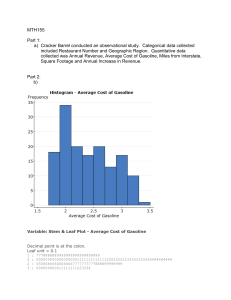
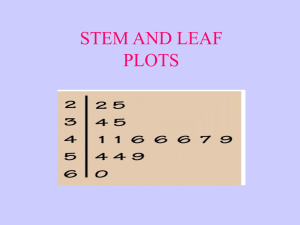
Logistic annotated example Some background: In simple linear regression the outcome variable Y is predicted from the equation: in which b0 is the Y intercept, b1 quantifies the relationship between the predictor and outcome, X1 is the value of the predictor variable and ε is an error term. For several predictors, a similar model is used in which the outcome (Y) is predicted from a combination of each predictor variable (X) multiplied by its respective regression coefficient (b) plus ε the error term in the model. Logistic regression is designed to use a mix of continuous and categorical predictor variables to predict a binomial/dichotomous categorical dependent variable. For a binary response taking the values 0 and 1 (e.g., absence of CHD and presence of CHD) the expected value is simply the probability, p, that the variable takes the value one, i.e., the probability of having CHD. We can’t model p directly as for linear regression two reasons 1. The observed values do not follow a normal distribution with mean p, but rather what is known as a Bernoulli distribution 2. The probability of an event occurring must be between 0 and 1. Using a linear regression model does not ensure that this is so. It is more appropriate to model p indirectly via logit transformation of p i.e., ln[p/(1-p)]. Remember that p/(1-p) is the odds of an event occurring, so we are in effect modelling the log-odds of an event as a linear function of the explanatory variables. The parameters in a logistic regression model are estimated by maximum likelihood. Loosely speaking, the likelihood of a set of data is the probability of obtaining that particular set of data given the chosen probability model. This expression contains the unknown parameters. Those values of the parameter that maximize the sample likelihood are known as the maximum likelihood estimates. Using MLE, the parameters are chosen to maximise the likelihood that the assumed model results in the observed data. Instead, the logistic equation predicts the log-odds of the event of interest occurring. Specifically, the general equation for logistic regression is: Which is usually written as below. The estimated regression coefficients in a logistic regression model give the estimated change in the log-odds corresponding to a unit change in the corresponding explanatory variable conditional on the other explanatory variables remaining constant. The parameters are usually exponentiated to give results in terms of odds. As an example, I have used a dataset consisting of the variables age (years), weight (kgs), gender (0=female, male=1), VO2max and coronary heart disease status (0=no, 1=yes). I have attached this dataset as an excel sheet “vo2max data.xlsx”. To perform logistic regression want to how well the incidence of coronary heart disease can be predicted based of age, weight, gender and VO2max in a sample of 100 persons. These are the steps I would perform. 1. Exploratory data analysis. This is to get a feel of your data e.g., what type are the variables (e.g., continuous, dichotomous, ordinal etc, how are the values of each are distributed, missing values, outliers). Part 1: Descriptive Statistics for Continuous Variables When summarizing a quantitative (continuous/interval/ratio) variable, we are typically interested in things like: How many observations were there? How many cases had missing values? (N valid; N missing) Where is the "center" of the data? (Mean, median) Where are the "benchmarks" of the data? (Quartiles, percentiles) How spread out is the data? (Standard deviation/variance) What are the extremes of the data? (Minimum, maximum; Outliers) What is the "shape" of the distribution? Is it symmetric or asymmetric? Are the values mostly clustered about the mean, or are there many values in the "tails" of the distribution? (Skewness, kurtosis) Descriptives Descriptives (Analyze > Descriptive Statistics > Descriptives) is best to obtain quick summaries of numeric variables, or to compare several numeric variables side-by-side. // Then use Statistics tab and choose the following You get this output: Explore Explore (Analyze > Descriptive Statistics > Explore) is best used to deeply investigate a single numeric variable, with or without a categorical grouping variable. It can produce a large number of descriptive statistics, as well as confidence intervals, normality tests, and plots. To explore statistics of by CHD statistic. EXAMINE VARIABLES=age weight gender vo2max BY chd /PLOT BOXPLOT STEMLEAF HISTOGRAM NPPLOT /COMPARE GROUPS /STATISTICS DESCRIPTIVES EXTREME /CINTERVAL 95 /MISSING LISTWISE /NOTOTAL. This is the output – some is more useful than other bits. I can go through this and explain and interpret any output you’re unsure about and let you know the main things you need to be looking for. Explore Notes Output Created 02-MAR-2022 12:19:14 Comments Input Active Dataset DataSet1 Filter <none> Weight <none> Split File <none> N of Rows in Working Data File Missing Value Handling Definition of Missing 98 User-defined missing values for dependent variables are treated as missing. Cases Used Statistics are based on cases with no missing values for any dependent variable or factor used. Syntax EXAMINE VARIABLES=age weight gender vo2max BY chd /PLOT BOXPLOT STEMLEAF HISTOGRAM NPPLOT /COMPARE GROUPS /STATISTICS DESCRIPTIVES EXTREME /CINTERVAL 95 /MISSING LISTWISE /NOTOTAL. Resources Processor Time 00:00:06.14 Elapsed Time 00:00:04.30 chd Case Processing Summary Cases Valid chd age 0 N Missing Percent 65 100.0% N Total Percent 0 0.0% N Percent 65 100.0% weight gender vo2max 1 33 100.0% 0 0.0% 33 100.0% 0 65 100.0% 0 0.0% 65 100.0% 1 33 100.0% 0 0.0% 33 100.0% 0 65 100.0% 0 0.0% 65 100.0% 1 33 100.0% 0 0.0% 33 100.0% 0 65 100.0% 0 0.0% 65 100.0% 1 33 100.0% 0 0.0% 33 100.0% Descriptives chd age 0 Statistic Mean 38.31 95% Confidence Interval for Lower Bound 36.51 Mean Upper Bound 40.11 5% Trimmed Mean 37.55 Median 36.00 Variance 7.271 Minimum 30 Maximum 63 Range 33 Interquartile Range 9 Skewness 1.532 .297 Kurtosis 2.643 .586 Mean 45.76 1.572 95% Confidence Interval for Lower Bound 42.56 Mean Upper Bound 48.96 5% Trimmed Mean 45.03 Median 43.00 Variance 81.564 Std. Deviation weight 0 .902 52.873 Std. Deviation 1 Std. Error 9.031 Minimum 35 Maximum 74 Range 39 Interquartile Range 12 Skewness 1.256 .409 Kurtosis 1.611 .798 76.9705 1.70114 Mean 95% Confidence Interval for Lower Bound 73.5720 Mean Upper Bound 80.3689 5% Trimmed Mean 76.5597 Median 77.3700 Variance 188.102 Std. Deviation 1 13.71503 Minimum 50.00 Maximum 115.42 Range 65.42 Interquartile Range 17.02 Skewness .401 .297 Kurtosis .389 .586 86.3697 2.63141 Mean 95% Confidence Interval for Lower Bound 81.0097 Mean Upper Bound 91.7297 5% Trimmed Mean 86.6962 Median 88.8300 Variance 228.502 Std. Deviation gender 0 15.11629 Minimum 53.43 Maximum 112.59 Range 59.16 Interquartile Range 22.18 Skewness -.470 .409 Kurtosis -.380 .798 .57 .062 Mean 95% Confidence Interval for Lower Bound .45 Mean Upper Bound .69 5% Trimmed Mean .58 Median 1.00 Variance .249 Std. Deviation .499 Minimum 0 Maximum 1 Range 1 Interquartile Range 1 Skewness Kurtosis 1 Mean -.286 .297 -1.980 .586 .79 .072 95% Confidence Interval for Lower Bound .64 Mean Upper Bound .94 5% Trimmed Mean Median .82 1.00 Variance .172 Std. Deviation .415 Minimum 0 Maximum 1 Range 1 Interquartile Range 0 Skewness Kurtosis vo2max 0 Mean .798 45.0000 1.14909 42.7044 Mean Upper Bound 47.2956 5% Trimmed Mean 44.9076 Median 45.0600 85.827 9.26427 Minimum 27.35 Maximum 62.50 Range 35.15 Interquartile Range 13.17 Skewness Kurtosis Mean .230 .297 -.801 .586 40.6242 1.03376 95% Confidence Interval for Lower Bound 38.5185 Mean Upper Bound 42.7299 5% Trimmed Mean 40.5282 Median 40.5800 Variance 35.266 Std. Deviation 5.93851 Minimum 28.30 Maximum 55.19 Range 26.89 Interquartile Range 8.95 Skewness .191 .409 -.022 .798 Kurtosis Extreme Values chd 0 .187 Lower Bound Std. Deviation age .409 95% Confidence Interval for Variance 1 -1.476 Case Number Highest Value 1 74 63 2 85 63 Lowest 1 Highest Lowest weight 0 Highest Lowest 1 Highest Lowest gender 0 Highest 3 65 55 4 76 51 5 35 50 1 67 30 2 63 31 3 57 31 4 43 31 5 41 31a 1 60 74 2 71 62 3 98 61 4 97 58 5 12 55b 1 9 35 2 32 36 3 25 36 4 4 36 5 45 37 1 48 115.42 2 15 111.80 3 5 103.00 4 83 101.62 5 43 97.75 1 58 50.00 2 49 51.96 3 88 53.00 4 67 55.00 5 35 58.07 1 59 112.59 2 94 111.98 3 90 103.53 4 64 103.23 5 40 101.25 1 25 53.43 2 17 56.18 3 32 62.59 4 55 64.00 5 62 68.29 1 1 1 2 2 1 Lowest 1 Highest Lowest vo2max 0 Highest Lowest 1 Highest Lowest 3 5 1 4 7 1 5 15 1c 1 91 0 2 88 0 3 86 0 4 78 0 5 74 0d 1 3 1 2 9 1 3 11 1 4 17 1 5 20 1c 1 87 0 2 62 0 3 60 0 4 55 0 5 31 0d 1 23 62.50 2 38 62.13 3 77 61.76 4 52 61.71 5 70 60.92 1 36 27.35 2 91 30.37 3 8 30.38 4 15 31.94 5 74 31.99 1 27 55.19 2 82 50.19 3 95 49.22 4 32 48.23 5 17 47.23 1 4 28.30 2 98 32.00 3 71 32.00 4 87 33.67 5 94 33.73 a. Only a partial list of cases with the value 31 are shown in the table of lower extremes. b. Only a partial list of cases with the value 55 are shown in the table of upper extremes. c. Only a partial list of cases with the value 1 are shown in the table of upper extremes. d. Only a partial list of cases with the value 0 are shown in the table of lower extremes. Tests of Normality Kolmogorov-Smirnova chd age weight gender vo2max Statistic df Shapiro-Wilk Sig. Statistic df Sig. 0 .148 65 .001 .854 65 .000 1 .165 33 .022 .893 33 .003 0 .082 65 .200* .981 65 .424 1 .200 33 .002 .952 33 .154 0 .375 65 .000 .630 65 .000 1 .483 33 .000 .505 33 .000 0 .067 65 .200* .967 65 .082 1 .074 33 .200* .991 33 .993 *. This is a lower bound of the true significance. a. Lilliefors Significance Correction age Histograms Stem-and-Leaf Plots age Stem-and-Leaf Plot for chd= 0 Frequency 26.00 16.00 13.00 5.00 Stem & 3 3 4 4 . . . . Leaf 01111112222222333333444444 5555666677789999 0000111233334 66888 2.00 5 . 3.00 Extremes Stem width: Each leaf: 01 (>=55) 10 1 case(s) age Stem-and-Leaf Plot for chd= 1 Frequency Stem & .00 3 8.00 3 10.00 4 6.00 4 2.00 5 4.00 5 2.00 6 1.00 Extremes Stem width: Each leaf: . . . . . . . Leaf 56667888 0011122233 566779 01 5558 12 (>=74) 10 Normal Q-Q Plots 1 case(s) Detrended Normal Q-Q Plots weight Histograms Stem-and-Leaf Plots weight Stem-and-Leaf Plot for chd= 0 Frequency 8.00 Stem & 5 . Leaf 01358889 11.00 6 23.00 7 13.00 8 6.00 9 2.00 10 2.00 Extremes Stem width: Each leaf: . . . . . 02223488899 00122234455567888899999 0112225567779 002457 13 (>=112) 10.00 1 case(s) weight Stem-and-Leaf Plot for chd= 1 Frequency 2.00 3.00 6.00 7.00 9.00 4.00 2.00 Stem width: Each leaf: Stem & 5 6 7 8 9 10 11 . . . . . . . Leaf 36 248 233445 7888889 013444577 1133 12 10.00 1 case(s) Normal Q-Q Plots Detrended Normal Q-Q Plots gender Histograms Stem-and-Leaf Plots gender Stem-and-Leaf Plot for chd= 0 Frequency Stem & 28.00 .00 .00 .00 .00 37.00 Stem width: Each leaf: 0 0 0 0 0 1 . . . . . . Leaf 0000000000000000000000000000 0000000000000000000000000000000000000 1 1 case(s) gender Stem-and-Leaf Plot for chd= 1 Frequency Stem & 7.00 Extremes 26.00 1 . Stem width: Each leaf: Leaf (=<.0) 00000000000000000000000000 1 Normal Q-Q Plots 1 case(s) Detrended Normal Q-Q Plots vo2max Histograms Stem-and-Leaf Plots vo2max Stem-and-Leaf Plot for chd= 0 Frequency 1.00 10.00 9.00 12.00 16.00 5.00 5.00 7.00 Stem width: Each leaf: Stem & 2 3 3 4 4 5 5 6 . . . . . . . . Leaf 7 0011333344 666667788 000011222344 5555577778899999 00124 55578 0001122 10.00 1 case(s) vo2max Stem-and-Leaf Plot for chd= 1 Frequency 1.00 4.00 9.00 12.00 5.00 1.00 1.00 Stem width: Each leaf: Stem & 2 3 3 4 4 5 5 . . . . . . . Leaf 8 2233 555577889 000012222444 55789 0 5 10.00 Normal Q-Q Plots 1 case(s) Detrended Normal Q-Q Plots Frequencies Part I (Continuous Variables) Frequencies (Analyze > Descriptive Statistics > Frequencies) is typically used to analyze categorical variables, but can also be used to obtain percentile statistics that aren't otherwise included in the Descriptives, Compare Means, or Explore procedures. Part 2: Descriptive Statistics for Categorical Variables When summarizing qualitative (nominal or ordinal) variables, we are typically interested in things like: How many cases were in each category? (Counts) What proportion of the cases were in each category? (Percentage, valid percent, cumulative percent) What was the most frequently occurring category (i.e., the category with the most observations)? (Mode) In Part 2, we describe how to obtain descriptive statistics for categorical variables using the Frequencies and Crosstabs procedures. Frequencies Part II (Categorical Variables) Frequencies (Analyze > Descriptive Statistics > Frequencies) is primarily used to create frequency tables, bar charts, and pie charts for a single categorical variable. Crosstabs The Crosstabs procedure (Analyze > Descriptive Statistics > Crosstabs) is used to create contingency tables, which describe the interaction between two categorical variables. This tutorial covers the descriptive statistics aspects of the Crosstabs procedure, including and row, column, and total percents. Multiple Response Sets / Working with "Check All That Apply" Survey Data Check-all-that-apply questions on surveys are recorded as a set of binary indicator variables for each checkbox option. Frequency tables and crosstabs alone don't capture the dependent nature of this data -- and that's where Multiple Response Sets come in.