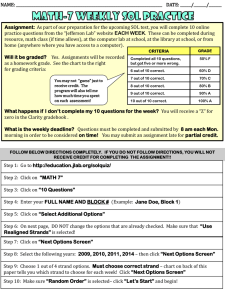
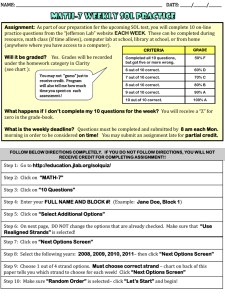
I. DNA Primer A. Significance of the ‘Size’ of the match. Not all matches are equal. Identifying a certain DNA segment as a “match” for two people doesn’t guarantee common ancestry. The vast majority of DNA shared by all humans is identical anyway. When science compares two people, they are really looking at the places where mutations have arisen and thus where people tend to be “different” from one another. If you are different in the same way, then you “match” over that segment. Generally, the smaller the matching segment, the more likely that the match is coincidental. The larger the matching segment, the more likely that the match is ancestrally derived. Some sources claim that a match measuring 7 cMs is ancestrally derived about 50% of the time. If we match because we both inherited that strand from a common ancestor, the match is referred to as “Identical By Descent” – abbreviated as IBD. Other sources claim that a 7 cM matching segment might only have a 30-40% chance of common ancestry – perhaps in part because the match can’t be proven to be from common ancestry (the match may be IBD, but the common ancestor may be so far back that it can’t be identified). In any event, while those quantifying the statistics around the size of match may differ in their conclusions about likelihood of IBD matches, one thing they will all agree on -- the smaller the match is, the less likely it is ancestrally derived.1 We all have coincidental matches if we look at segments that are really small. It is possible to have coincidental matches for even longer segments, but the odds that a match is coincidental decreases as the size of the match increases. That said, when you KNOW you have a common ancestor, even smaller matches may reflect legitimate matches, especially with higher SnPs (defined below). Because we KNOW we have some common ancestry, the odds are much higher that our matching DNA (even on smaller segments) came from the same source. But generally we are looking for matches of at least 7cMs or more, preferably with 700 SnPs or more, and frankly we prefer dealing with much larger matches. Using larger matches can be important -- ftDNA may report that you have 80 cMs matching a distant cousin. On Ancestry, they may report that you have 54 cMs matching. How can they be different? Well, one may cumulate matches of 5cMs or more. The other may cumulate matches of 7 cMs or more. Only in looking more carefully at the matches can you really see the quality of the matches. For example, in theory you could have twelve different 5cM matches with someone (called ’60 cM’ matching), and they could ALL be coincidental matches. On the other hand, you could have ONE consecutive 35cM match with another distant cousin (called ‘35cM’ matching). Which is the better match? At first glance 60 seems like it would be better than 35, but the 35 may be FAR more important. The first could be nothing at all, the second is statistically almost certain to be a ‘real’ match – derived from a common ancestor. This is why the total matches can mean different things, and why the quality of your longest matches is really the more important consideration. 1 Pick a random person on gedmatch.com—someone related to your spouse, but not you, and compare your kits using 7.0 cMs and 700 SnPs. If it shows NO matches, then run it again using 2.0 and 200 – you may see many ‘matches’ but odds are these are not ancestrally derived, they are simply coincidental. When you see a match of 30 cM, however, that is almost certainly ancestrally derived. It is just ‘too’ much to be a coincidence. B. Visualizing the Chromosome. DNA in a human cell is basically 46 strands (22 “autosomal” strands from each parent, plus the XX or the XY), and the mitochondrial material included in each cell. 2 Each strand then has millions of incremental points along the strand. These points are typically referenced in increments of one millon ‘spots’ – so segment 1 represents the first 1 million spots. Mom gives you one strand of Chr-1 through Chr-22, each strand being a recombination of her two strands for that chromosome. She selectively passes on, for each spot on your chromosome, either what she got from her father or what she got from her mother. You end up with a beautifully mixed hodge-podge (“recombination”) of what she got from her parents, all mixed onto one single strand for each chromosome, from her to you. She gives you half of her genetic material, but she also leaves the other half on the cutting room floor. Your Dad does the same thing, recombining his two chromosome strands into one to pass on to you, so you have one strand from each parent, and now YOU have two strands for each chromosome. Each strand is a long stretch of genetic material containing millions of individual ‘spots’ along the length of the chromosome, joined by a bonding material… if stretched out, a strand might look a little like a ladder, long sides with rungs in the middle. Imagine letting go of this long strand of stretched out material, and it springs back into its normal position…twisted pairs, coiled to form a double helix. Each spot on a strand is called a “nucleotide” and it has genetic material that can be represented by either the letter ACG or T. 3 For example, one part of a strand might look like this: ACCTGAGTCAGTAC. And remember that you have two strands, so there will be a corresponding series of letters for the other strand. There are literally MILLIONs of these spots (each represented by a letter) making up each strand, so a pair of these spots (along with the bonding material) making up each chromosome. Different chromosomes have varying number of spots. Chr-1 may have around 247,000,000 spots on each strand, while Chr-21 may have only around 47,000,000. Don’t be surprised if you find more Chr-1 DNA matches than Chr-21 matches. Matches are identified from one spot, to another where the match ends. So, it might be from spot 9,124,232 to spot 19,896,974. These spots can be referenced as sections comprised of one million spots each; so, if someone says they match from Section 12 to Section 44 of Chr-4, they really mean that the match starts somewhere in the 12,000,000 range, and it ends somewhere in the 44,000,000 range of that chromosome. Each group of a million spots can be thought of as one segment or section. 2 Actually, there is also a mirror image of the base pairs also included, so you really have 4 strands of each chromosome, but for our purposes, we’ll stick with the part that matters, and ignore the mirror images. 3 Each letter references a different protein, but for our purposes, all we care about are the letters. For the most part, humans have the same genetic code…. We are all almost identical! BUT there are places in each chromosome where mutations have occurred. A single-nucleotide polymorphism, abbreviated SnP and pronounced snip, is a single spot in the genome where, due to mutation, there is a relatively high degree of variation between different people. These are the spots where they test. In addition to the twisted pairs of autosomal chromosomes (1-22), Mom gives a recombined X chromosome strand to all of her children, and Dad passes on the exact X chromosome he received from his mom to his daughters only (this is not recombined… it is EXACTLY what his mom passed out to him). So, the girls have a recombined X from Mom (with DNA material from both her maternal and paternal strands), and an UNRECOMBINED X from Dad – as a male he had only one X strand, so he passed it along unchanged. Note that X chromosome material can come from Mom or Dad, if you are female, but only from Mom if you are male. When tracing back the contributor for a chr-X match, it NEVER flows through two men in a row on a family tree. Dad doesn’t give the sons his chr-X, instead he passes on the Y-chromosome that he received from his Dad. The Y from the Dad does not recombine (his mom didn’t have a Y, so Dad had only one Ystrand and it passes on from father to son, intact… always without recombining because there is nothing to recombine with. Aside from mutations along the way, the Y is preserved along the MALE line. For this reason, Y-Testing can be done by men to help confirm a common male line. In families with very common surnames, this can help a Smith line know what Smith line it ties back into. Mom has genetic material that came only from her mother the (mT material). She gives it to all of her children (so they all have this info about their own mom), but males do not pass it on. A female will have the same mT material from her mom, but she WILL pass it on. It does not recombine. Y-material passes only from male to male. The ‘mitochondrial’ DNA passes from Mom to ALL children without recombining, but it won’t pass on from the male children to further descendants. ONLY the daughters pass it on. Since it comes from only one parent and does not recombine, the mT DNA material from mom in theory is passed on intact at each generation, identical from her maternal line since the beginning… mutations along the way may however result in small changes. A nice link to see how DNA recombines (or not, as to the Y and the mT), is available at this site. CentiMorgans. A CentiMorgan (cM) is a measurement of how likely an area of DNA is to recombine from one generation to the next. A single centiMorgan is considered equivalent to a 1% (1/100) chance that a segment of DNA will crossover or recombine within one generation. For humans, one million base pairs (bp) average about one centiMorgan. (I copied that definition from the FT-DNA website). I prefer to think of a centimorgan (cM) is a unit of length, over which a match occurs. Just for visualization purposes. SNPs. Defined above, GedMatch reports the number of “SnPs” that match within a matching segment. This means the number of unique spots on a matching string of DNA that match and are different from the prototype. You can have a match that is 10 cMs long, with only 400 SNPs (matching points of departure in the mutated area). You can also have a 5 cM strip where you have 1200 SNPs in common. These are both factors in likelihood of common ancestor. Segments. A third factor is the number of segments a matching strip crosses. Spots 1-1,000,000 is treated as segment 1. Segments are referenced by dividing the area by 1M. So, I will refer to the ‘area’ of a match by taking the GedMatch numbers for Start and End and dividing those by 1,000,000. The more segments per cM also has some bearing. C. Distinguishing the Strands. If AncestryDNA or Gedmatch, or any other cite actually looking at the raw data says you ‘match’ someone else… that means that for that area of that chromosome you have the same genetic material… and typically you can do a one-to-one comparison of the two kits on gedmatch and see the cMs, the SNPs, and the segments involved. This lets us map those matches if you like, and see who ELSE matches you in the same areas… If (A) matches (B), and if (A) matches (C) in the same segment of a chromosome, will (B) also match (C) in the EXACT same area of the same chromosome? Maybe, maybe not. It is important to realize that the testing sights don’t separately test and report on each of your two strands of each chromosome— they simply report BOTH letters present at each spot. It’s up to you to separate the match info. So, you match two people over the same segments on Chr-10. Sometimes each of the three of you do indeed match both of the others on one of your two chromosome strands for the involved chromosome. BUT, it could also be that you (A), match (B) on your paternal strand, and (C) on your maternal strand, so it would make perfect sense that (B) and (C) don’t match at all! This can help you. If you have multiple matches in a section of a chromosome, run them all against each other and they will begin to fall into two groups,4 one corresponding to each of your strands. If you KNOW the common ancestry with one match, then this tells you which strand that group matches on. THEN, you can mark the other group as belonging to the other strand. One group will be matching your Maternal (M) line, or the other your Paternal (P) line. You may not know the common ancestry for the second group but at least you have cut in half the tree you much work from to find it! As you map your matches, you’ll be working on a maternal and a paternal list. Your matches will help you know the source of your DNA. D. False Positives. Sometimes, a match isn’ t a match at all. You are looking for others who have a strand that matches EITHER of your strands. But a false positive can arise when by coincidence you could go back and forth between your strands and find that ONE fo the two strands matches ONE of the two strands of the other person over a particular stretch of a chromosome. These coincidental matches are more common in short matches, and the longer the match the less likely that it is a false positive. But it can happen. You could be mixing and matching and have a false match reported, when in fact the other person didn’t match 4 There may be a few in the group that match you, but don’t seem to fall into either group. These will be false matches, discussed below. either of your strands, but did coincidentally have matching letters across the various strands. Just as an example, let’s say you have two strands (for one particular section of Chr-1) ACCTGAGTCAGTAC CCTGAGTCAGTACA Now, let’s say Z had a single strand that had the bolded letters from your two strands: ACCGGGGCCGGAAA. That might read as a match, but it is just a coincidence. You don’t have a strand that matches his strand, your ‘match’ is nothing more than a coincidental set of letters – his on one strand matching spots on your two strands. The false positive doesn’t have to follow a single strand for Z. The coincidence (going back and forth over your two strands) could also be matching back and forth over Z’s two strands. False positives can happen. That is why we look for triangulation and use more sophisticated tools to confirm the matches. Likewise, you might have a LEGITIMATE match from a common ancestor, but you might have some of this coincidental matching on either or both ends of the matching segment that might make the match look a little bit longer than it is. 5 Okay, that is your DNA primer. It is included to help us all help each other. Anyone who can correct any misunderstandings I have, or better explain something, please do send me the updated info! I may be able to include it with future reports. Now onto the important part. How do we match, and what do we learn from those matches. 5 I struggled with this at first. I might run two cousins, and toss in the child of one cousin, and find the child’s match was larger than his or her parent. Usually the parent’s match will be longer, or the same if ALL of the match was passed on to the child. After all, for a match on the maternal side, everything I got came from Mom, so how could my match be larger? Well, it could be if there was some of the coincidental matching on the ends.