Success stories at CSCS
advertisement

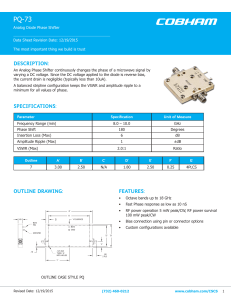
Best Practices Supporting Science at CSCS Michele De Lorenzi (CSCS) Case 1 – Simplify the usage of HPC resources GPU Accelerated In-Situ Visualization § Peter Messmer (NVIDIA Co-Design Lab for Hybrid Multicore Computing at ETH Zurich) HPC Workflow Workstation Setup Analysis, Visualization Supercomputer Viz Cluster Dump, Checkpointing Visualization, Analysis File System Best Practices Supporting Science at CSCS 3 Traditional Workflow: Challenges Lack of interactivity prevents “intuition” Workstation Setup Supercomputer High-end viz neglected due Analysis, Visualization to workflow complexity Viz Cluster I/O becomes main simulation bottleneck Dump, Checkpointing File System Best Practices Supporting Science at CSCS 4 Viz resources need to scale with simulation Visualization, Analysis Eliminating the IO Bottleneck: In-Situ ViZ Workstation Setup Analysis, Visualization Supercomputer Viz Cluster Best Practices Supporting Science at CSCS 5 Eliminating the IO Bottleneck: In-Situ ViZ Workstation Setup Analysis, Visualization Supercomputer Viz Cluster Best Practices Supporting Science at CSCS 6 Different Visualization Scenarios § 1) Legacy workflow § Separate compute & Viz system § Communication via filesystem Comput e GPU HPC System § 2) In-situ partitioned or integrated Filesyste m § Different nodes for viz & compute § Communication via High Performance Network Viz GPU Viz System § 3) In-situ co-processing § Compute and visualization on same node Comput e GPU Network HPC System Comput e+Viz nodes HPC System Best Practices Supporting Science at CSCS 7 Viz GPU Typical Scientific Visualization system § Visualization is more than Rendering Simulation Filtering Rendering Compositing Visualization § Simulation: Data as needed in numerical algorithm § Filtering: Conversion of simulation data into data ready for rendering § § Typical operations: binning, down/up-sampling, iso-surface extraction, interpolation, coordinate transformation, subselection, .. Sometimes embedded in simulation § Rendering: Conversion of shapes to pixels (Fragment processing) § Compositing: Combination of independently generated pixels into final frame Best Practices Supporting Science at CSCS 8 Visualization Dataflow Rendering Filtering CPU Simulation GPU Best Practices Supporting Science at CSCS 9 Fully GPU accelerated Visualization CPU Simulation Filteri ng Renderi ng GPU Best Practices Supporting Science at CSCS 10 FuLLy GPU accelerated Visualization CPU Simulation Filteri ng Renderi ng Speedup GPU Best Practices Supporting Science at CSCS 11 Bonsai With In-Situ Viz On Daint Best Practices Supporting Science at CSCS 12 Acknowledgement § Bonsai development: Evghenii Gaburov (SURFsara), Jeroen Bédorf (CWI) § OpenGL on Daint: Gilles Fourestey (CSCS), Nina Suvanphim (Cray) § OpenGL on Titan: Don Maxwell (ORNL) Thomas Schulthess (CSCS) and Jack Wells (ORNL) for providing access to Daint and Titan for these experiments Best Practices Supporting Science at CSCS 13 Case 2: Software as a service Supporting the MARVEL community 14 MATERIALS’ REVOLUTION: COMPUTATIONAL DESIGN AND DISCOVERY OF NOVEL MATERIALS MARVEL EPFL-ETHZ-UNIBAS-UNIFR-UNIGE-USI-UZH-IBM-CSCSEMPA-PSI Best Practices Supporting Science at CSCS 15 Access Pattern MARVEL Researcher Research Communit y EPFL Repository access AiiDA Server Traditional access (batch jobs) Access through AiiDa CSCS External Login Access (ELA) /store Piz Dora Best Practices Supporting Science at CSCS 16 Scientific Community access Case 3: Supporting scientific communities The Platform for Advanced Scientific Computing (PASC) 17 Swiss Platform for Advanced Scientific Computing § Promote joint effort to address key scientific issues in different domain sciences exploiting the potential of the next generation of HPC architectures § Interdisciplinary collaboration between § domain scientists, § computational scientists, § software developers, § computing centres § hardware developers. § Builds on the principle of co-design involving all these actors Best Practices Supporting Science at CSCS 18 PASC Organization Best Practices Supporting Science at CSCS 19 PASC instruments Organized in four pillars: § Application Support Network (ASN) § Co-design projects § Institutional computing system pillar supporting the procurement of institutional computing systems § The training pillar supporting the Swiss Graduate Program Foundations in Mathematics and Informatics for Computer Simulations in Science and Engineering (FOMICS). Best Practices Supporting Science at CSCS 20 Application Support Network (ASN) § ASN are intended to coordinate and support the effort in different domain science areas to pursue the objectives of the project § The following Domain Science Networks are active today: § Climate - Climate and Atmospheric Modelling § Earth - Solid Earth Dynamics § Life Sciences - Life Sciences Across Scales § Materials - Materials Simulations § Physics - Plasma Physics, Astrophysics and Multiscale Fluid Dynamics Best Practices Supporting Science at CSCS 21 Co-design projects § Interdisciplinary projects involving several (or all) actors § address the following priorities for optimal exploitation of new and emerging supercomputing platforms: § Refactoring of key application codes and re-engineering of their algorithms. § Development of numerical libraries and their integration into application codes. § Incorporations of innovative sub-systems (e.g. I/O, data streams, etc.) into existing simulation codes § Development of programming environments or components, performance tuning and analysis/monitoring tools. Best Practices Supporting Science at CSCS 22 Supported projects Physics and Astrophysics: § Particles and fields; PI: Laurent Villard (EPFL) § DIAPHANE Radiative Transport in astrophysical simulations; PI: Lucio Mayer (University of Zurich) Material Science § ANSWERS: nano-device simulations; PI: Mathieu Luisier (ETH Zurich) § Electronic Structure Calculations: electronic structure calculations; PI: Jürg Hutter (University of Zurich) § ENVIRON A Library for Electronic-structure Simulations; PI: Stefan Goedecker (University of Basel) Best Practices Supporting Science at CSCS 23 Supported projects (continued) Life Science § Angiogenesis in Health and Disease: In-vivo and in-silico; PI: Petros Koumoutsakos (ETH Zurich) § Coupled Cardiac Simulations: HPC Framework for Coupled Cardiac Simulations; PI: Rolf Krause (University of Lugano), Alfio Quarteroni (EPFL) § Genomic Data Processing; PI: Ioannis Xenarios (University of Lausanne) § HPC-ABGEM: agent-based general ecosystems models; PI: Christoph Zollikofer (University of Zurich) Best Practices Supporting Science at CSCS 24 Supported projects (continued) Geo-physics and Climate § GeoPC: hybrid parallel smoothers for multigrid preconditioners; PI: Paul Tackley (ETH Zurich) § GeoScale: A framework for multi-scale seismic modelling and inversion; PI: Andreas Fichtner (ETH Zurich) § Grid Tools: Towards a library for hardware oblivious implementation of stencil based codes; PI: Oliver Fuhrer (Meteosuisse) Computer Science § Heterogeneous Compiler Platform for Advanced Scientific Codes; PI: Torsten Hoefler (ETH Zurich) § Multiscale applications: Optimal deployment of multiscale applications on a HPC infrastructure; PI: Bastien Chopard (UNIGE) Best Practices Supporting Science at CSCS 25 PASC conference § PASC organizes yearly a Platform of Advanced Scientific Computing Conference. § The goal is to bring together research groups within diverse scientific domains to foster interdisciplinary collaborations and to strengthen (HPC) knowledge exchange. § PASC15 – June 1-2, 2015 hosted by ETH Zurich: http://www.pasc15.org/ Best Practices Supporting Science at CSCS 26 Best Practices Supporting Science at CSCS 27 Thank you for your attention.