RNA known as the pregenome, as well as the viral enzyme reverse transcriptase, an RNA-dependent DNApolymerase. • The pregenome is used as a template for the reverse transcriptase to produced minus-strand DNA genomes, with a small piece of pregenome used as a primer to produce the double-stranded region of the genomes. Example: Hepadnaviruses 21 pregenome, Reverse transcriptase 22 RNA viruses (Class III, IV, V, VI) • RNA Genome strategies • RdRp: virus encoded for RNA dependent RNA polymerase • RT: Virus encoded Reverse Transcriptase 23 RNA viruses (Class III, IV, V, VI) Class III: dsRNA • Double-stranded RNA viruses infect bacteria, fungi, plants, and animals, such as the rotavirus that causes diarrheal illness in humans. • Cells do not utilize dsRNA in any of their processes and have systems in place to destroy any dsRNA found in the cell. Thus the viral genome, in its dsRNA form, must be hidden or protected from the cell enzymes. • Cells also lack RNA-dependent RNA-polymerases, necessary for replication of the viral genome so the virus must provide this enzyme itself. The viral RNA-dependent RNA polymerase acts as both a transcriptase to transcribe mRNA, as well as a replicase to replicate the RNA genome. 24 Class III: dsRNA (Continue..) • Messenger RNA is transcribed from the minus-strand of the RNA genome and then translated by the host ribosome in the cytoplasm. • Viral proteins aggregate to form new nucleocapsids around RNA replicase and plus-strand RNA. • The minus-strand RNA is then synthesized by the RNA replicase within the nucleocapsid, once again insuring protection of the dsRNA genome. 25 RNA viruses (Class III, IV, V, VI) • Class IV: +ssRNA • Viruses with plus-strand RNA can use their genome directly as mRNA with translation by the host ribosome occurring as soon as the unsegmented viral genome gains entry into the cell. • One of the viral genes expressed yields an RNA-dependent RNApolymerase (or RNA replicase), which creates minus-strand RNA from the plus-strand genome. • The minus-strand RNA can be used as a template for more plusstrand RNA, which can be used as mRNA or as genomes for the newly forming viruses. such as coronoviridae, poliovirus 26 Class IV: +ssRNA 27 RNA viruses (Class III, IV, V, VI) • Class V: -ssRNA • Minus-strand RNA viruses include many members notable for humans, such as influenza virus, rabies virus, and Ebola virus. • Since the genome of minus-strand RNA viruses cannot be used directly as mRNA, the virus must carry an RNA-dependent RNA-polymerase (RdRp) within its capsid. • Upon entrance into the host cell, the plus-strand RNAs generated by the RdRp are used as mRNA for protein production. • When viral genomes are needed the plus-strand RNAs are used as templates to make minus-strand RNA. 28 29 RNA viruses (Class III, IV, V, VI) • Class VI: +ssRNA, retroviruses (With ds DNA intermediate) • Despite the fact that the retroviral genome is composed of +ssRNA, it is not used as mRNA. Instead, the virus uses its reverse transcriptase to synthesize a piece of ssDNA complementary to the viral genome. • The reverse transcriptase also possesses ribonuclease activity, which is used to degrade the RNA strand of the RNA-DNA hybrid. • The reverse transcriptase is used as a DNA polymerase to make a complementary copy to the ssDNA, yielding a dsDNA molecule. This allows the virus to insert its genome, in a dsDNA form, into the host chromosome, forming a provirus. • Unlike a prophage, a provirus can remain latent indefinitely or cause the expression of viral genes, leading to the production of new viruses. 30 31 PART 3: Viral genetic Variations • Viruses grow rapidly. • A single particle produces a lot of progeny. • DNA viruses seem to have access of proof reading enzymes, RNA viruses do not have Variations (or even appearing a new virus can occur as a result of): 1. Mutation 2. Recombination 32 1- Mutations Origin of mutations 1- Spontaneous mutation • Polymerase errors (Mutations rates are higher in RNA viruses because of the lack of proof reading enzymes) • Tautomeric form of bases 2- Physically or Chemically induced • Types of mutations: • Point (Silent, missense, nonsense mutation) • Insertion (Frameshift mutation) • Deletion 33 Phenotypic changes seen in virus mutants • Plagues size, may be larger or smaller than in the wild type virus • Drug resistance: the possibility of drug resistant mutant arising must always be considered • Enzyme-deficient mutant: some viral enzymes are not always essential and so we can isolate viable enzyme-deficient mutants. • They may be more virulent as a result of mutations • Attenuated mutants (cause milder symptoms and may be used to develop vaccine) 34 2- Recombination • Viral recombination occurs when viruses of two different parent strains coinfect the same host cell and interact during replication to generate virus progeny that have some genes from both parents. • Exchange information between two genomes 35 Thank you Questions 36