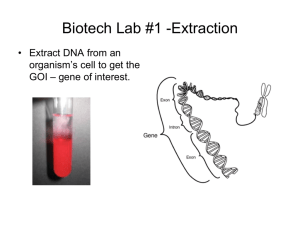
Agenda • Learn why biotechnology is important in modern-day medicine, agriculture, and environmental remediation • Learn about about plasmids & restriction enzymes as tools in molecular biology/biotechnology/genetic engineering • Learn about precise measurement of ultra-small volumes using a micropipette • Watch and become familiar with the procedures for plasmid isolation and restriction enzyme digest Important Vocabulary/Terms -Plasmid -Vector -Insert -Construct -Digest -Ligase -Palindromic -Restriction Enzyme -Sticky Ends -Complementary Bases -Origin of replication -Promoter -Antibiotic resistance gene -Regulatory gene What is a plasmid? • Comparatively small circular double-stranded DNA molecules of bacterial origin • Range in size from 1,000 to 200,000 base pairs (bp) • Independent of bacterial chromosome, carrying “nonessential” genes Plasmid Features: • Origin of replication • Promoter • Antibiotic resistance • Multiple cloning site (polylinker) • Regulatory element(s) Function: - Region responsible for initiating the copying of plasmid DNA - Site to which RNA polymerase binds to begin transcription - Gene coding for a product that confers antibiotic resistance - Unique restriction sites allow for the digestion of plasmid & introduction of insert (foreign gene) - Gene coding for a product that regulates transcription of the insert promoter regulatory gene MCS (with insert) antibiotic resistance gene origin of replication (Ori) Plasmids as Vectors What are restriction enzymes? • Catalytic proteins that function like molecular scissors, cutting double-stranded DNA at distinct recognition sites that are usually unique to a particular enzyme. What are restriction enzymes? Characteristics: • Recognition sites are palindromic sequences, usually 4-8 nucleotides in length Cut sites 5’ – G A A T T C – 3’ are NOT aligned 3’ – C T T A A G – 5’ • Cleave covalent bonds of sugar-phosphate backbone • If enzyme is a staggered cutter, generates sticky ends (unpaired overhangs capable of hydrogen bonding with complementary bases) 5’ – G A A T T C – 3’ 3’ – C T T A A G – 5’ • Nonemclature based on source bacterial species & strain 1st letter roman numeral designates E co R I of genus order of discovery 1st two strain (Escherichia) letters of species (coli) What about aligned cuts? Characteristics: • Recognition sites are still palindromic sequences, usually 48 nucleotides in length Cut sites 5’ – C C C G G G – 3’ ARE aligned 3’ – G G G C C C – 5’ • Covalent bonds of sugar-phosphate backbone are cleaved as before • In the above example, the enzyme is now a blunt cutter, generates blunt ends (NO overhangs – any two blunts ends can match up…not based on complementary/matching sequence) 5’ – C C C G G G – 3’ 3’ – G G G C C C – 5’ Application of These Molecular Tools – Scientists can build designer plasmids that contain specific restriction sites – This allows scientist to cut out and recombine genes to allow for cloning and gene expression (requires cutting each DNA sample with same restriction enzyme(s)) Gene Cutting & Splicing Simulator • Visit the following simulation activity site: • http://glencoe.mheducation.com/sites/dl/free/007880284 9/383937/BL_22.html • Perform two simulations by following the instructions provided by the simulator: 1. Splice the human insulin gene into E. coli bacteria 2. Splice the firefly luminescence (“glow-in-the-dark”) gene into tobacco plant Tip: Make sure you select an enzyme (A-J; descriptions of where each will cut are provided by the simulator) that will cut outside of the highlighted gene sequence Our Recombinant Plasmid (rpARA) Regulatory gene – codes for repressor protein that controls the expression of rfp/tomato (insert) gene Promoter for insert gene Restriction Enzymes Ampicillin resistance gene – codes for b-lactamase enzyme that destroys the antibiotic called ampicillin Insert rfp gene – codes for Red Fluorescent Protein Why “Tomato”? • The name “tomato” is a nickname derived from the shared color of tomatoes and the gene’s protein product, Red Fluorescent Protein (RFP) • The tomato gene (a.k.a. rfp gene) is NOT derived from tomato plants; it originates from a species of sea anemone that glows/fluoresces red in the ocean upon exposure to UV light from the Sun. • With this gene as our insert in the recombinant plasmid, rpARA, we can give it to any living cell and cause that cell to make red fluorescent protein and glow/fluoresce red upon expression of the gene Extracting & Digesting rpARA (isolating and cutting the recombinant plasmid) • Purpose: to collect plasmid DNA from bacteria, perform a restriction enzyme digest, and produce DNA fragments of appropriate size that will confirm the presence of the rfp/tomato gene. – Will need to perform a plasmid isolation protocol • Miniprep using alkaline lysis – Will use two restriction enzymes on the isolated plasmid, liberating the rfp/tomato insert from the remainder of the plasmid (a.k.a. vector): • BamH I • Hind III Plasmid Isolation from E. coli • First, read the introduction on pg. 89 of your lab manual and then watch the tutorial on how to use the various micropipettes:https://www.youtube.com/watch?v=uEy_NGDfo_ 8 • Complete the virtual micropipette activity & take the quiz at the end as practice: http://www.virtual-labs.leeds.ac.uk/pres/micropipettes/ • Next, view the procedure, outlined on pages 94-96, executed in a video format (follow along as you watch) – Note: steps 11-18 in the actual lab protocol require a DNA precipitation and washes…this has been replaced in the video with a spin column purification (video time index = 7:25) : https://www.youtube.com/watch?v=8xEDEJ0DHFA&t=181s Plasmid Double Digestion • Lastly, read the introduction on pg. 97 and watch a similar procedure to that on pgs. 98-99 demonstrated in the following video – Note: the actual lab protocol is for a “double digest” of bacterial plasmid DNA (rpARA) where the enzymes, BamH I & Hind III are combined in the same reaction at the same time rather than individually/separately: https://www.youtube.com/watch?v=GsWo8dCivWs&t=128s How will we know if we were successful in our digestion of the plasmid? 1. We must confirm the double digestion of the rpARA plasmid produced fragments of expected size. 2. We must confirm that the negative control did not cut the rpARA plasmid. We will confirm these two points next week with a methodology know as agarose gel electrophoresis; Please answer the questions on page 100 of your lab manual. www.amgenbiotechexperience.com