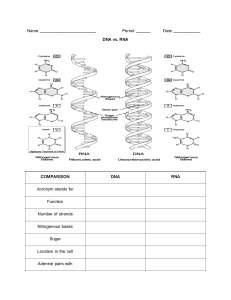
AP Biology Study Guide Table of Contents Table of Contents 2 Biochemistry 4 Water Properties 4 Macromolecules 4 Enzyme Kinetics 6 Thermodynamics 6 Cell Biology 8 Cell Structure 8 Cell Membrane 11 Cell Metabolism 12 Cell Signaling 16 Cell Cycle 17 Genetics 18 Heredity 18 DNA Replication 19 Genetic Expression/Regulation 20 Genomics 22 Viruses 23 Cancer 26 Embryonic Development 26 Biotechnology 27 Evolution 30 Evidence of Evolution 30 Microevolution 30 Speciation 32 Macroevolution 33 Biosystematics 34 Abiogenesis 34 Prokaryotic Diversity 35 Eukaryotic Diversity 39 Ecology 42 Ecosystem Ecology 42 Community Ecology 45 Population Ecology 47 Human Anatomy/Physiology 49 Organ Systems 49 Nervous System 49 Endocrine System 50 Digestive System 51 Excretory System 52 Respiratory System 52 Circulatory System 53 Immune/Lymphatic System 53 Biochemistry Water is polar & can form hydrogen bonds between its δ+ H and δ- O. Water Properties ● ● ● ● ● ● ● ● Co/Adhesion: hydrogen bonding to itself/other substances to hold a substance together Surface Tension: hydrogen bonded layers of water molecules increase tension on surface Capillary Action: Water moves up tubules opposing gravity due to cohesion/adhesion High Specific Heat/Heat of Vaporization: 1 cal/g⋅°C; able to conserve temperature (more energy used to break hydrogen bonds), stabilized internal temperatures for organisms & external environments & climate Evaporative Cooling: As water evaporates, the liquid remaining cools down (net loss of average kinetic energy); this preserves temperature & prevents organismal overheating Expansive Freezing: As water freezes, it forms a crystalline structure (hydrogen bonds), which has a lower density than liquid water; preserves aquatic environments (ice floats) Universal Solvent: Water’s polarity can dissolve polar substances, forming hydration shells around charged particles Neutral pH: 7 ([H+] = 10-7 at 25°C; Neutral); 2H2O → H3O+ + OH- (Dissociation) Essential elements are elements an organism needs to live and reproduce (20-25% of natural elements; O, C, H, N – 96%, Ca, P, K, S, etc. – 4%). Trace elements are required by organisms in smaller quantities (Fe, I [thyroid gland], etc.) Organic compounds consist of hydrocarbons: most(4)/strongest(electronegativity) covalent bonds. Macromolecules Monomer/Functional Groups Polymer Carbohydrates: Instant Energy, Structure (Polar), C/H/O [CH2O], Carbonyl (>C=O) & Hydroxyl (-OH) Groups, Monosaccharides (Ketone/Aldehyde if carbonyl in/at the end of C-skeleton) ● G3P ● (Deoxy)Ribose ● Glucose, Fructose, Galactose Bonded by glycosidic linkages Disaccharides ● Sucrose (G-F) ● Maltose (G-G) ● Lactose (gl-G) Oligosaccharides ● Glycolipids ● Glycoproteins Polysaccharides ● Cellulose (plant walls) ● Glycogen (animal energy) ● Starch (plant energy) ● Chitin (animal/fungi structure) Lipids: Stored Energy, Insulation, Structure (Nonpolar), C/H/O Triglycerides: Glycerol, 3 Fatty Acids - Methyl Groups Phospholipids: Choline, Phosphate (PO4) Group, Glycerol, Hydrophobic Tail (2 Fatty Acids - Methyl Groups) Steroids: 4 Carbon-fused rings (3 6-C, 1 5-C: 17 total C atoms) w/ Methyl (-CH3) Groups Bonded by ester linkages Triglycerides (Energy) ● Saturated (Solid) ● Unsaturated (Liquid) ● Trans (Hydrogenated) Phospholipids (Structure) ● Lipid Bilayer Steroids (Structure/Ligand) ● Cholesterol ● Estrogen ● Progesterone ● Androgens (Testosterone) Proteins: Catalysis, Storage, Hormonal, Contractile/Motor, Defensive, Transport, Receptor, Structural, C/H/O/N/S Amino Acids: Amino Group (NH3), H, R-Group, & Carboxyl Group (OH>C=O) bonded to Cα. ● Cysteine (contains Sulfhydryl (-SH) Groups) ● Methionine (begins polypeptides) Bonded by peptide bonds Primary: Amino Acid Sequence (20 amino acids) Secondary: 2D Structure (α-helix, β-sheets, h-bonds) Tertiary: 3D Structure (Hydrogen, Disulfide, Ionic Bonds, Hydrophobic/philic, Van der Waals Interactions) Quaternary: Multiple protein subunits (Noncovalent bonds) Globular (spherical), Fibrous (long fiber shaped) ● Pancreatic Lipase ● Casein, Ovalbumin ● Insulin/Glucagon ● Actin/Myosin ● Antibodies ● Aquaporins ● Tyrosine Kinase Receptors ● Collagen/Keratin Nucleic Acids: Store Genetic Information, Energy, Catalysis, C/H/O/N/P Nucleotide Monomer (5-Carbon Sugar, Phosphate (-PO4) Group, Nitrogenous Base) Nucleoside is sugar ([Deoxy]ribose) and Nitrogenous Base Purines have two N-rings (Adenine, Guanine) Pyrimidines have one N-ring Bonded by phosphodiester bonds ● DNA (Deoxyribose) ● RNA (Ribose) ● ATP (Adenine) ● GTP (Guanine) Methyl (-CH3) Groups bonded to DNA decrease expression, Acetyl Groups bonded to histones increase expression (Thymine, Uracil, Cytosine) (O=C-CH3) Enzyme Kinetics ● ● Anabolic pathways build polymers and use energy (condensation/dehydration synthesis) ○ Endergonic: Nonspontaneous; ΔG > 0 (Energy Absorbed) ○ Dehydration Synthesis/Condensation reactions remove H2O, form covalent bond Catabolic pathways break polymers and release energy (hydrolysis) ○ Exergonic: Spontaneous; ΔG < 0 (Energy Released) ○ Hydrolysis reactions add H2O, cleave (break) covalent bond Catalysts reduce reaction activation energy (main biological catalysts are ribozymes & enzymes) of specific reactions (enzyme-substrate complex forms as a transition state (substrate-induced fit) on the active site) in 4 ways: 1. 2. 3. 4. Orient substrates Strain substrate bonds Create a favorable microenvironment Covalently bond w/substrate Local Conditions on Enzyme Activity ● ● ● ● ● ● ● Temperature: Increases reaction (more kinetic energy) until enzyme denatures ○ Denaturation: Tertiary/Secondary structure (noncovalent interactions) is broken pH: Moderate, optimal pH, variation causes denaturation of the enzyme Compartmentalization: Enzymes are grouped with other enzymes for reactions in favorable environments (temperature, pH, etc.) Cofactors/Coenzymes: Metals (Inorganic)/Vitamins (Organic), assist in the reaction Enzyme Inhibitors: Covalent is irreversible, Noncovalent Interactions are reversible ○ Competitive Inhibitors: Mimic the substrate, compete for the active site ○ Noncompetitive Inhibitors: Bind to another site, modify active site shape ■ Allosteric Regulation: Allosteric Activators stabilize the active form, Allosteric Inhibitors stabilize the inactive form ● Positive Feedback: Reaction product stimulates enzyme ● Negative Feedback: Reaction product inhibits enzyme Substrate Concentration: Affects enzyme activity logarithmically ○ Cooperativity: One substrate activates multiple enzyme subunits’ active sites Enzyme Concentration: Affects enzyme activity linearly Phosphorylation reduces molecular stability, promoting reactions. Thermodynamics ● 0th Law: Transitive property of heat ● ● ● 1st Law: Conservation of energy 2nd Law: Entropy (chaos) increases with spontaneous reactions 3rd Law: Entropy is 0 at 0 K (no movement/uncertainty/chaos) Cell Biology Cell Structure All cells contain plasma membrane, ribosomes, cytosol, & DNA (nucleus/nucleoid). Prokaryotes (Bacteria/Archaea) are smaller (0.1-5 μm) w/o membrane-bound organelles, Eukaryotes (Eukarya) are larger (10-100 μm) w/ membrane-bound organelles Endomembrane, Endosymbionts, Cytoskeleton, Eukaryotes, Plants, Animals, Prokaryotes, All Organelle Description/Function Nucleus DNA/Nucleolus surrounded by double membrane nuclear envelope Nucleoid Region in prokaryotes where DNA is found (no compartmentalization) Nucleolus Synthesis site for rRNA and ribosomal subunits Ribosomes Protein synthesis sites (on ER (secretion) or in cytosol (for cell use)) Vesicles Sacs of membranes, transport chemicals through the cell Endoplasmic Reticulum (ER) Smooth (w/o ribosomes) synthesizes lipids, metabolizes carbs, detoxifies drugs, stores Ca+; Rough synthesizes secretory proteins & membranes Golgi Apparatus Modifies proteins, synthesizes glycans/carbs, and sorts/secretes products Lysosome Hydrolytic enzymes surrounded by membrane; breaks down ingested substances (food vacuole) and recycles damaged organelles (apoptosis) (Central) Vacuole Large vesicle; digestion, storage, waste disposal, osmoregulation Peroxisome Single membrane w/ enzymes that produce hydrogen peroxide and water Mitochondrion Double membrane (w/ cristae); aerobic cellular respiration Chloroplasts Double membrane around stroma and thylakoids; photosynthesis (plastids) Amyloplasts Colorless, Stores starch (amylose) in roots Chromoplasts In fruits/flowers, give orange/yellow color Cell Wall Polysaccharide outer layer, exerts turgor pressure on the cell, keeps shape; primary cell walls of adjacent cells connected by middle lamella pectins (sticky polysaccharide) (w/ cell junctions called plasmodesmata); after maturation, cells may strengthen or add secondary cell wall behind primary Cytoplasm Fluid within the cell (mostly water - cytosol) Cytoskeleton Fibers in cytoplasm; support cell structure, tracks for vesicle movement, manipulates membrane for endocytosis & motility; Microfilaments (actin) bear tension for cell structure & w/ myosin provide motility; Intermediate Filaments are more permanent for cell shape & organelle anchorage Centrosomes Microtubule organizing complex (2 centrioles, duplicate for cell division) Flagella/Cilia Microtubules (2*9+2 pattern connected by motor protein dyneins) for movement (few, long, undilating/many, short, dynamic); single nonmotile primary cilium for signaling; anchored by basal body (3*9+0) (in animal sperm, flagellum’s basal body becomes centriole); Bacterial flagella different Extracellular Matrix Fibrous collagen glycans support cell structure, adhesion, movement, and regulation (fibronectin connects complex to membrane integrins for control) Cell Membrane Lipid bilayer surrounding the cell w/ proteins & glycans for homeostasis Microvilli Membrane projections that increase cell surface area Cell Junctions Gap Junctions/Plasmodesmata allow macromolecules to pass between cells, Tight Junctions prevent extracellular fluid leakage, Desmosomes allow for maintaining structure in stretching tissue (Anchoring Junctions) Cell Membrane Cell Membrane is a selectively permeable (small nonpolar molecules pass easiest (N2, O2, CO2, Hydrocarbons), ionic/large/polar molecules utilize transport proteins) fluid mosaic (dynamic structure with moving phospholipids/proteins laterally). ● ● ● ● Phospholipids are amphipathic: both hydrophobic/hydrophilic ○ Cholesterol maintains fluidity in animal cells ■ Restrains movement at high temperatures (Lowering Fluidity) ■ Disrupts packing at lower temperatures (Lowering Viscosity) Integral Proteins span the membrane Peripheral Proteins are outside the membrane Protein Functions: ○ Transport (Aquaporins, Na+/K+ Pump) ■ Channel Proteins act as a tunnel, no conformational change ■ Carrier Proteins change conformation for transport ○ Enzymatic Activity (Metabolism, ATP Synthase) ○ Extracellular Matrix/Cytoskeleton Integration (Coordination) ○ Cell-Cell Recognition (Oligosaccharide Recognizers) ○ Intercellular Joining (Gap/Tight Junctions) ○ Signal Transduction (Tyrosine Kinase Receptors) Passive Transport does not expend energy ● ● ● Diffusion is the net movement along a concentration gradient (high to low concentration) ○ Tonicity is a solution’s solute concentration relative to another ■ Hypertonic is greater solute concentration (net diffusion inside) ■ Hypotonic is lower solute concentration (net diffusion outside) ■ Isotonic is similar solute concentration (no net diffusion) Facilitated Diffusion is diffusion with transport proteins Osmosis is the diffusion of water (water potential gradient) ○ Water Potential is the potential energy of water (Ψ=Ψp+Ψs+Ψg+Ψm) ■ Ψp is pressure potential (closed systems only, only to cells) ■ Ψs=-iCRT is solute/osmotic potential ● i is Ionization constant (how many resulting molecules) ● C is solution molarity (mol/L) ● R is pressure constant (0.0831 (bar∙L)/(mol∙K)) ● T is temperature/kinetic energy (K) ■ Ψg is gravity potential ■ Ψm is matrix potential (adhesive intermolecular forces) ○ Osmoregulation is the control of solute concentrations/water balance ■ Osmoconformers: Isosmotic with environment (marine animals) ■ Osmoregulators: Control internal osmolarity (freshwater/terrestrial) ○ Osmolarity: Total solute concentration (Molarity, Moles per Liter of Water) ■ Isosmotic: Same osmolarity (no net osmosis) ■ Hypoosmotic: Lower osmolarity (net osmosis outside) ■ Hyperosmotic: Higher osmolarity (net osmosis inside) Net Diffusion Animal Cell Plant Cell Outside the Cell Crenated (Shriveled) Plasmolysis No Net Diffusion Normal Flaccid Inside the Cell Cytolysis (Lysed) Turgid (Normal) Active Transport expends energy, net movement against a concentration gradient (low to high) ● ● Membrane Potential is potential energy difference across a membrane ○ Electrochemical Gradient is both concentration and membrane potential forces Electrogenic Pumps generate voltage (membrane potential) ○ Na+/K+ Pump exchanges Na+ for K+ creating an electrochemical gradient ○ H+ Pump actively transports protons creating an electrochemical gradient ○ Cotransport uses the active transport of one molecule to power that of others ■ Sodium/Glucose Transporters (Na/K Pump) ■ Sucrose/H+ Transporters (Proton Pump) Bulk Transport moves large particles ● ● Exocytosis is the fusion of vesicles from inside the cell to membrane (bulk transport out) Endocytosis is the fusion of vesicles from outside the cell to membrane (bulk transport in) ○ Phagocytosis is endocytosis of particles with the use of pseudopodia (food vacuole) ○ Pinocytosis is endocytosis of extracellular fluids resulting in coated vesicles ○ Receptor-Mediated Endocytosis is pinocytosis of specific molecules (bind to receptors on the membrane triggering vesicle formation) Cell Metabolism Primary Catabolism (Glycolysis/Fermentation) 1. Glycolysis: Glucose is split; the products are oxidized/rearranged to result as pyruvates (net 2 ATP by substrate level phosphorylation, occurs in cytoplasm) a. Energy Investment Phase: Glucose is phosphorylated twice (2 ATP) to form Fructose 1,6 Bisphosphate which is split into G3P and DHAP b. Energy Payoff Phase: G3P is oxidized/dephosphorylated forming 2 Pyruvates, 2 H2O, 4 ATP, & 2 NADH/2 H+ c. Regulation: Phosphofructokinase is stimulated by AMP, inhibited by ATP/Citrate 2. Lactic Acid Fermentation: directly reduces 2 pyruvates into 2 lactates (cytoplasm) a. Oxidizes 2 NADH/2 H+ into 2 NAD+ (recycled to glycolysis) b. Lactic Acid is a toxin, bonds with O2 and is transported to the liver 3. Alcoholic Fermentation: directly reduces 2 pyruvates into 2 ethanol (cytoplasm) a. Releases 2 CO2 b. Oxidizes 2 NADH/2 H+ into 2 NAD+ (recycled to glycolysis) Secondary Catabolism (Aerobic/Anaerobic Cellular Respiration) Aerobic Carbohydrate Metabolism: C6H12O6 + 6O2 → 6CO2 + 6H2O + ~33 ATP (686 kcal/mol) 1. Glycolysis: Glucose is split; the products are oxidized/rearranged to result as pyruvates (net 2 ATP by substrate level phosphorylation, occurs in cytoplasm) 2. Pyruvate Oxidation: Pyruvate is transported into the mitochondrial matrix, oxidized releasing CO2 and reducing NAD+ into NADH + H+, and bonded to a CoA group forming Acetyl-CoA (2x) 3. Krebs (Citric Acid) Cycle: Full oxidation of pyruvates releasing 2 CO2 per pyruvate (2 ATP by substrate level phosphorylation, occurs in the mitochondrial matrix, 2x) a. Primary Oxidation: Acetyl-CoA loses the CoA-SH and is oxidized releasing 2 CO2 and reducing 2NAD+ into 2 NADH + 2 H+ b. ATP Formation: CoA group is readded and removed from α-ketoglutarate phosphorylating GDP into GTP which can be used to phosphorylate ADP→ATP c. Regeneration: Succinate is further oxidized to form oxaloacetate, reducing NAD+ into NADH + H+ & FAD into FADH2 4. Oxidative Phosphorylation: Electron transport chain of carrier electrons, chemiosmosis generates energy (32-34 ATP by oxidative phosphorylation, occurs in inner membrane) a. Electron Transport Chain: e- from NADH/FADH2 are transferred through the cytochrome complex pumping H+ (electrochemical gradient) and finally binding to O­2­, producing H­­2O b. Chemiosmosis: The flow of H+ through ATP Synthase along their electrochemical gradient, generating (~32-34) ATP from ADP + Pi Aerobic Lipid Metabolism: 1. Triglyceride Formation: Bile/Lipase hydrolyze lipids to form triglycerides 2. Glycerol is substituted into glycolysis (~38-40 ATP produced from the entire metabolism because glycolysis energy input phase missed) 3. Beta oxidation: Converts fatty acids into 2 Acetyl CoA (mitochondrial matrix) a. Reduces 2 NAD+ into 2 NADH + 2 H+ b. Releases 2 CO2 c. Requires 2 CoA (Recycled from Krebs Cycle) 4. Carbohydrate Metabolism: CTC/ETC, ~28-30 total ATP produced Aerobic Protein Metabolism: 1. Amino Acid Formation: Polypeptides are hydrolyzed to amino acids (pepsin/peptidases) a. Breaking the peptide bonds results in products that can be metabolized for ~28-30 total ATP produced (added in the end of glycolysis) 2. Amine Metabolism: Amine groups (hydrolyzed by aminopeptidase) are converted into urea at the liver and excreted at the kidneys a. Requires CO2 b. Some molecules enter the CTC and are further metabolized (~22-24 ATP) 3. R-Group Metabolism: R groups directly enter the Krebs Cycle a. ~22-24 total ATP produced 4. Carboxyl Metabolism: Carboxyl group enters pyruvate oxidation a. ~28-30 total ATP produced Anaerobic Cellular Respiration: 1. Glycolysis: Splits glucose into two pyruvates 2. Krebs (Citric Acid) Cycle: Full oxidation of pyruvates releasing CO2 3. Nonoxidative Phosphorylation: Electron transport chain of carrier electrons, chemiosmosis generates energy (32-34 ATP by oxidative phosphorylation, occurs in inner membrane) a. Electron Transport Chain: e- from NADH/FADH2 are transferred through the cytochrome complex pumping H+ (electrochemical gradient) and binding to the oxidizing agent (final electron acceptor): i. Nitrate Reduction: NO3 → NO2 ii. Nitrite Reduction: NO2 → N2 iii. Sulfate Reduction: SO4 → H2S iv. Sulfur Reduction: S → H2S b. Chemiosmosis: The flow of H+ through ATP Synthase along their electrochemical gradient, generating (~32-34) ATP from ADP + Pi Cellular Respiration Regulation: Phosphofructokinase (enzyme in Glycolysis) is allosterically stimulated by AMP, inhibited by ATP/Citrate (from Oxidative Phosphorylation/Krebs Cycle). Primary Anabolism (Chemo/Photosynthesis) Photosynthesis: 6CO2 + 6H2O + Solar Energy (686 kcal/mol) → C6H12O6 + 6O2 1. Light Reactions: Solar energy excites electrons in pigments which are used in an electron transport chain to generate chemical energy, splitting water/releasing oxygen (thylakoid) a. Linear Electron Flow: Electrons are finally transferred to NADP+, forming NADPH i. Photosystem II (PSII): Photons excite pigments in PSII’s light-harvesting complex, nearby pigments are simulated as others return to their ground state, P680 chlorophyll a pigments finally lose their electrons to a primary electron acceptor, triggering photolysis of H2O to compensate for the lack of electrons, releasing O2 and H+ for the electron transport chain/chemiosmosis ii. Electron Transport Chain: e- from the primary electron acceptor are transferred through the phytochrome complex pumping H+ (electrochemical gradient) iii. Chemiosmosis: The flow of H+ through ATP Synthase along their electrochemical gradient, generating ATP from ADP + Pi iv. Photosystem I (PSI): Photons excite pigments in PSI’s light-harvesting complex, nearby pigments are simulated as others return to their ground state, P700 chlorophyll a pigments finally lose their electrons to a primary electron acceptor, P700+ accepts the electrons from the electron transport chain v. Second Electron Transport Chain: e- from PSI’s primary electron acceptor are transferred through the phytochrome complex vi. NADP+ Reduction: NADP+ reductase reduces NADP+ into NADPH w/ ETC 2 e-/H+ b. Cyclic Electron Flow: Electrons are recycled in the photosystems/ETC, generating ATP i. Photosystem I (PSI): Photons excite pigments in PSI’s light-harvesting complex, nearby pigments are simulated as others return to their ground state, P700 chlorophyll a pigments finally lose their electrons to a primary electron acceptor, P700+ accepts the electrons from the electron transport chain ii. Electron Transport Chain: e- from the primary electron acceptor are transferred through the phytochrome complex pumping H+ (electrochemical gradient) iii. Chemiosmosis: The flow of H+ through ATP Synthase along their electrochemical gradient into the thylakoid lumen, generating ATP from ADP + Pi 2. Calvin Cycle: Chemical energy from the light reactions reduces 3 CO2 to G3P (stroma) a. Carbon Fixation: Rubisco bonds 3 CO2 to 3 RuBP forming 6-PGA i. C3 Plants: CO2 is fixed into 6-PGA (3 Carbon, Calvin Cycle) ii. C4 Plants: CO2 is fixed into oxaloacetate (moved to bundle-sheath cells) iii. CAM Plants: CO2 is fixed into malic acid (used during day: Calvin Cycle) b. Reduction: 6-PGA is reduced to 6-G3P using 6 ATP and 6 NADPH to 1-G3P c. Regeneration: 5-G3P are rearranged to form 3 RuBP using 3 ATP 3. Photorespiration: Rubisco binds to O2 rather than CO2, releasing CO2 and using ATP Chemosynthesis: CO2 + 2H2S → [CH2O] + H2O + 2S; sulfur/methanogens Cell Signaling Signaling Summary Examples Synaptic (Neuro) Neurotransmitters at synapses Nervous system synapses Paracrine Neighboring Cells Growth hormone following injury Endocrine Through blood Pituitary gland hormones Exocrine Through ducts Pancreas Autocrine Self-signaling Apoptosis (programmed cell death) Pheromones Outside the organism to another Fruits promoting maturation Cell-Cell Recognition Membrane glycans recognition Immune system recognition 1. Reception: ligand (lipid/protein) is first recognized and triggers pathway a. Lipids are membrane soluble; bind to intracellular receptors; hormone-receptor complex binds to nuclear DNA (Transcription Factor) causing gene expression b. Proteins aren’t membrane-soluble; bind to membrane receptors which trigger internal response i. G-Protein Coupled Receptor: ligand binds, G-protein activates, moves, and activates membrane enzyme (i.e. adenylyl cyclase): response (i.e. cAMP) ii. Tyrosine Kinase Receptor: 2 ligands bind, monomers dimerize, 6 ATP phosphorylate 6 tyrosines, relay proteins bind and trigger responses iii. Gated Ion Channel: ligand binds, channel opens/ions diffuse, relay response c. Signaling specificity to only cells expressing the receptor gene protein 2. Transduction: relays protein reception to cellular targets (by phosphorylation cascade) a. Secondary Messengers are relay molecules that trigger transduction i. Adenyl Cyclase (ATP → Cyclic AMP [cAMP]) activates Protein Kinase A ii. Phospholipase C cleaves PIP2 leaving Diacylglycerol (DAG) [second messenger] and Inositol Triphosphate (IP3) which bond to an ER Gated Ca2+ Channel, releasing them into cytosol as secondary messengers b. Secondary messengers can activate protein kinases (transfer Pi from ATP to other proteins), creating a phosphorylation cascade (secondary messenger → PK1 → PK2 → … → Target Response Protein) i. Protein Phosphatases dephosphorylate kinases, regulating response ii. Signal Amplification: enzymes use many substrates (i.e. 1PK1→10PK2) iii. Signaling Efficiency affected by Scaffolding Proteins holding relay proteins 3. Response: transcription factors (gene expression/protein synthesis) & cellular responses (activation of enzymes, other signaling pathways, cell division, ligand secretion, etc.) Cell Cycle Controlled by specific cyclin-dependent kinases bonding to cyclin (CDK-Cyclin complex can prevent/progress cell cycle by triggering cellular responses such as being transcription factors). 1. Interphase: Cell Growth/Development a. G1 Phase: First Gap, Growth i. G1 Checkpoint: Usually signals whether cell division will occur or not 1. Growth Factors stimulate cell division via signal transduction 2. Density-Dependent Inhibition: Crowded cells stop dividing 3. Anchorage Dependence: Cells need substratum to divide 4. G0 Phase: Non Dividing phase; fails first checkpoint (neurons) b. S Phase: Synthesis, DNA Replication c. G2 Phase: Second Gap, Growth/Preparation of Division, Organelle Division i. G2 Checkpoint: Checks for DNA damage/repair & other things 2. Mitotic Phase: Cellular/Nuclear Division (promoted by MPF & CDK-Cyclin abundance) a. Mitosis: Division of Genetic Material (Somatic Cells) i. Prophase: Chromosome condensation, spindle formation 1. Condensin II creates a central pillar scaffold for DNA to loop around and coil; Condensin I binds outside the middle scaffold and coils those DNA loops further ii. Prometaphase: Nuclear envelope disintegrates, kinetochore attachment iii. Metaphase: Chromosomes lineup on Spindle Metaphase Plate 1. M Checkpoint: Cells are arrested in metaphase until all chromosomes attach to the spindle (controlled by anaphase promoting complex protein, which triggers move to anaphase) iv. Anaphase: Cohesins cleaved, microtubules pull chromosomes/elongate cell v. Telophase: Endomembrane system reforms, chromosome uncoiling b. Cytokinesis: Cytoplasmic division; Cleavage Furrow (Animals)/Cell Plate (Plants) c. Meiosis: Genetic Material Reduction (Germ Cells) i. Prophase: Mitosis Prophase/Prometaphase, Synapsis/Crossing Over, Tetrads (w/ Chiasmata, crossover points hold homologs together) ii. Metaphase: Homologs Lineup on Spindle Metaphase Plate Independently iii. Anaphase: Cohesins are cleaved (except centromere to keep chromatids together), Microtubules separate & pull chromosomes/elongate cell iv. Telophase/Cytokinesis: Complete duplicated haploid genome (nuclear envelope & decondensation occur in some organisms), Cytokinesis v. Another meiotic division similar to mitosis (4 non-identical gametes) d. Fertilization: Random fusion of 2 gametes & their nuclei to form a zygote e. Binary Fission: Chromosome replication, membrane cleavage/division (Bacteria) Genetics Heredity Characters/Genes: heritable features with traits/alleles as variants for the phenotype/genotype Testcross is between unknown dominant phenotype (homo/heterozygous) and recessive phenotype Mendel’s Laws of Inheritance: ● ● ● Dominance: Dominant alleles determine the phenotype if alleles at the gene locus differ Segregation: Alleles separate during gamete formation (meiosis) into separate gametes Independent Assortment: Segregation of alleles on different chromosomes is an independent event; genes aren’t required to inherited together and do so by probability ○ Linked genes are on the same chromosome (AB/ab); lead to more parentals in F1 testcrosses, recombinants arise from crossing over, shuffling the linked alleles. ○ Recombination frequency is recombinants/total offspring; genetic maps (1%/mu) ■ Genetic map frequencies don’t add up because of interference (multiple crossovers on a chromosome); don’t represent actual distances (relative) Nonmendelian Inheritance Degrees of Dominance 1. Complete: Dominant completely masks recessive (mendelian traits like pea color) a. Dominant alleles code for proteins, recessive alleles are nonfunctional proteins 2. Incomplete: Traits are mixed and expressed (blending hypothesis; snapdragon color) a. Both dominant alleles code for proteins, phenotype varies in smaller groups 3. Codominance: Traits are expressed distinctly from each other (MN/AB Blood Group) a. Both dominant alleles code for proteins, phenotype varies in larger groups Multiple alleles: More than 2 alleles (ABO blood system has alleles IA, IB, & i) Pleiotropy: A gene has multiple phenotypic effects (Sickle-Cell Disease & Cystic Fibrosis) Epistasis: One gene affects the expression of another (Black/Brown/Yellow Labradors) Polygenic Inheritance: Multiple genes affect one phenotype, opposite of pleiotropy (human height/skin color) Multifactorial Traits: genes and environment influence phenotype (human nutrition & height) Genomic Imprinting: Phenotype variation depending on gene parent (gene silencing, methylation) Organelle (Cytoplasmic/Extranuclear) Genes are found in plastids/mitochondria in egg cytoplasm only (those from sperm are destroyed in zygote); therefore, show maternal inheritance patterns. Wild Type is the most common trait in natural populations (mutant first letter (white); w+, w) Chromosomal Theory of Inheritance: Genes housed on chromosomes, anaphase I in meiosis is segregation, metaphase I independent assortment, random fertilization in crosses. Chromosomes consist of DNA coiled around 8 histones (20% of amino acids are positively charged (lysine/arginine), H2A/B/3/4; H1 on +30nm level) twice to form a nucleosome. Nucleosomes are separated by linker DNA. Chromosomal Systems of Sex Determination ● ● ● ● ● XY System: Mammal Males (XY), Females (XX); sex determined sperm carries X or Y ○ Y Chromosome contains SRY gene which triggers male testicular development; Female development begins with gene WNT4 (Chromosome 1) which triggers female ovary development ○ In somatic cells, 1 X chromosome is randomly inactivated to form a Barr Body (DNA methylation, XIST becomes active on the future Barr Body, lncRNA coded from that gene attach/cover the chromosome, preventing expression) X0 System: Insect Males (X0), Females (XX); sex determined by sperm carries X or not ZW System: Bird Males (ZZ), Females (ZW); sex determined by egg carries Z or W Haplo-Diploid System: Bees/Ants Males (Haploid), Females (Diploid); sex determined by fertilization (females) or not (males) Other: ratio of sex chromosomes to autosomes or the environment, etc. X linked traits (on X-chromosome) in males are hemizygous; females: homozygous & heterozygous Nondisjunction (failure for chromosomal separation in anaphase I/II) results in aneuploidy, an abnormal number of a chromosome (monosomy for missing 1, trisomy for 1 extra chromosome) Chromosomal Abnormalities ● ● ● ● Deletions: Chromosomal fragment is lost Duplication: Chromosomal fragment is repeated in the same orientation Inversions: Chromosomal fragment is repeated in a different orientation Translocations: Chromosomal fragment is moved between nonhomologous chromosomes ○ Reciprocal translocation is an exchange & transfers 2 segments ○ Nonreciprocal translocations just transfer one segment w/o another in return Central Dogma of Molecular Biology: DNA (Replicates) → RNA → Proteins DNA Replication 1. Topoisomerase unwinds, helicase separates leading/lagging strands at recognized origin(s) of replication, single-strand binding proteins keep strands separate 2. RNA Primase synthesizes RNA, DNA Polymerase III (ε) continually synthesizes the leading (5’- 3’) [uses nucleotides (dNTP), releases a Pyrophosphate (2Pi), and hydrolyzes it to 2Pi for energy] 3. RNA Primase synthesizes RNA, DNA Polymerase 𝛼 synthesizes first stretch, DNA Polymerase III (𝛿) synthesizes an Okazaki fragment until it reaches the prior RNA primer, DNA Polymerase I (RNAse/𝛼) replace RNA Primer w/ DNA, DNA Ligase binds Okazaki fragments lagging strand (3’ - 5’) 4. The DNA replicated “bubbles” are bonded together by DNA Ligase, forming two daughter DNA molecules; Telomerase extends telomeres (has RNA molecule for DNA synthesis) DNA Repair ● ● ● Direct Reversal: Dimers by UV light, reversed photoreactivation by photolyase, not in humans, in bacteria, fungi, some animals. Excision: Nuclease cuts, DNA Polymerase I (𝛼) fill in the missing nucleotides, DNA Ligase bonds strands, in most organisms Postreplication (Translesion Synthesis): Gap is left at Okazaki fragment, filled in through recombinant homology directed repair (sequence from homolog) or error-prone non-homologous end joining repair (damaged strand as template) Genetic Expression/Regulation Bacteria do transcription/translation simultaneously, no RNA processing ● Operons: Genes in bacteria, consist of the promoter region, operator (where repressor, from regulatory gene, binds), and genes of interest typically all related to a single metabolic pathway (e.g., tryptophan synthesis). ○ Lac Operon: regulate lac genes (z,y,a), inducible (norm off, repressor binds to allolactose to inactivate it), positive regulation by cAMP bonded to CAP (more w/ low glucose, CAP/CRP is an activator & binds to an upstream site, used in many more operons) ○ Trp Operon: regulate trp genes (a-e), repressible (norm on, trp binds to repressor to activate it) Eukaryotes do RNA processing compartmentalized by nucleus ● ● Epigenetic Inheritance: Histone Acetylation (uncharged lysine residues) increases expression (DNA is more accessible); CpG site (Cytosine-Phosphate-Guanine) Methylation (5M-Cytosine) decreases expression (DNA less accessible, more tightly coiled, hydrophobic interactions) TADs (topologically associated domains: regions of chromatin loops) may extend from individual chromosome territories into specific sites in the nucleus (transcription factories inside the middle of nucleus for expressed genes, unexpressed genes on the outside of the nucleus) Transcription (RNA Polymerase synthesizes mRNA 5’-3’) TATA Box: Eu/Archaea Promoter (TATAWAW); W = A/T Pribnow Box: Bacterial Promoter: -10 (TATAAT); -35 (TTGACA) (σ TF binds) Specific Transcription Factors are activators or repressors, bind to DNA (homeodomain), general transcription factors and mediator proteins connect them to the promoter site (DNA Bending) ● ● ● ● ● ● RNA Poly I - All Eukaryotes - large r RNA Poly II - All Eukaryotes - pre-m, some sn/sno/mi RNA Poly III - All Eukaryotes - t, small r, some sn/mi RNA Poly IV - Plants - Some si RNA Poly V - Plants - for heterochromatin formation Bacteria have only 1 Type RNA Polymerase Rho Termination: Rho protein binds at RNA rut site, releases; Non-Rho: Rich C-G (3 H-Bonds), forms hairpin that stalls strand, downstream codes for U-A (weaker, 2 H-Bonds), strand separates RNA Processing (Post-Transcriptional, pre-mRNA→mRNA, Only Eukaryotes): ● ● ● ● 5’ Cap: Modified Guanine, 20-40 nucleotides 3’ Poly-A-Tail: 50-250 Adenine nucleotides ○ Polyadenylation Sequence: AAUAAA, polyadenylation tail in RNA processing RNA Splicing: spliceosomes (snRNP/snRNA) splice introns, leaving exons (code for protein domains) ○ Alternative Splicing: exon shuffle, new protein ○ U1 snRNP and BBP bind to the mRNA ○ U2 snRNP displaces Branchpoint Binding Protein (BBP) ○ Branch site (nucleophile) is extruded ○ snRNP complex creates a lariat shape to initiate a cut RNA Editing: Alters mRNA nucleotide sequence Small Noncoding RNAs: ● ● ● ● Micro RNA (miRNA): in exons/introns, multiple target mRNAs Small Interfering (siRNA): argonaute RISC complex, 1 target Piwi-interacting RNA (piRNA): heterochromatin transposons, reestablish appropriate DNA methylation in germ cells Long Noncoding RNAs: Example is XIST Gene, transcripts lead to lncRNAs which bind back to the X chromosome, causing heterochromatin formation and the chromosome inactivation Translation (mRNA to Polypeptide) 1. Initiation (Met, AUG, A site, 1 GTP, Small then large subunit bind) 2. Elongation (rRNA (made w/ snoRNA) 5’-3’, tRNA moves through A-P-E sites adding AA to chain, 2 GTP, one to recognize AA, other to translocate tRNA) a. Prokaryotes have 70s (50s/30s), Eukaryotes have 80s (60s/40s) ribosomes b. Wobble: G-U, I-A/U/C (assuming X-Y, Y-X pairing prior), flexible final pairing c. Aminoacyl-tRNA Synthetases bond tRNA to Amino Acids (hydrolyzing 1 ATP) 3. Termination (Stop Codon, Release Factor, peptide bond H2O, 2 GTP) Signal Peptide recognized by SRP, binds ribosome to ER (all translation begins in the cytosol); also targets polypeptides, to mitochondria, chloroplasts, the interior of the nucleus, and other organelles that are not part of the endomembrane system. Polyribosome: Multiple ribosomes translating an mRNA (bacteria couple transcription/translation) Post-Translational Modifications: carbs, lipids P-groups, fold, cleave/add, edit aa, quaternary structure, proteins tagged with ubiquitin are targeted by proteasomes and degraded Mutations: Silent: Same AA; Nonsense: Stop; Missense: Diff AA; Suppressor: fixes AA (Mutagens are physical/chemical elements that cause mutations like high radiation (X-Ray/UV), base analogs (like DNA bases but don’t pair properly), chemicals that interfere w/ DNA structure, etc.; many mutagens are carcinogens (cancer-causing chemicals)); Insertion/Deletions are frameshift mutations; have greater impact than nucleotide-pair substitution (one switched pair). Spontaneous Mutations: random errors in DNA replication that aren’t fixed (around 1 in 1010) Genomics Bacterial/Archaeal Genomes ~ 1-6 Mb (1,500 to 7,500 genes, high gene density); Eukaryotes much larger (5,000 genes for unicellular fungi (yeasts) to 40,000+ genes for multicellular; low gene density because of introns, alternative splicing instead, more DNA control regions, noncoding RNAs, etc.) Pseudogenes: genes that gained mutations and became nonfunctional over time (unique noncoding) Transposable Elements: jumping genes (cut-paste or copy-paste; both w/ transposases coded by the element), 75% of human repetitive DNA, found in prokaryotes/eukaryotes (Barbara McClintock: variegations color of the kernels on a corn cob); Transposons: DNA intermediate; Retrotransposons: RNA intermediate (transcription, reverse transcriptase creates dsDNA copy, enzyme catalyzes insertion; only copy-paste method, DNA is not cut at original site). Alu elements (transposable) are transcribed into RNA to maybe help regulate gene expression L1 retrotransposon may be critical for 1-2 cell embryo development by modifying chromatin structure Simple Sequence DNA consists of copies of tandemly repeated short sequences (STRs included) found in centromeres (structural/cell division) & telomeres (protects ends from joining to other chromosomes and degradation) (also organizes chromatin in the nucleus when found in other regions) Human Chromosome 2 is fusion of Chimpanzee chromosome 12/13 (telomere/unused centromere in center are evidence for this) Unequal Crossing Over leads to duplications and deletions for each chromosome (Transposable elements provide homologous regions for translocations/crossing over to occur) Template slippage during replication could lead to variable STRs (multiplicated regions) Multigene families: 2+ identical (tandemly clustered, RNA products such as repeated rRNA genes) or similar genes (in the same chromosomal region such as embryo/fetus/adult version of globin genes); arise from duplications, transpositions, & mutations (both enhancing functions for a new type of gene and inhibiting function resulting in a pseudogene) Exon duplication (multiple of one exon in one gene)/shuffling (multiple different exons from different genes) can lead to new enhanced proteins (such as TPA, which controls blood clotting) (Transposable elements can move genes/exons throughout the genome, resulting in genes scattered (combinatorial control in Eukaryotes)) Transcription factor genes are evolving the fastest as they control genetic expression Copy-Number Variants (CNVs): loci where some individuals have one or 3+ copies of a particular gene; Short Tandem Repeats: repeated units of 2-5 nucleotide sequences in specific regions of the genome; Single Nucleotide Polymorphism: base pair variation site in a species (genetic marker in disease alleles); Human genome evolution seen in CNVs, STRs, & SNPs. Viruses Viruses: single/double stranded DNA/RNA packaged in a protein coat (capsid, built from capsomeres); no metabolism/reproduction by itself (borrowed life) Types of Capsids: ● ● ● Helical Virus: Capsomeres arranged in helical shape forming a rod, nucleic acid coiled inside Adenovirus/Icosahedral Viruses: Infect respiratory tracts, 252 identical capsomeres in icosahedron formation, protein spikes at vertices, nucleic acid found inside Influenza Viruses: have phospholipid membranes with glycoproteins/proteins from host cell; eight double-helical RNA-protein complexes, each associated with a viral polymerase ● Bacteriophages (phages): infect bacteria, elongated icosahedral heads (w/ DNA) & have tail sheath/fibers that help with cellular attachment; T-even phages infect bacteria w/ their DNA with their tail injected Host range is the limited range of host species viruses can infect (membrane receptor specificity) DNA/RNA Viral Replicative Cycle 1. Viral Entrance (T-even inject DNA, others enter by receptor-mediated endocytosis or plasma membrane fusion, receptor proteins bind to virus glycoproteins, digestion of capsid leaves RNA) 2. Genome Replication (host cell uses DNA Polymerase/Primase/Ligase/etc. to replicate DNA genomes; RNA viruses use virally encoded RNA polymerases to replicate RNA genomes) 3. Protein Synthesis (host cell uses enzymes to transcribe/translate host cell DNA/RNA forming capsid proteins (cytosol)/glycoproteins (ER/Golgi), concurrent with genome replication) 4. Viral Assembly (viral genomes/capsid proteins (w/ viral RNA Polymerase) self-assemble; vesicles transport glycoproteins to cell membrane; capsid assemblies bud from studded cell membrane) Lytic Cycle: After viral assembly, viruses exit and lyse the cell (break open), virulent only lytic Lysogenic Cycle: Replication of the viral genome without cell lysis (both lysogenic/lytic viruses called temperate; prophage: viral DNA is incorporated in bacterial genome; genes inhibit transcription of each other (may cause different host phenotype); may become lytic if exposed to certain chemicals/radiation) Prokaryotic defense: restriction enzymes hydrolyze foreign DNA (not methylated like genome) Prokaryotic CRISPR Immune System: clustered regularly interspaced short palindromic repeats; genome consists of DNA from previous infections spaced by spacer DNA (reads the same backwards/forwards), Cas (CRISPR-associated) nucleases use processed RNA from CRISPR region to detect and cut foreign phage DNA, leading to degradation (genome adds copy). Most Plant Viruses are RNA helical/icosahedral capsid. Horizontal Transmission: external source to plant (i.e., herbivore, human, etc.); Vertical Transmission: Parent to progeny Retroviruses use reverse transcriptase to generate dsDNA copy from ssRNA genome (incorporated into host genome as provirus, similar to lysogenic cycle, then gene expression of viral RNA, virus form). Viruses cause cellular damage by releasing hydrolytic lysosomal enzymes, stimulating the release of toxins, etc. Symptoms result from body defense and cellular replication of infected tissue. Vaccine: a harmless derivative of a pathogen (killed or weak pathogen, toxin, protein, or specific DNA/RNA) that stimulates the immune response. Prions: infectious misfolded versions of brain proteins that cause neurodegenerative diseases by causing functioning brain proteins to misshapen and aggregate (Alzheimer’s and Parkinson’s disease). Pandemic: Global Epidemic: widespread disease outbreak (mutation, easy human transmission) Emerging viruses may arise because RNA has a high mutation rate (viral RNA polymerases don’t proofread in replication, new viruses may affect people immune to the previous virus), due to high human travel throughout the world, or because virus originates from animals (75% viruses). Cancer Uncontrolled cell division; caused by loss of dependence on growth factors/abnormal cell cycle control system (genetic mutations); Benign Tumors don’t spread, Malignant Tumors metastasize Proto-oncogenes: regular growth ● ● ● Tumor-suppressor genes proteins prevent uncontrolled growth Ras Gene G-Protein sends signal from growth factor to transduction p53 Gene tumor suppressor (ATM protein kinase activates), halts cycle (activates p21 gene), repair, apoptosis (programmed cell death by autocrine cell signaling) Oncogenes: cancer mutated, changed by epigenetic changes, translocations, gene amplification, point mutations, or viral DNA integration (abnormally high activity of normally low expressed genes or knockout of tumor-suppressor genes) Small molecules can combat cancer by suppressing activity of oncogenes; Cultured cells can mass produce proteins that can act as steroids lacking in disorders (Insulin/Growth Hormone/TPA) Embryonic Development Cell Differentiation: cells are more specialized in structure/function (reversible)” ● ● ● ● Stem Cells are unspecialized, replicate indefinitely, and can differentiate into multiple types Totipotent: cell can differentiate into all embryonic/extraembryonic cells (new organism, only Embryonic Stem Cells before blastula/blastocyst and in zygote/morula) ○ Nuclear potential (ability for a cell to develop into a new organism) decreases with age (embryonic development/cell differentiation) in animals Pluripotent: cell can differentiate into all somatic/germ cells (Embryonic Stem Cells at blastula/blastocyst phase) Multipotent: cell can differentiate into a limited number of cell types (Adult Stem Cells) Determination: point at which the cell is irreversibly committed to becoming a specific type of cell: ● ● Cytoplasmic Determinants: Cytoplasmic molecules from the egg of the zygote that regulate genetic expression (i.e., Transcription Factors, Proteins, RNA, etc.) ○ Maternal Effect Genes: Cytoplasmic determinants involved in axis establishment coded from the mother’s genome ○ Morphogen Gradient Hypothesis: gradients of substances called morphogens (such as Bicoid mRNA) establish an embryo’s axes and other features of its form (such as Anterior end of Drosophila) Induction: Cell signaling causes changes in genetic expression & phenotype ● Master Regulatory Proteins control cell differentiation as they act as transcription factors and activate tissue specific genes/TFs while also activating their own gene (Positive Feedback, MyoD stimulates itself in myoblast determination/differentiation) Morphogenesis: the development of an organism’s structure/form ● ● Pattern Formation: spatially organizing the tissues and organs in an organism during embryonic development, controlled by homeotic (hox in animals) genes contain a homeobox coding for a homeodomain (which is the DNA binding region of the final transcription factor) in the final transcription factor (differing hox gene expression and regions → major anatomical differences) In animals w/ true tissues, embryo layers into Ectoderm (integumentary & possibly central nervous system), Endoderm (digestive system and other organs such as liver/lungs), &/or Mesoderm (muscles and organs between digestive/integumentary system, in bilaterians) Biotechnology Genetic Engineering: the direct manipulation of genes for practical purposes Genetic Therapy: introducing new genes; primarily used to cure tough-to-cure diseases Nucleic Acid Hybridization: complementary base pairing between similar/different nucleic acids Polymerase Chain Reaction (PCR): 1. Denaturation: Heat DNA to separate hydrogen bonds 2. Annealing: Cool for DNA primers (40-60% CG, not complementary, 15+ nucleotides) 3. Extension: Taq/Pfu Polymerase; f(n) ~ 2n = 2[f(n-1) + n-2]; f(3) = 2 Sanger Sequencing: smaller DNA splices (<900); expensive/inefficient for large splices; reliable 1. Denature DNA, add primers, polymerase, nucleotides, and fluorescent chain-terminating dideoxynucleotides (end strand) 2. Primer binds, Polymerase extends until ddNTP is inserted, many diff length strands are created w/ last colored ddNTP 3. Strands are separated through gel electrophoresis and sequenced based on ddNTP’s fluorescent color Next-Generation Sequencing (NGS): 1. 2. 3. 4. Fragment gDNA (400-1000 base pairs) Isolate fragments in aqueous solution w/ a bead Copy fragment (PCR, 106 copies), 5’ ends captured by bead Bead placed in well w/ DNA Polymerase/Primers (primer 3’) 5. 2 million wells on a multiwell plate, A/C/G/T solution added/washed 6. If light flashed (dNTP → dNMP + PPi), nucleotide is recorded Third Generation Sequencing: one DNA moved through a nanopore in a membrane, identifying the bases one by one electrical current interruptions (distinct per base) Gene/DNA Cloning: Identical copying of DNA/genes by adding the target gene to a cloning vector (e.g., plasmids (which yeasts have), including an expression vector, which has an active bacterial promoter, then restriction site to eukaryotic cDNA gene (R Transcriptase recoded w/o introns) is in correct reading frame; protein may also need to be modified in eukaryotes or mammalian culture cells (vector can be virus)) and allowing organisms with the vector to replicate it, producing multiple copies of the gene. Electroporation: Cells are electrocuted creating cell membrane pores for DNA to enter Restriction Enzymes cut DNA at specific, typically symmetric restriction sites (used by bacterial immune system, their genome is appropriately methylated at adenines/cytosines), gene of interest with same cut side can be added to a cloning vector and ligated to form recombinant DNA DNA Profiling/Fingerprinting uses gel electrophoresis (runs current through polymer (agarose) gel w/ DNA (-); smaller molecules move away from the cathode wall; DNA is organized by size (nicked, linear, covalent, supercoiled, circular), Ethidium bromide to stain). Transforming Bacteria: 1. Extract plasmid DNA from bacteria a. Lysis, Phase separation, clear proteins, precipitate DNA 2. Cut plasmid DNA using restriction enzymes 3. Hydrogen bond plasmid DNA to foreign DNA 4. Use DNA ligase to seal the DNA fragments 5. Transform bacteria with recombinant DNA Nucleic Acid Probe: short ssD/RNA fluorescent complementary to mRNA of interest RT-PCR Analysis: 1. cDNA Synthesis from each mRNA using reverse transcriptase/RNA polymerase/nucleases 2. PCR Amplification using primers from gene of interest on cDNAs (if gene is present) 3. Gel Electrophoresis reveals more DNA from mRNAs that originally had the gene qRT-PCR: dye lights/fluoresces when bound to dsDNA, no gel electrophoresis, quantitative data RNA Sequencing: 1. 2. 3. 4. mRNAs isolated/cut using nucleases Reverse transcribed into cDNAs cDNAs are sequenced (using another method, Sanger/NGS/TGS) Data mapped into genome by computer, frequency of certain sequences determines expression DNA Microarrays: ssDNA on a glass slide, represent the genome, need mRNA sequences prior, mRNA to fluorescently labeled cDNA by reverse transcriptase, hybridization and color intensity reveals expression Gene Drive: new engineered allele is more favorable for inheritance than the wild type Genes can be knocked out with RNA interference, complementary RNAs to inhibit translation Genetic markers: DNA sequences that vary in the population (genome-wide association studies) Blotting: Southern: DNA from film, Northern: RNA from film, Western: Proteins from film, Eastern: Post-transcript changes in proteins FISH: Finding specific DNA complementary sequences CRISPR-Cas9 Gene Editing: Clustered Regularly Interspaced Short Palindromic Repeats: E.coli defense, Cas genes followed by interspaced repeating DNA (spaced by viral DNA samples), viral DNA is recognized and added to the genome, transcribed into crRNA, crRNA + tracrRNA → gRNA (guide); Cas9 enzyme (nuclease) uses gDNA to recognize target DNA (PAM; NGC), target is cut, replaced with either a mutation of random sequence (non-homologous end joining translesion synthesis, knocks out gene) or homologous gene used as a template (homologous recombination/homology directed repair, translesion synthesis); use inserted gene for gene editing. Nuclear Transplantation: replace the nucleus in a cell with another (most likely a differentiated) Variations in cloned animals can arise from random events such as X-inactivation, environmental exposure, and epigenetic variations (differentiated cells are more methylated than zygotes, improper genetic expression can lead to defects and abnormalities halting development). Induced Pluripotent Cells: differentiated cells were introduced (retrovirus used) with 4 ES genes (Oct3/4, Sox2, Klf4, & c-Myc), causing dedifferentiation, restoring pluripotency Evolution Evolution: change in the genetic composition of populations over time (perfect species not formed due to mutation limitations, adaptation compromises, and dynamic environment). Aristotle: species are unchanging; Linnaeus: grouped species from patterns of their creation; binomial nomenclature; Lamarck: Individuals evolve through their life, use and disuse (more used anatomy evolves, less used deteriorates), inheritance of acquired characteristics (used/disused transferred to offspring); Darwin/Wallace: Populations evolve over time (descent w/ modification/natural selection), variations exist, more offspring than can carry are produced, favorable variations provide a survival/reproductive advantage, causing the accumulation of those traits in the gene pool over time (only occur w/ non fixed genes, fixed genes have only one possible allele and therefore can’t be selected for/against). Evidence of Evolution ● ● ● ● ● ● Paleontology: Shows differing species from the past compared to current life (sedimentary rock/strata, relatively dated by depth, radiometrically dated with radioactive isotopes/half-life) Anatomy: Comparing anatomical structures to determine ancestry/convergent evolution ○ Homologous Structures: Similar anatomy (different physiology), showing divergent evolution (vertebrate forelimbs) ○ Analogous Structures: Similar physiology (different anatomy), showing convergent evolution (wings in birds/bats/insects) ○ Vestigial Structures: Anatomy present in ancestor but serves no current purpose (ostrich wings, human tail bone, whale/snake pelvis/leg bones) Biochemistry: Comparison of DNA/RNA/Proteins and similarities implies common descent (universal genetic code, t/rRNA, pseudogenes, etc.) Embryology: Most animal embryos contain pharyngeal arches, tails, etc. Biogeography: Ancestor species range compared to similar species range & analyzing geographic isolation/similar environments (convergent/parallel evolution) ○ Endemic Species: Species only found in one geographic region Direct Observation: Directly observing evolution occurring (bacteria antibiotic resistance) Microevolution Change in the allelic frequencies of a population; can be due to 5 factors: 1. Mutations, introduce new alleles/change current alleles in the gene pool a. New beneficial mutations in alleles (DNA mutations) or differing number/position of alleles (chromosomal abnormalities) can enhance survival/reproduction & 2. 3. 4. 5. expand genomes (mammal olfactory genes); mutations happen more frequently with rapid asexual reproduction (prokaryotes/viruses) i. Mutations in developmental homeotic genes or their expression can cause massive anatomical/physiological changes (pattern formation/morphogenesis) and heterochrony (change in developmental event timings in an organism) b. Sexual reproduction also produces genetic variation/possible mutations through crossing over (recombination of paternal/maternal genes), independent assortment (random segregation of maternal/paternal chromosomes), and random fertilization (new allelic combinations from each sperm/egg). c. Genetic Recombination can also enhance prokaryotic variation: transformation (Geno/phenotype altered by integrated foreign DNA), transduction (viruses exchange DNA between prokaryotes), & conjugation (DNA transfer by plasmids through pili) Natural Selection, differential reproductive success results in some alleles manifesting in a population over time (relative fitness compared to others, higher contribution to future gene pool, they pass on their genes and respective traits, called adaptations, which can be in anatomy (changes in shape), physiology (changes in function), camouflage (blend into surroundings), or mimicry (Batesian: safe mimics harmful, Mullerian: harmful mimic each other)) a. Stabilizing Selection: Favors intermediate phenotype b. Directional Selection: Favors one extreme phenotype c. Disruptive/Diversifying Selection: Favors both extreme phenotypes d. Sexual Selection: females’ mate for traits, differential mating/reproduction success (intersexual competition: one sex choosing mating; intrasexual competition: members of a sex competing) i. Sexual Dimorphism: differences in secondary characteristics to fit traits e. Balancing Selection: Favors all phenotypes to an extent i. Frequency-Dependent Selection: Phenotype favor depends on frequency (sided-eating fish, less left frequent gives them an advantage) ii. Heterozygote Advantage/Balanced Polymorphism: Heterozygous phenotype is advantageous, maintaining both alleles in the gene pool (sickle-cell/malaria) f. Artificial Selection: Humans choosing favorable phenotypes (larger, sweeter crops) Nonrandom Mating, interbreeding/specific breeding patterns doesn’t shuffle the gene pool Gene Flow, Migrations into and out of the population introduce and take out alleles (reduces genetic differences between populations and may cause populations to merge) Small Population (Gene Drift), smaller populations are subject to significant random changes in the gene pool due to chance events (e.g., natural disasters or mass extinctions) i. ii. Founders Effect, few individuals of a population colonize a new habitat, new population gene pool consists only of those individuals, not the original population Bottleneck Effect, few individuals of a population survive a natural disaster event, new population gene pool consists only of those individuals, not the original population Hardy Weinberg Equilibrium models this; if a population is not evolving/fits the 5 factors: 𝑝 + 𝑞 = 1(Allelic frequencies p & q consist of the entire gene pool, only 2 alleles) 2 2 𝑝 + 𝑞 + 2𝑝𝑞 = 1(Entire population consists of the 3 possible genotypes) Species are separate populations that can’t interbreed to yield viable, fertile offspring. Factors that prevent interbreeding between different species include: Prezygotic ● ● ● ● ● Habitat: Differing habitats/biogeography Temporal: Differing mating seasons/timings Behavior: Differing mating rituals/preferences (Sexual Selection) Mechanical: Differing reproductive anatomy Gametic: Fertilization doesn’t occur (sperm die in reproductive tract or penetrate egg) Postzygotic ● ● ● Inviability: Hybrid organism may be lacking essential genes for development Infertility: Hybrid organism may have odd diploid number/abnormal chromosomes: sterile Breakdown: Hybrid inviability/infertility not seen in first gen, seen in future generations Speciation Development of multiple new species from an original species; occurs through either gradualism (slow, steady rate of adaptations) or punctuated equilibrium (rapid burst of evolution followed by long static periods); 4 main methods: ● ● Allopatric: geographic isolation, independent mutations, and evolution Sympatric/Parapatric: no geographic isolation, other factors prevent interbreeding: ○ Intense competition leads to resource partitioning, individuals evolve for their resource ○ Sexual Selection: females’ mate for certain traits, variants don’t interbreed/speciate ○ Hybridization: Species interbreed after speciation in hybrid zones ■ ■ ■ ■ ● Reinforcement: Hybrids produced less; reproductive barriers strengthen Fusion: Hybrids produced more; reproductive barriers weaken Stability: Hybrids continually produced; reproductive barriers maintained New Species: Hybrids produced more but evolve viability/fertility ● Balanced Polymorphism: Hybrids advantaged over parent species ● Polyploidy: Extra sets of chromosomes prevent parent species breeding (as long as there is one functional gene, other copies can mutate/evolve over time to serve a novel purpose) ○ Autoploidy: Polyploid from one species (asexual reproduction preserves polyploidy) ○ Alloploidy: Polyploid from 2 species (asexual reproduction preserves aneuploidy until fertilization creates fertile polyploid) Adaptive Radiation: species evolve to fit new niches (new habitat/extinction) Evolution can be classified depending on the resulting species as: ● ● ● ● Divergent Evolution: One species evolved into many distinct species Convergent Evolution: Unrelated species evolve similar traits due to similar pressures Parallel Evolution: Related species evolved similar characteristics from similar pressures Coevolution: Species affect each other’s evolution (such as predator-prey relationships) Macroevolution Evolution above species level (domain, kingdom, phylum, class, family, order, genus) Biosystematics Hadean Eon: 4.6 - 3.85 Billion years ago: Origin of Earth, Rock Formations, Water/Oceans Form Abiogenesis 1. Abiotic synthesis of organic monomers a. Hydrothermal vents release alkaline (basic) fluids, causing the formation of lipids which escape the vent as a blob w/ a basic fluid in an acidic environment (gradient) b. Celestial objects contain organic molecules (due to high UV radiation/low temps) c. Oparin/Haldane theorized early reducing atmospheric molecules (NH3, CO2, H2O, H2, & CH4) and lightning/UV radiation could form organic compounds (Primordial Oceanic Soup/Electric Spark Theory, tested by Miller/Urey and succeeded, neutral environment could also produce organic compounds or in reducing volcanoes/hydrothermal vents) 2. Polymerization of these monomers to form polymers a. Dripping monomers onto hot clay/sand results in spontaneous polymerization (Iron Sulfides may have been catalysts through energy from coupled redox reactions) 3. Polymers surrounded by protocells (lipid membrane to restrict internal chemistry) a. Phospholipid vesicles/bilayers originate from hydrothermal vents/self-assembly i. Reproduce (bubble off other smaller vesicles) ii. Grow (absorb other smaller vesicles or phospholipids) iii. Absorb clay particles (containing polymers like RNA/Proteins) iv. Perform chemical/metabolic reactions with external reagents 4. Polymers develop self-replicating abilities (life’s inheritance capabilities) a. RNA molecules can catalyze reactions (ribozymes, including RNA replication) due to 3D structure, functional groups, & hydrogen bonding (reaction specificity), RNA World i. Genotype: sequence; Phenotype: shape; competition for NMP, natural selection b. RNP World: RNA replication to proteins as proteins were more efficient catalysts c. Central Dogma: DNA replication to RNA to proteins as DNA is more chemically stable & accurately replicated compared to RNA for expanding genome d. Origin of horizontal gene transfer: genes transferred between genomes by plasmids, transposons, viruses, & endosymbiosis. e. Viruses may have begun evolving here as catalytic nucleic acids (mobile genetic elements such as transposons/plasmids) that move between hosts (protocells), eventually evolved to DNA/Proteins for increased cell infiltration (coevolution) i. Viruses have scavenged host genes such as DNA repair, translation, protein folding, & polysaccharide synthesis, supporting this hypothesis. 5. Primary heterotrophic prokaryotes develop (vesicle w/ nucleic acids) a. Heterotrophic hypothesis predicts life began as anaerobic (glycolysis) heterotrophs 6. Primary autotrophic prokaryotes develop (cells w/ primitive photosystems in membrane) a. Produce O2/Sugars (Photosynthesis), ancestors of cyanobacteria b. Atmospheric oxygen increased (selection against obligate anaerobes towards facultative/obligate aerobes, including developments in cellular respiration), ozone layer formed (blocked UV light ending abiogenesis of organic molecules) Prokaryotic Diversity Bacteria: ● ● ● ● Structure: Peptidoglycan cell walls (large layer is gram-positive, small layer w/ outer glycolipid membrane is gram-negative) covered by a sticky capsule, fimbriae, and/or pili (F factor genes) Motility: Flagella enable movement (proton gradient turns curved fibers for motion) Genome: circular/histone-less in nucleoid w/ extra plasmids & replicative/expressive machinery Metabolism: Photo (certain prokaryotes)/Chemoheterotrophs (many prokaryotes, obligate & facilitative (an)aerobes perform fermentation or anaerobic respiration/aerobic respiration); Photo (photosynthesis)/Chemoautotrophs (chemosynthesis) perform organic carbon fixation; some cyanobacteria/methanogens have nitrogen fixation (N2 to NH3) for amino acids/n-bases Gram-Negative: ● ● ● ● Proteobacteria (alpha – eukaryotic symbiosis (mitochondria), beta – nitrogen recycling, gamma – sulfur/pathogenic, delta – slime-secreting myxobacteria, epsilon – pathogenic) Chlamydia (lack peptidoglycan in cell walls, rely on animal hosts (for ATP), pathogenic) Spirochetes (helical heterotrophs (move through rotation like large flagella), pathogenic) Cyanobacteria (photoautotrophs (chloroplast), oxygen release, make up phytoplankton) Gram-Positive: Diverse, can be pathogenic, free-living/solitary, Mycoplasmas only w/o cell walls Archaea: Extreme halophiles (salt) or crenarchaeota (mostly thermophiles), euryarchaeota (mostly methanogens, H2 + CO2 to CH4, obligate anaerobes), korarchaeota (Koron - young man, aren’t in previous two clades), & nanoarchaeota (smaller, symbiotic archaea, in other archaea in hot springs or hydrothermal vents) 7. Primary eukaryotes (protists) develop (cells w/ compartmentalization) a. Endosymbiotic Theory: i. Infoldings of the plasma membrane (endomembrane system engulfing DNA) ii. Engulfing of aerobic bacterium (mitochondria performs aerobic respiration) iii. Engulfing of photosynthetic bacterium (plastid performs photosynthesis) b. Evidence for Endosymbiotic Theory (similarities to bacteria): i. Independent reproductive process similar to binary fission ii. Circular, histone-less, intron-less DNA (simultaneous protein synthesis) iii. Smaller ribosomes than Eukaryotes found in Prokaryotes (30S/50S/70S) iv. Membranes include structures homologous to those in prokaryotes v. Double membrane structure (bacterial membrane & vacuole membrane) c. Secondary Endosymbiosis: i. Autotrophic eukaryotic diversified into green/red algae, which underwent endosymbiosis again. Red algae endosymbionts diversified into dinoflagellates and stramenopiles; depending on the heterotroph, green algae endosymbionts were euglenids or chlorarachniophytes. d. Evidence for Secondary Endosymbiosis i. Plastids surrounded by 4 membranes: 2 inner membranes from original gram-negative cyanobacterium, 1 from algae plasma membrane, & 1 from eukaryote food vacuole where endosymbiosis occurred ii. Vestigial nucleus/DNA/expressive machinery from primary eukaryote exists 8. Complex multicellularity develops (colonies of differentiated cells) a. Originally colonies of cells which cell walls/membranes held together b. Complex multicellularity arose by reordering genes (protein domains) & a few novel genes involved in cell junctions/connections & signaling (which led to cell differentiation by cytoplasmic determinants/induction) i. Animals are sister taxa with choanoflagellates, which have genes for protein domains found in only animals such as for cadherins (cell attachment) and signaling c. Mostly marine life 9. Diversification/Colonization of Land by Plants, Animals, & Fungi a. Land provided unfiltered sunlight, CO2, and soil nutrients but had scarce water/lack of support against gravity b. Plants (from Charophytes, a green algae) derived many novel traits for land: i. Altering Generations: Diploid (sporophyte) & haploid gens (gametophyte) arise from zygote (2n, from gamete fertilization) and spores (n) mitotically dividing ii. Walled Spores: Spores from sporophytes (made in sporangia organs) coated with sporopollenin to survive adverse conditions 1. Charophytes living near edge of water developed a layer of sporopollenin to prevent drying out, enabling 1st land movement iii. Apical Meristems: Regions on roots/shoots for cell division to elongate iv. Cuticle Layer: Epidermis covered with layer of wax/polymers, prevents excessive transpiration/protects from microbial attacks v. Stomata: Pores which allow gas exchange for photosynthesis, can be closed to minimize water losses in hot/dry conditions vi. Transport Systems: moved nutrients and water up and carbohydrates down c. Fungi (from unicellular nucleariids) derived many novel traits to move to land: i. Absorptive Nutrition: Fungi bodies are unicellular or multicellular filaments (hyphae, together form mycelium, which increase SA/V for absorption) with chitin walls (strong/flexible, more nutrients/water can be absorbed w/o lysis) ii. Plant Mutualism: Specialized hypha (haustoria) exchange absorbed nutrients with plant roots for carbohydrates (sym genes for mycorrhizal formation in plants). Ectomycorrhizal don’t penetrate cells, penetrate extracellular space & form layer outside roots; Arbuscular mycorrhizal penetrate root cell walls (not plasma membrane) for molecular exchanges; endophytes live in plants and benefit them by giving them pathogen resistance, deterring predators, etc. iii. Sexual/Asexual Reproduction: Sexual reproduction/fertilization of spores when cytoplasm (plasmogamy) then nucleus fuse (karyogamy), meiosis then mitosis for building haploid filaments, yeasts can also asexually reproduce as single cells d. Plants began as nonvascular bryophytes and developed: i. Vascular tissue to grow taller (outcompete for sunlight, disperse spores farther, directional selection for taller plants) ii. Roots to absorb soil nutrients/anchor plants iii. leaves to increase photosynthetic output with a greater SA/V ratio iv. Seeds (multicellular w/ protected embryo supplied w/ food for longer lifetime, seeds can transfer sperm w/o water by pollination and pollen tube which is better for dry land environments) v. Flowers (sexual organs specifically for reproduction) vi. Fruit (more viable medium for spreading seed through heterotrophic nutrition and excretion) e. Animals began as sponges, mollusks, & cnidarians (grazers/scavengers/filter feeders, marine); Cambrian Explosion led to rapid diversification of predators/prey (coevolution), bilaterians, & hard-bodied, large animals (could be due to natural selection of predator traits, oxygenation selected for larger, metabolically stronger organisms, or Hox genes regulating body structure develop/mutate) f. Animals colonized land in many events: i. Arthropods first: cuticle prevented dehydration; Insects are most diverse clade (radiated in response to new plant foods, resource partitioning), developed flight (extensions of the cuticle, still have walking legs, evolutionary advantage for land) ii. Tetrapods developed limbs from lobe-fins, Amphibians lived on both land/water, Amniotes developed amniotic egg (reduced aquatic dependence for reproduction, slowed dehydration, similar to plant seeds) Summary Eukaryotic Diversity Nucleus/membrane-bound organelles, cytoskeleton for shapeshifting/motility, cilia, complex multicellularity (cell differentiation), sexual life cycles, photosynthesis, large body. 4 Supergroups ● ● ● ● Excavata: Parabasalids/Diplomonads: modified mitochondria (mitosomes/hydrogenosomes); Euglenozoans: modified flagella (rod w/ spiral/crystalline structure) “SAR” Clade: Stramenopiles/Alveolates (sacs under plasma membrane), (both photosynthetic from secondary endosymbiosis), Rhizaria (amoeba w/ pseudopodia) Archaeplastida: Red/Green Algae & Land Plants (Photosynthetic, one/colony/multicellular) Unikonta: Amoeba w/ lobe/tube pseudopodia, animals, fungi, choanoflagellates, & other animal or fungi like protists (may have diverged first based on DHFR-TS gene divergence/fusion) Protists refer to all but plants/animals/fungi. Fungi Diversity ● ● ● ● ● Chytrids: Multicellular branched hyphae body, flagellated spores, lakes/soil Zygomycetes: Rapid hyphae growth in certain mediums, can be decomposers/parasites Glomeromycetes: Form arbuscular mycorrhizae with plants roots for commensalism Ascomycetes: Marine/freshwater/terrestrial habitats, sac fungi Basidiomycetes: Decomposers/Ectomycorrhizal fungi, long heterokaryotic stage Plant Diversity ● ● ● Bryophytes (liverworts, mosses, hornworts): Lacked vascular tissue (height constraint), moist habitats, flagellated sperm, gametophytes are dominant generation (sporophytes depend on gametophyte for nutrition) Seedless Vascular Plants (lycophytes: mosses/monilophytes: ferns): Dominant sporophyte generations (gametophytes are independent/free-living), well-developed roots (for water and mineral absorption and anchoring)/leaves (for photosynthesis & SA/V increase; lycophytes have microphylls with one vascular tissue, others have megaphylls with branched vascular system), taller growth, flagellated sperm, vascular tissue (xylem moves water/minerals by tracheid cells (tube shaped with lignin-strengthened walls), phloem moves organic products) Seeded Vascular Plants (Gymnosperm/Angiosperm): Dominant sporophyte generations (gametophytes are reduced/dependent on sporophyte for nutrition), seeds (embryo & food protected w/ coat) ○ ○ megasporangium (2n) covered by integument tissue (2n) contain megaspore (n, egg) making up ovule, megaspore develops into gametophyte w/ egg; microspores (n, sperm) develop into pollen grain (gametophyte with sporopollenin wall), gametophyte germinates in ovule, develops pollen tube to discharge sperm into egg (water not needed for fertilization, dry environmental advantage), zygote develops into embryo enclosed with food supply (female gametophyte remnants) by seed coat derived from the integument tissue Angiosperms developed flowers (specialized reproductive organs) with four leaf layers: ■ Sepal (green, sterile, enclose flower before blooming) ■ Petals (colored, sterile, attract pollinators) ■ Stamens (fertile, produce sperm/pollen) ■ Carpels (fertile, produce eggs, pollen enters through stigma into ovary where multiple ovules can be fertilized into a seed, ovary becomes fruit) Animal Diversity ● ● ● Radial Symmetry - no left/right sides; have ectoderm & endoderm Bilateral Symmetry - dorsal/ventral (top/bottom), left/right, anterior/posterior (front/back); have ectoderm, mesoderm, & endoderm w/ body cavities All animals are part of the monophyletic clade Metazoa ○ Sponges Porifera are basal taxon, Eumetazoa have true tissues (germ layers); basal Eumetazoa (Ctenophora/Cnidaria) are radians/have 2 germ layers, rest are Bilateria (Deuterostomia (Chordates - only vertebrates, most animals are invertebrates), Ecdysozoa (Nematodes/Arthropods), & Lophotrochozoa (Molluscs, Brachiopoda). ○ Chordates have a notochord (flexible rod for skeletal support), dorsal/hollow nerve cord (brain-spine), pharyngeal slits/clefts (filter feeding, gills, or part of the head), & post-anal tail (ends after anus) ■ Jawed vertebrates proliferated (Gnathostomes) into Chondrichthyans (Cartilage Skeleton, sharks/rays), Ray-Finned Fish (Bony Skeleton, Lungs), & Lobe Fins (rod bones/muscles around pelvic/pectoral bones, coelacanths, tetrapods) ● Tetrapods consist of Amphibians (Salamanders, Frogs, & Caecilians) and Amniotes (Reptiles/Mammals, have amniotic egg - 4 membranes, Embryo protected w/ yolk food source, excretion collection (Allantois), & Chorion gas exchange) ○ Amniotes diversified into Reptiles (Crocodilians, Dinosaurs, & Birds; keratin scales, shelled eggs, mostly ectotherms external heat is main body heat source) & Mammals (Monotremes (egg-laying), Marsupials (pouch), & Eutherians (placenta); mammary glands produce milk for ○ offspring, hair, fat layers, endotherms - high metabolic rate to maintain body heat, varying teeth) Arthropods are segmented w/ a hard exoskeleton (cuticle, made from protein/chitin) & jointed appendages Ecology Ecology: study of interactions between organisms/environment Ecosystem Ecology Biosphere (Global)/Connected Ecosystems (Landscape)/Ecosystem (Abiotic & Biotic Factors) Climate: long term abiotic weather conditions (temperature, precipitation, sunlight, wind; from sun which hits tropics more directly than poles; Earth’s tilted axis causes seasons (December: north tilts away; June: north tilts towards), water bodies moderate temperature (high specific heat, currents transport moisture, as air rises over mountain ranges, it cools, mountain backside w/o moisture)). Biomes – major life zones characterized by vegetation/physical environment (Ecotone is overlap area; in water, Benthic is seafloor, Pelagic consists of Photic (surface for photosynthesis) & Aphotic; warm upper water separated by thermocline from colder deeper water): ● ● ● ● ● ● ● ● ● ● ● ● Savannah: Equatorial/subequatorial, seasonal rainfall (common fires), grasses are most common, large herbivores/insects & predators make up animal biodiversity Dessert: 30° N/S latitudes/in-between, seasonal/daily temperature (low rain), few, widely scattered plants (C4/CAM photosynthesis), animals: insects, reptiles/birds, mammals Tropical Forests: Equatorial/subequatorial, little seasonal changes, plants compete for light, high animal biodiversity Temperate Broadleaf Forests: North/South midlatitudes, seasonal temperatures, deciduous forests, hibernating/migrating mammals/birds Northern Coniferous Forests: North America/Eurasia (taiga), seasonal temperatures, mostly conifers, moose/bears/birds/Siberian tigers Chaparral: Midlatitude coastal regions, seasonal temp/precipitation, fire-adapted trees/shrubs, insects, amphibians, small mammals, birds, large grazing herbivores Temperate Grassland: Midlatitude interior regions, seasonal temp/precipitation, grasses, grazing large mammals/burrowing animals Tundra: Arctic/high mountains, low seasonal temp, mostly mosses/grasses, large grazing animals & predators (bears/wolves/foxes/owls) Wetlands/Estuaries: River/Sea zone & flooded areas, cattail/sedges/saltmarsh grasses, aquatic insects/crustaceans, & other insects/frogs/birds/fish/crabs/oysters Lakes: Standing bodies of water, oligotrophic are oxygen/nutrient rich/poor, eutrophic is opposite, rooted aquatic plants/phytoplankton, zooplankton/invertebrates/fish Streams/Rivers: As salt/nutrients increase oxygen decreases, rocky/silty bottom, phytoplankton & aquatic rooted plants, fishes/invertebrates Intertidal Zones: rock/sand periodically submerged by tides (high nutrient/oxygen), seagrass & algae, invertebrates/small fishes/clams/crustaceans ● ● ● Coral Reefs: photic zone of oceans (border land), fringing to barrier to atoll, algae, corals/fish & other invertebrates (high diversity) Ocean Pelagic Zones: open ocean, phytoplankton, zooplankton/squid/fish/turtle/mammals Marine Benthic Zones: sandy/rocky seafloor, cold/high water pressure, some seaweed, algae & chemoautotrophs near deep-sea hydrothermal vents, some invertebrates (Solar) Energy flows through ecosystems; matter cycles through it (law of conservation of mass); laws of thermodynamics state energy is transformed inefficiently (entropy); trophic levels: ● ● ● Primary Producers: Autotrophs, light/chemical energy to synthesize organic compounds ○ Gross/Net Primary Production: represent solar energy converted to chemical energy (net, includes subtraction of autotrophic metabolism/respiration, ~GPP/2) ○ Primary Production is limited by light for photosynthesis & nutrients (i.e.: N & P) ■ Nutrients can promote cyanobacteria/algae proliferation, death, & decomposition using O (anoxia, called eutrophication (well-nourished)) ■ Land plants adapted symbiosis with nitrogen-fixing bacteria and mycorrhizal fungi for obtaining nitrogen/phosphates from the soil Primary/Secondary/Tertiary/etc. Consumers: Heterotrophs of Producers/Consumers ○ Net Ecosystem Production: biomass gain (GPP – respiration of all organisms) ○ Secondary Production: Chemical energy converted to biomass (not excreted or respired) ■ Production Efficiency: Net Secondary Product (used for growth, not cellular respiration)/Assimilated Primary Product (food - feces); Birds/Mammals are low because of endothermy, insects/microorganisms are higher ■ Trophic Efficiency: energy passed on levels, approximately 10% per level ● Typically, biomass pyramids represent a sharp spike up; aquatic habitats may be inverted be as phytoplankton have high turnover Decomposers/Detritivores: Heterotrophs of detritus (dead organic compounds), convert it into inorganic compounds for producers for energy by chemically recycling; Biogeochemical Cycles for Water (All), Carbon Dioxide (All), Phosphorous (NA/L), Nitrogen (NA/P), & Sulfur (P) [NA - Nucleic Acids; P - Proteins; L - Lipids; C - Carbs]: Disturbances – Storm, fire, drought, or human activity that removes organisms/reduces resources Nonequilibrium model states communities as changing after disturbances; Intermediate disturbance hypothesis states most biodiverse communities arise from moderate disturbance frequency/intensity. Ecological succession: after disturbance, an ecosystem is rebuilt, and species replace each other Primary succession: lifeless area w/o soil: Pioneer species are first to colonize (alter soil properties for future species: prokaryotes, protists, etc.), plants colonize (lichens/mosses first, then come grass/trees), more species enter the ecosystem and replace/add to the niches of others. Secondary succession: previous life, left w/ soil, similar to primary succession in ecosystem rebuilding. Latitudinal Gradients: more diversity in tropics than poles because less disturbance events and higher evapotranspiration (total water evaporated, represents precipitation/temperature/sunlight, richness) Island equilibrium model states dynamic equilibrium in island habitats when migration = extinction; species diversity then is correlated to island area & distance from mainland (resource availability) Biogeography of a species can be limited by dispersion (migrations away from dense areas), biotic factors (negative symbiosis, disease, lack of pollinators/food), & abiotic factors (salinity, water/oxygen availability, pH, temperature (protein denaturing), light availability, rocks/soil, etc.) Community Ecology All biotic factors in an ecosystem (many populations of all species) Interspecific Interactions: Species interactions in a community ● ● ● Competition: -/-, compete for niche – ecological role (i.e., habitat, food), limits both species Intraspecific (same species), Interspecific (different species) ○ Competitive Exclusion Principle states best fit for niche outcompetes other ■ Resource Partitioning can result in coexistence of similar species ● Scramble (universally available resource), Contest (specific access) ■ Fundamental (potential) differs realized (actual) niche proves competition Predation/Herbivory: +/-, predator/herbivore kills/eats prey/producer ○ Adaptations such as predator fitness for hunting and prey colors/toxins/etc. Symbiosis: Species are in direct permanent contact and interact with each other ○ Parasitism: +/-, parasites (endo/ecto – inside/outside host) use host nutrients ○ Mutualism: +/+, Ex. Flowering plants nectar/fruit attract pollinators (food) ● ○ Commensalism: +/0, Ex. hitchhiker organisms, eating food exposed by another ○ Amensalism: -/0, Antibiosis: anything -, Allelopathy: secreted chemicals influence ○ Altruism: benefits another at its own expense, sacrificing warning organisms Facilitation: +/+(0), Positive effects without direct/intimate contact (Black Rush makes New England marshes more habitable for other plants by preventing salt buildup and storing oxygen) ○ Antagonism: opposite, interference without direct contact Species Diversity: Species richness: species #; Relative Abundance: individuals per species Shannon Diversity Index: − (𝑝𝐴𝑙𝑛(𝑝𝐴) + 𝑝𝐵𝑙𝑛(𝑝𝐵)...)where pN is individuals of species N 𝑛 2 Simpson’s Diversity Index: 1 − Σ( 𝑁 ) ; where n is # relative abundance and N is total individuals. More diversity increases community stability from disturbances (more genetic variation for changing conditions, can also prevent invasive species from outside their native range from manifesting) and increases consistency in biomass (total mass of all individuals in a population). Diversity threatened by: ● ● ● ● Habitat loss, resulting in less resources/space for growth/proliferation Introduced/Invasive Species, which alter communities drastically mostly negatively Overharvesting, which results in net loss of species if the rate is higher than the birth rate Global Change, altered climate/chem affects biogeochemical cycles, rupturing ecosystems ○ Humans altering global dynamics by: ■ Nutrient Movement (moved from farms to cities, centralized in new areas, more fertilizer use, leads to enrichment past critical load resulting in killed organisms, blooms, eutrophication, etc.) ■ Toxins (arise from pharmaceuticals, manufacturing wastes, & synthetic compounds, which persist in ecosystems, are recycled, and harm species) ■ Climate Change (more CO2 emissions from fossil fuel burning increases temperature by absorbing more light and reflecting it back to Earth (greenhouse effect); changing temperatures reduce habitat, extinction) ○ Human population is no longer exponentially growing, ecological footprint is resources needed per unit of people to live (can be used to estimate carrying capacity); sustainable development meets current needs and future needs Feeding relationships in a community are trophic structure: connected food chains in webs Bottom-Up model suggests lower trophic levels influence higher levels (changing predators doesn’t affect producers); Top-down model suggests higher trophic levels influence lower levels (limit individuals, which increases next species, etc.; trophic cascade model). Dominant species are the most abundant/biomass (best fit, best resource exploitation, best avoid predators/disease); Keystone species aren’t abundant but important due to their niche; Foundation species dramatically alter the environment (ecosystem engineers, beavers: forests into flooded plains). Pathogens cause diseases (zoonotic are animal to human, may be by vector – intermediate species); can kill off species in an ecosystem causing unbalance Species classification on the path to extinction: Threatened, Endangered, Extinct Minimum Viable Population is smallest size population can be before entering extinction vortex (fewer fit individuals, less traits passed on, even fewer fit, continued till extinction) Effective population size is breeding potential (for proliferation): 𝑁𝑒 = 4𝑁𝑓𝑁𝑚 𝑁𝑓+𝑁𝑚 where Ns is number of breeding individuals of sex s. Population Ecology Population (Species living in same area/time) Demographics: Population density affected by birth (affected by reproductive rates & age structure – proportion of males/females in a population; life history is age beginning reproduction, cycles, and offspring per cycle), immigration, emigration, & mortality (natural selection, more reproduction decreases survival rate; time/resource investment per offspring). Dispersion: ● ● ● Clumped: may grow near areas w/ food/resources Uniform: evenly spaced for developmentally inhibiting pheromones/territoriality Random: adequate resources everywhere, location doesn’t affect life as significantly Type I/II are K-selected species (iteroparity (periodic reproduction), few large offspring/late maturity, population density dependent traits selected for; high densities approaching K due to resource/territory competition, more predation, disease spread, secreted toxins, and intrinsic adaptations to maximize survival); Type III are r-selected species (semelparity (single reproduction), more small offspring/early maturity, population density independent traits selected for; low densities with high r) 𝑑𝑁 𝑑𝑡 = 𝑟𝑁 = (𝑏 − 𝑚)𝑁 Where N is population size, t is time, r/b/m are rates of increase/birth/death per capita. J-curves arise from exponential growth (unlimited resources/no competition/maximum reproduction) Carrying Capacity (K) is the max N an environment can sustain, modeled by sigmoidal S-curves: 𝑑𝑁 𝑑𝑡 = 𝑟𝑁(𝐾 − 𝑁)/𝐾 = (𝑏 − 𝑚)𝑁(𝐾 − 𝑁)/𝐾 Metapopulation is linked local populations by migrations (promoted by high N densities) Human Anatomy/Physiology Organ Systems Organs (Anatomy) Function (Physiology) Skeletal Bones, Tendons, Ligaments, Cartilage Support/Structure (Protect Organs) Muscular Skeletal Muscles Locomotion Integumentary Skin, Hair, Claws Stop Injury/Infection, Homeostasis Nervous Brain, Spine, Sense Organs Coordination, Stimulus Response Endocrine Glands (Pituitary, Thyroid, Pancreas) Coordination Digestive Mouth, Stomach, Intestine, Liver, etc. Food Processing Excretory Kidney, Uterus, Bladder, Urethra Waste Removal, Osmoregulation Respiratory Mouth/Nose, Lungs, Trachea Gas Exchange Circulatory Heart, Capillaries, Veins, Arteries Nutrient/Waste Transport Immune & Lymphatic Bone Marrow, Lymph Nodes, Spleen, Thymus, White Blood Cells Body Defense [Text] Nervous System Neurons: Dendrites for input information, cell body, & axon for output information (synapses) Action Potential: Axon begins negative w/ more Na+/K+ outside/inside, stimulus opens Na+ channels and Na+ diffuses in (axon more positive, triggers more Na+ voltage-gated channels), at peak Na+ channels close & K+ channels open (K+ diffuses out, axon more negative), original negative membrane potential established, Na+/K+ pump resets gradients, potential spreads along the axon as Na+ in the cell move down it and K+ stay outside the back side (stopping double flow). Synapse: As an action potential moves down the axon, Ca+ voltage-gated channels open, Ca+ influx causes exocytosis of neurotransmitters (i.e.: acetylcholine, amino acids, norepinephrine, serotonin, dopamine, neuropeptides/endorphins, & nitric oxide), they bind to ligand-gated ion channels (for Na+ in, K+ out, etc.) in the receiving neuron’s dendrite, allowing action potential to propagate (memory and learning is changed in chemical synapses rather than electrical axons). ● Sensory Neurons: Input senses (Light (sight), Chemical (smell/taste), Mechanical (hearing/touch), Thermal), output many axon ends (by Peripheral Nervous System). ○ ● ● Mechanoreceptors (ion channels connected to cilla/cytoskeleton) detect mechanical energy (pressure, touch, stretch, motion, sound) ■ Audition (sound) - ear, air pressure waves cause tympanic membrane (eardrum) & 3 inner ear bones to vibrate oval window causing waves in cochlea fluid (perilymph), which move basilar hairs on fixed tectorial membrane, detected by mechanoreceptors. ■ Vestibulation (balance) - ear, utricle/saccule have otoliths in gel to sense gravity & linear acceleration, angular acceleration in 3 planes sensed by bent hairs in semicircular canals w/ perilymph pushing cupula. ○ Electromagnetic Receptors detect light, electricity, & magnetism ■ Vision ○ Thermoreceptors detect heat/cold ○ Chemoreceptors detect chemicals (ligand-gated ion channels, etc.) ■ Olfaction (smell) - nose, 1000+ genes for different receptors of new smells ■ Gustation (taste) - taste buds on tongue papilla, taste cells have one receptor for either sweet, salty, sour, bitter, or umami (delicious, glutamate) ○ Nociceptors detect pain (extreme pressure, temperature, chemicals, etc.) Interneurons: Input from many sensory neurons, integrate information, output many axon ends signaling response (Central Nervous System consists of the brain (brainstem: information to/from the brain, diencephalon: sorts senses, endocrine system, cerebellum: movement/balance/learning/memory/coordination, cerebrum: occipital lobe is vision, parietal lobe is touch & integration, temporal lobe is hearing, frontal lobe is skeletal muscle control, decision-making, speech, etc.) & spine). Motor Neurons: Input from interneurons (transmitted from CNS through PNS), output to muscles for contraction (Autonomic (smooth/cardiac muscles & glands) Sympathetic (fight-or-flight), Parasympathetic (rest and digest), Enteric (digestive system), & Nonautonomic Motor (skeletal muscles) Systems) Endocrine System Glands across the body secrete hormones into the circulatory system to regulate body processes. ● ● ● Pineal Gland (Mid-Back Brain) ○ Melatonin: Regulates biological rhythms (i.e. circadian rhythm, sleep) Hypothalamus (Lower Brain) ○ Releases hormones regulating anterior pituitary secretion Anterior Pituitary Gland (Under Hypothalamus, Front) ○ Follicle-Stimulating/Luteinizing Hormone (FS/LH): Reproductive gland secretion ○ Thyroid-Stimulating Hormone (TSH): Thyroid gland secretion ○ Adrenocorticotropic Hormone (ACTH): Adrenal cortex glands secretion ○ Prolactin: Mammary gland secretion ● ● ● ● ● ● ● ○ Melanocyte-Stimulating Hormone (MSH): Melanocytes (skin epidermis, melanin) ○ Human Growth Hormone (HGH): Cell cycle progression/division in many tissues Posterior Pituitary Gland (Under Hypothalamus, Back) ○ Oxytocin: Uterus contractions, stimulates mammary glands ○ Vasopressin (Antidiuretic Hormone - ADH): Kidney water retention, sociality ■ Activates more aquaporins in nephron distal tubules & collecting ducts Thyroid Gland (Neck, Butterfly Shape) ○ Thyroid Hormone (T3/T4): Stimulates/Maintains metabolic processes ○ Calcitonin: Lowers blood Ca+ level Parathyroid Gland (Circles Within Thyroid) ○ Parathyroid Hormone (PTH): Raise blood Ca+ level Adrenal Cortex Glands (Atop Kidneys, Outside Surface) ○ Glucocorticoids: Raise blood glucose by liver gluconeogenesis (non-carbs → carbs) ○ Mineralocorticoids: Na+ reabsorption, K+ excretion Adrenal Medulla Glands (Atop Kidneys, Inside Core) ○ Epinephrine/Norepinephrine: Raise blood glucose level, increase metabolism, vasoconstriction/vasodilation of certain blood vessels (fight-or-flight response) Pancreas (Behind Stomach) ○ Insulin/Glucagon: Lower/Raise blood glucose level Reproductive Glands (Ovaries - Females, Testes - Males) ○ Estrogens/Progestins: Uterine lining/growth, female sexual traits (estrogens only) ○ Androgens (i.e. Testosterone): Sperm formation, male sexual traits ■ Note: all hormones found in both sexes, only play a major role in one Digestive System Alimentary Canal: complete mouth-anus extracellular digestive tract (peristalsis - muscles lining canal push food; sphincters - muscle layers regulating food passage through the canal) 1. Ingestion: Mechanical digestion a. Oral Cavity (Mouth): Teeth/saliva (salivary gland) break & hydrolyze food b. Food in ball shape (bolus) transported into throat (pharynx) & esophagus 2. Digestion: Chemical digestion a. Stomach: Gastric gland secretes HCl, pepsin, & mucus for polypeptide hydrolysis b. Small Intestine: HCO3- (secretin to pancreas) buffer for stomach acid in duodenum i. Pancreatic trypsin/chymotrypsin/carboxypeptidase (proteins), amylases (carbohydrates), nucleases (nucleic acids), lipase (lipids) with bile (lipid detergent from liver), (CCK to pancreas/gallbladder, lipids slow peristalsis) 3. Absorption: Nutrients enter body a. Small Intestine (Jejunum/Ileum): digested monomers transported through epithelial tissue villi/microvilli and to blood vessels (lipids coated into water soluble chylomicrons and transported through the lacteal of the lymphatic system) i. Blood vessels lead to liver next (can modify/store/detoxify blood nutrients) b. Large Intestine: Cecum (fermenting undigested material), Colon (Na+ pumped in body/osmosis for water retention), & Rectum (undigested food/bacteria stored) 4. Excretion: Undigested material/bacteria (feces) excreted a. Anus: 2 sphincters (voluntary/involuntary) control feces excretion, bacterial colon metabolism releases gases (CH4, H2S, etc.) through anus Excretory System Nitrogenous Wastes: Ammonia (High Toxic, Dilute Urine, Low Energy), Urea (Low Toxicity, Moderate Water-Urine & Energy), & Uric Acid (Nontoxic, Concentrated Urine, High Energy) Nephron: Functional unit of the kidney - processes urine from renal artery/vein (Cortical: short extension into renal medulla, most; Juxtamedullary: deep extension into renal medulla) 1. Afferent Arteriole (Renal Artery) brings blood to Glomerulus (ball of capillaries) where filtrate diffuses into the Bowman’s Capsule due to blood pressure, blood leaves through Efferent Arteriole (Filtration) 2. Filtrate moves through Proximal Convoluted Tubule; water, salt, nutrients, potassium, & HCO3- are reabsorbed by Peritubular Capillaries; protons (H+) and ammonia are secreted 3. Filtrate moves through the Descending Loop of Henle; water is reabsorbed by Vasa Recta; osmolarity increases in Renal Medulla 4. Filtrate moves through the Ascending Loop of Henle; salt is reabsorbed by active transport (restoring renal osmolarity; Countercurrent Multiplier System forms salt chemical gradient) 5. Filtrate moves through the Distal Convoluted Tubule; water, salt, & bicarbonate are reabsorbed; potassium/ammonia are secreted 6. Filtrate moves through the Collecting Duct; some salt/water is reabsorbed, filtrate is transported to Renal Pelvis, Urinary Bladder (by the Ureter Tubule), Urethra (Excretion) Renin-Angiotensin-Aldosterone System (RAAS) regulates blood pressure/volume; when low, Juxtaglomerular Apparatus secretes Renin which ultimately secretes Angiotensin II (triggers vasoconstriction (increasing blood pressure) & stimulates the Adrenal Glands releasing Aldosterone which causes more salt & water reabsorption in the distal tubules/collecting ducts). Respiratory System Inhaling: O2 in air (160 mm Hg) moves down pressure gradient to the nasal/oral cavity down to the alveoli (circular tips of bronchioles in lungs, where increased pressure because expanding rib cage and lower diaphragm, contracting muscles); O2 diffuses into oxygen-poor blood, CO2 diffuses out of it (CO2 rich); CO2 and oxygen-poor air expelled as diaphragm moves up and rib cage contracts (relaxing muscles, less V, more pressure, exhaling). Regulation: More blood pH (7.4 normally, CO2 → H2CO3 + H+ increases it) detected in cerebrospinal fluid by Medulla oblongata → faster/deep breathing, restoring pH w/ less CO2. Hemoglobin (4 protein subunits/heme iron group) can bond to and transport O2 (releases it in acidic, CO2 rich blood)/CO2 (bonds to it after releasing O2 in acidic blood) in the blood. Circulatory System Immune/Lymphatic System