Recombinant DNA Simulation: Insulin Production
advertisement

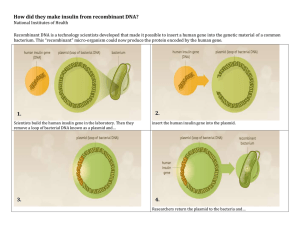
NAME __________________________ LAB Recombinant DNA Simulation (Modified from Boulay/Joslin) INTRODUCTION Genetic engineering is possible because of special enzymes that cut DNA. These enzymes are called restriction enzymes. Restriction enzymes are proteins produced by bacteria used to prevent (or restrict) the invasion of viruses. They act as “DNA scissors”, cutting viral DNA into pieces so that it cannot function. Restriction enzymes recognize and cut at specific places along a DNA molecule called restriction sites. Each restriction enzyme has its own restriction site. In general, a restriction site is a 4–8 base pair sequence of DNA. The DNA sequence below is the restriction site for the restriction enzyme known as EcoR1. Its name comes from the bacterium in which it was discovered: Escherichia coli (E. coli). 5’ – GGATCC – 3’ 3’ – CCTAGG – 5’ The EcoR1 enzyme scans a DNA molecule, and each time it finds the sequence above, it makes one cut between the G and G in each of the DNA strands (see below). After the cuts are made, the DNA is held together only by weak hydrogen bonds between the four bases GATC. These bonds are easily broken apart. Cut Sites 5’ – GGATCC – 3’ 3’ – CCTAGG – 5’ Notice the EcoR1 cut sites are not directly across from each other on the DNA molecule. When EcoR1 makes its cuts, it leaves single-stranded “tails” on the new ends (see above). This type of end is called a STICKY END because complementary DNA sticky ends can be easily rejoined. In this activity, we will simulate the production of INSULIN – a hormone made by cells of the pancreas that regulates carbohydrate metabolism. Individuals with diabetes mellitus require supplements of insulin to make up for their body's inability to produce sufficient amounts. To achieve our goal, we need to collect plasmids from E. coli cells. Plasmids are small circular pieces of DNA found in prokaryotic (bacterial) cells. Next, we need to extract the normal insulin gene from a human chromosome (chromosome 11). Insulin is made up of TWO polypeptide chains (51 amino acids). 153 DNA nitrogen bases constitute its gene sequence. MATERIALS Paper strip of bacterial plasmid DNA (blue) & paper strip of human insulin gene (yellow) Scissors, tape, highlighter, pencil METHODS PLASMID DNA (Bacteria) 1. Cut out the paper plasmid DNA along the rectangular border. 2. Tape the ends together so the plasmid is circular and the DNA bases face outward. INSULIN GENE EXTRACTION (Human) 3. Cut out the paper human insulin gene along the rectangular border. 4. Locate the TWO EcoR1 restriction sites on the gene and mark the cuts that will be made with a pencil. (See Figure 1) The restriction site is: Figure 1: 5’ – GGATCC – 3’ 3’ – CCTAGG – 5’ 5. Cut the human DNA with scissors at the 2 restriction sites (see the Introduction for proper cut sites). The human insulin gene is the DNA found BETWEEN the 2 restriction sites. INSULIN GENE INSERTION into the PLASMID 6. Scan the plasmid for the EcoR1 restriction site and highlight it. 7. Cut the plasmid open at this site in the same way as done in the above human insulin gene. If you make an error, tape up your plasmid and try again. 8. Using the rules for base pairing, insert the human insulin gene into the plasmid. Use tape to seal the ends together. RESULTS Your finished product is known as recombinant DNA since it contains DNA from more than one organism. If this were a real experiment, the recombinant plasmid would then be inserted into bacteria and the bacteria would be allowed to reproduce asexually (cloning). Show your teacher your recombinant plasmid and have him/her initial your lab handout. DISCUSSION 1. What is genetic engineering? ----------------------------------------------------------------------------------------------------------------------2. What is recombinant DNA (hybrid DNA)? ----------------------------------------------------------------------------------------------------------------------3. What chemical did the “scissors” represent in this experiment? ----------------------------------------------------------------------------------------------------------------------4. DNA ligase is an enzyme used to fuse two DNA fragments together. What material represented DNA ligase in this experiment? ----------------------------------------------------------------------------------------------------------------------5. How do bacteria use restriction enzymes to protect themselves against viral infections? --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------6. Why was the human insulin gene able to be so easily inserted into the cut bacterial plasmid? --------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------7. Imagine your biology teacher gave you another bacterial plasmid with TWO restriction sites that EcoR1 would recognize/digest (instead of just the one seen in this lab). How many DNA fragments would you get? ________ 8. If scientists wanted to produce human insulin, where should the recombinant plasmid you created go next? ----------------------------------------------------------------------------------------------------------------------9. What process must occur in the bacteria in order to make thousands of copies of the recombinant plasmid? ----------------------------------------------------------------------------------------------------------------------- 10. What will the bacteria be able to produce once this recombinant plasmid is inserted? ----------------------------------------------------------------------------------------------------------------------Watch the video https://www.youtube.com/watch?v=dRW9jIOdBcU then Label the process of creating recombinant DNA below: bacterial plasmid, human cell, restriction enzyme cut open plasmid, sticky end, recombinant DNA, plasmid transformation of bacteria, translation, mRNA/transcription, human/donor gene