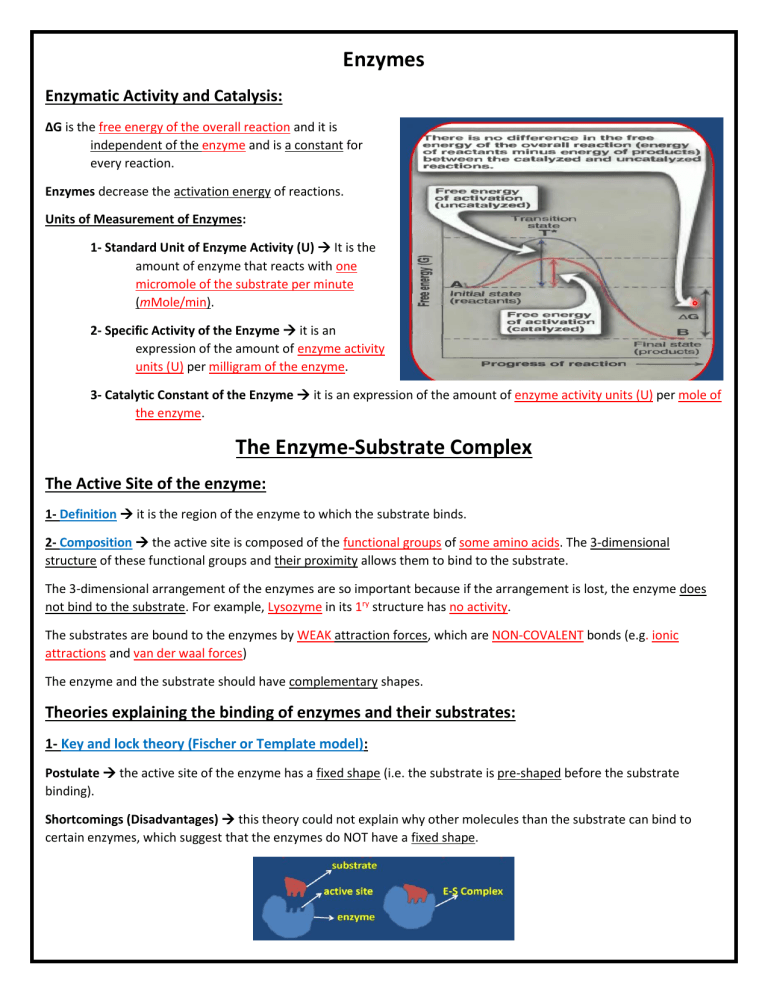
Enzymes Enzymatic Activity and Catalysis: ΔG is the free energy of the overall reaction and it is independent of the enzyme and is a constant for every reaction. Enzymes decrease the activation energy of reactions. Units of Measurement of Enzymes: 1- Standard Unit of Enzyme Activity (U) It is the amount of enzyme that reacts with one micromole of the substrate per minute (mMole/min). 2- Specific Activity of the Enzyme it is an expression of the amount of enzyme activity units (U) per milligram of the enzyme. 3- Catalytic Constant of the Enzyme it is an expression of the amount of enzyme activity units (U) per mole of the enzyme. The Enzyme-Substrate Complex The Active Site of the enzyme: 1- Definition it is the region of the enzyme to which the substrate binds. 2- Composition the active site is composed of the functional groups of some amino acids. The 3-dimensional structure of these functional groups and their proximity allows them to bind to the substrate. The 3-dimensional arrangement of the enzymes are so important because if the arrangement is lost, the enzyme does not bind to the substrate. For example, Lysozyme in its 1ry structure has no activity. The substrates are bound to the enzymes by WEAK attraction forces, which are NON-COVALENT bonds (e.g. ionic attractions and van der waal forces) The enzyme and the substrate should have complementary shapes. Theories explaining the binding of enzymes and their substrates: 1- Key and lock theory (Fischer or Template model): Postulate the active site of the enzyme has a fixed shape (i.e. the substrate is pre-shaped before the substrate binding). Shortcomings (Disadvantages) this theory could not explain why other molecules than the substrate can bind to certain enzymes, which suggest that the enzymes do NOT have a fixed shape. 2- The Induced-fit model of Koshland: Postulate the active site of the enzymes have shapes that are complementary to that of the substrate only after the binding of the substrate (i.e. the enzyme is NOT pre-shaped.) Mechanism the substrate induces the active site. Advantages It explains how more than one substrate can bind to an enzyme. The Specificity of Enzymes: Enzymes are highly specific both in the reaction catalyzed and their choice of substrates. The specificity is determined by the functional groups of the enzyme, cofactors & the substrates and the physical proximity between these groups. Types of Enzyme specificity: 1- Relative Specificity the enzyme works on many substrates but has a ↑ affinity for a specific substrate. - Examples Lipases and Proteases. 2- Structural (positional) Specificity the enzyme works on a specific bond in a very specific position. - Examples Pepsin (works between the carbonyl of aspartic acid and the Nitrogen of tyrosine) 3- Absolute Specificity the enzyme works on only one substrate. - Examples Ureases & Sucrase 4- Optical Specificity the enzyme works on only one substrate in a specific stereo form. - Examples Glycolytic enzymes & amino acid oxidases. Enzyme Kinetics Factors affecting enzymatic activity (measuring enzymatic activity): 1- Enzyme Concentration: 2- Substrate Concentration: - At ↓ concentrations of the substrate, the reaction has a first-order reaction kinetics. At ↑ concentrations of the substrate, the reaction has a zero-order reaction kinetics. Km (Michaelis constant) Value: - Definition it is the amount of substrate needed for the reaction rate to reach ½ Vmax. Characters Km is inversely proportional to the affinity of the enzyme to the substrate. Km is independent of the enzyme concentration. If an enzyme acts on several substrates, it will have more than one Km for each substrate. Michaelis-Menten Equation: Lineweaver-Burk Plot: Uses Determination of Km and VMAX & Studying the mechanism of action of Enzyme Inhibitors. Graph and Formula N.B. The value of VMAX reflects the amount of active enzyme present and it is independent of the substrate’s concentration. 3- Temperature: At ↓ temperatures Energy < Eact At ↑ temperatures Denaturation due to cross-links destruction. Enzymes need very specific temperatures (usually 37oC) 4- pH: Effective concentration of an enzyme (or a substrate) It is the concentration of the enzyme that can actually bind to the substrate (has the required 3-D shape and charge). The pH of the medium causes alterations in the charges of both the Enzyme and the substrate, which changes their effective concentrations. Enzyme denaturation occurs at high pH. 5- Time of the reaction & Product of the reaction 6- Light and other physical factors Coenzymes Apoenzyme + Coenzyme = Holoenzyme Apoenzyme Determines the substrate. It’s the protein part that connects to the substrate. Coenzyme Determines the biochemical reaction. It accepts or donates groups removed or added to the substrates, thus is regarded as 2ry substrate sometimes. Some of the coenzymes are vitamin B derivatives and are of nucleotide structure. Coenzymes are classified according to the group they transfer. Enzyme Coenzyme NAD + NADP Dehydrogenases FAD FMN Lipoic Acid Pyruvate Dehydrogenase Amino Transferase (Trans-aminase Reactions) Ligases (Carboxylases) Transferase TPP (Thiamine pyrophosphate) Group Transferred Hydrogen (As HYDRIDE) Coenzyme Reaction Hydrogen (As HYDRIDE) Hydrogen (As ATOM) Hydrogen (As ATOM) Hydrogen NADP + 2H NADPH + H+ FAD + AH2 FADH2 +A Vitamin Precursor NAD++ AH2 NADH + H+ + A + Niacin (Vit B3) Riboflavin (Vit B2) S—S SH , SH PLP (Pyridoxal Phosphate) Pyridoxine (Vit B6) Biotin CO2 Vitamin H Acyl group Nicotinamide + Adenine + Dinucleotide (2 Ribose + 2 Phosphate) Nicotinamide + Adenine + Dinucleotide Phosphate Flavin + Adenine + Dinucleotide Flavin + Mono Nucleotide 6,8-dithio-octanoic acid Aldehyde (Acetal) Aldehyde (Acetal) Amino Group Co-enzyme A Structure Components Thiamin (Vit B1) Pantothenic Acid (Vit B5) Thiamine + 2 Phosphate groups Pyridoxine (Vit B6) + Phosphate Nitrogenous base + Sugar + P + P + Pantothenic Acid (Has a thioethanol amine which is the active moiety) Inhibition of Enzyme Activity An Inhibitor is any substance that can diminish the velocity of an enzyme-catalyzed reaction. Enzyme Inhibitors Reversible Inhibitors Competitive Inhibitors Uncompetitive Inhibitors Irreversible Inhibitors Non-competitive Inhibitors Significance of Enzyme Inhibition Study: 1- Enzyme inhibition is a major control mechanism in the biological systems. 2- Many drugs and toxic chemicals act by inhibiting enzymes. 3- It is useful in studying the mechanism of enzyme action. Group-Specific Irreversible Inhibitors: 1Irreversible Inhibitor DIPF (Diisopropyl fluorophosphates) Enzyme Acetylcholinesterase It reacts with a critical Serine (-OH) group. 2Irreversible Inhibitor Iodoacetamide Enzyme Enzymes containing (-SH) group of cysteine. It reacts with a critical cysteine (-SH) residue. Competitive Non-competitive (Enzyme Poison) Reversible Inhibitors Competitive Inhibitors Uncompetitive Inhibitors Mechanism The inhibitor resembles the substrate and binds at active site The inhibitor binds to the ES complex Non-Competitive Inhibition The Inhibitor and the substrate bind on different sites on the enzyme. The inhibitor can bind to Enzyme or ES complex. Diagram for the Mechanism Reaction Equation Overcome by ↑↑ Substrate Concentration Dialysis via semipermeable membrane Direct Plots LineweaverBurk Plot Effect on Enzym e Kinetic s Km Vmax ↑↑ No change [G.R.] Available amount of enzyme has not changed. If [S] is ↑↑ then Vmax can be reached. ↓↓ No Change ↓↓ ↓↓ [G.R.] because the number of active enzymes is decreased. Competitive Inhibitors Examples 1C.I. Malonate Enzyme Succinase Dehydrogenase Substrate Succinate 2Sulfa drugs (Sulfanilamide) is a C.I. for PABA, which is required by M.O. for folic acid synthesis. N.B. Sulfa drugs do not harm humans [G.R.] Humans cannot produce folate. 3Folate Analogs (e.g. Aminopterin, Methotrexate) acts as antimetabolites. They are used as anti-tumor agents in Childhood leukemia. C.I. Methotrexate and Aminopterin Enzyme Dihydrofolate Reductase Substrate Folic Acid N.B: Methotrexate bounds 1000 fold better than folate to DHF reductase. S.E. of Methotrexate Bone marrow suppression, GIT mucosal damage and drug resistance. 4Lovastatin Is a hypercholesteremic drug that inhibits endogenous cholesterol synthesis. C.I. –> Lovastatin Enzyme HMG Co-A Reductase Substrate HMG Co-A (Hydroxymethylglutyryl Co-A) Uncompetitive Inhibitors NonCompetitiv e Inhibition Extract of the parasite (Ascaris) secretes pepsin and trypsin inhibitors so that the parasite escapes digestion in intestine.