Chapter 1 Basic Concepts - Perception and Cognition Lab
advertisement

Chapter 1
Basic Concepts
If it can go wrong, it will. —Murphy
The phenomenon of toast falling from a table to land butter-side
down on the floor is popularly held to be empirical proof of the
existence of Murphy’s Law. —Matthews, 1995, p. 172
In this chapter, we present a few basic concepts about statistical modeling. These concepts, which form a cornerstone for the rest of the book,
are most easily discussed in the context of a simple example. We use an
example motivated by Murphy’s Law. As noted by Matthews in the above
quote, the probability toast falls butter-side down is an important quantity
in testing Murphy’s law. In order to estimate this probability, toast can be
flipped; Figure 1.1 provides an example of a toast flipper. The toast is first
buttered and then placed butter-side up at Point A of the device. Then the
experimenter pushes on the lever (Point B) and observes whether the toast
hits the floor butter-side up or butter-side down. We will call the whole act
flipping toast as it is analogous to flipping coins.
1.1
Random Variables
1.1.1
Outcomes and Events
The basic ideas of modeling are explained with reference to two simple experiments. In the first, Experiment 1, a piece of toast is flipped once. In
1
2
CHAPTER 1. BASIC CONCEPTS
B
A
11111111111
00000000000
00000000000
11111111111
00000000000
11111111111
00000000000
11111111111
00000000000
11111111111
Toast
Figure 1.1: Flipping toast. A piece of toast is placed butter-side at Point A.
Then, force is applied to Point B, launching the toast off the table.
the second, Experiment 2, a piece of toast is flipped twice. The first step in
analysis is describing the outcomes and sample space.
Definition 1 (Outcome) An outcome is a possible result of
an experiment.
There are two possible outcomes for Experiment 1: (1) the piece of toast
falls butter-side down or (2) it falls butter-side up. We denote the former
outcome by D and the latter by U (for “down” and “up,” respectively). For
Experiment 2, the outcomes are denoted by ordered pairs as follows:
• (D, D) : The first and second pieces fall butter-side down.
• (D, U) : The first piece falls butter-side down and the second falls
butter-side up.
• (U, D) : The first piece falls butter-side up and the second falls butterside down.
• (U, U) : The first and second pieces fall butter-side up.
3
1.1. RANDOM VARIABLES
Definition 2 (Sample Space) The sample space is the set of
all outcomes.
The sample space for Experiment 1 is {U, D}. The sample space for
Experiment 2 is {(D, D), (U, D), (D, U), (U, U)}.
Although outcomes describe the results of experiments, they are not sufficient for most analyses. To see this insufficiency, consider Murphy’s Law
in Experiment 2. We are interested in whether one or more of the flips is
butter-side up. There is no outcome that uniquely represents this event.
Therefore, it is common to consider events:
Definition 3 (Events) Events are sets of outcomes. They are,
equivalently, subsets of the sample space.
There are four events associated with Experiment 1 and sixteen associated
with Experiment 2. For Experiment 1, the four events are:
{U}, {D}, {U, D}, ∅.
The event {U, D} refers to the case that either the toast falls butter-side
down or it falls butter-side up. Barring the miracle that the toast lands on
its side, it will always land either butter-side up or butter-side down. Even
though this event seems uninformative, it is still a legitimate event and is
included. The null set (∅) is an empty set; it has no elements.
The 16 events for Experiment 2 are:
{(D, D)},
{(U, U)},
{(D, D), (U, U)},
{(U, D), (U, U)},
{(D, D), (D, U), (U, U)},
∅.
{(D, U)},
{(D, D), (D, U)},
{(D, U), (U, D)},
{(D, U), (U, D), (U, U)},
{(D, D), (D, U), (U, D)},
{(U, D)},
{(D, D), (U, D)},
{(D, U), (U, U)},
{(D, D), (U, D), (U, U)},
{(D, D), (D, U), (U, D), (U, U)}
4
CHAPTER 1. BASIC CONCEPTS
1.1.2
Probability
Definition 4 (Probability) Probabilities are numbers assigned to events. The number reflects our degree of belief in
the plausibility of the event. This number ranges from zero (the
event will never occur in the experiment) to one (the event will
always occur). The probability of event A is denoted P r(A).
To explain how probability works, it is helpful at first to assume the
probabilities of at least some of the events are known. For now, let’s assume
the probability that toast falling butter-side down. is .7; e.g., P r({D}) = .7,
It is desirable to place probabilities on the other events as well. The following
concepts are useful in doing so:
Definition 5 (Union) The union of sets A and B, denoted
A ∪ B, is the set of all elements that are either in A or
in B. For example, if A = {1, 2, 3} and B = {3, 4, 5}, then
A ∪ B = {1, 2, 3, 4, 5}.
Definition 6 (Intersection) The intersection of sets A and
B, denoted A ∩ B, is the set of all elements that are both in A
and in B. For example, if A = {1, 2, 3} and B = {3, 4, 5}, then
A ∩ B = {3}.
Probabilities are placed on events by applying the following three rules
of probability:
5
1.1. RANDOM VARIABLES
Event
{D}
{U}
{U, D}
∅
Pr
.7
.3
1
0
Table 1.1: Probabilities of events in Experiment 1.
Definition 7 (The Three Rules of Probability) The three
rules of probability are:
1. P r(A) ≥ 0, where A is any event,
2. P r(sample space) = 1, and
3. if A ∩ B = ∅, then P r(A ∪ B) = P r(A) + P r(B).
The three rules are known as Kolmogorov Axioms of probability (Kolmogorov, 1950). It is relatively easy to apply the rules to the probability of
events. Table 1.1 shows the probabilities of events for Experiment 1.
For Experiment 2, let’s assume the following probabilities on the events
corresponding to single outcomes: P r({(D, D)}) = .49, P r({(D, U)}) = .21,
P r({(U, D)}) = .21, and P r({(U, U)}) = .09. Using the above rules, we can
compute the probabilities on the events. These probabilities are shown in
Table 1.2.
1.1.3
Random Variables
Suppose we are interested in the number of butter-side-down flips. According
to Murphy’s Law, this number should be large. For each experiment, this
number varies according the probability of events. The concept of random
variable captures this dependence:
6
CHAPTER 1. BASIC CONCEPTS
Event
{(D, D)}
{(D, U)}
{(U, D)}
{(U, U)}
{(D, D), (D, U)}
{(D, D), (U, D)}
{(D, D), (U, U)}
{(D, U), (U, D)}
{(D, U), (U, U)}
{(U, D), (U, U)}
{(D, U), (U, D), (U, U)}
{(D, D), (U, D), (U, U)}
{(D, D), (D, U), (U, U)}
{(D, D), (D, U), (U, D)}
{(D, D), (D, U), (U, D), (U, U)}
∅
Probability
.49
.21
.21
.09
.70
.70
.58
.42
.30
.30
.51
.79
.79
.91
1
0
Table 1.2: Probabilities of events in Experiment 2.
7
1.1. RANDOM VARIABLES
Outcome
D
U
Value of X
1
0
Table 1.3: Definition of a random variable X for Experiment 1.
Outcome
(D, D)
(D, U)
(U, D)
(U, U)
Value of X
2
1
1
0
Table 1.4: Definition of a random variable X for Experiment 2.
Definition 8 (Random Variable) A random variable (RV)
is a function that maps events into sets of real numbers. Probabilities on events become probabilities on the corresponding sets
of real numbers.
Let random variable X denote the number of butter-side down flips. X
is defined for Experiments 1 and 2 in Tables 1.3 and 1.4, respectively.
Random variables map experimental results into numbers. This mapping also applies to probability: probabilities on events in the natural word
transfer to numbers. Tables 1.5 and 1.6 show this mapping.
Value of X
1
0
Corresponding Event
{D}
{U}
Probability
.7,
.3,
Table 1.5: Probabilities associated with random variable X for Experiment
1.
8
CHAPTER 1. BASIC CONCEPTS
Value of X
2
1
0
Corresponding Event
{(D, D)}
{(D, U), (U, D)}
{(U, U)}
Probability
.49
.42
.09
Table 1.6: Probabilities associated with random variable X for Experiment
2.
Random variables are typically typeset in upper-case; e.g., X. Random
variables take on values and these values are typeset in lower-case; e.g., x.
The expression P r(X = x) refers to the probability that an event corresponding to x will occur. The mappings in Tables 1.5 and 1.6 are typically
expressed as the function f (x) = P r(X = x), which is called the probability
mass function.
Definition 9 (Probability Mass Function) Probability
mass function, f(x), provides the probability for a particular
value of a random variable.
For Experiment 1, the probability mass function of X is
For Experiment 2, it is
.3 x = 0
f (x) = .7 x = 1
0 Otherwise
.09
.42
f (x) =
.49
0
(1.1)
x=0
x=1
x=2
Otherwise
(1.2)
Probability mass functions always sum to 1, i.e., x f (x) = 1 as a consequence of the Kolomogorov Axioms in Definition 7. As can be seen, the
above probability mass functions sum to 1.
P
1.1. RANDOM VARIABLES
9
As discussed in the preface, we use the computer package R to aid with
statistical analysis. R can be used to plot these probability mass functions.
The following code is for plotting the probability mass function for Experiment 2, shown in Eq. (1.2). The first step in plotting is to assign values to x
and f . These assignments are implemented with the statements x=c(0,1,2)
and f=c(.09,.42,.49). The symbol c() stands for “concatenate” and is
used to join several numbers into a vector. The values of a variable may seen
by simply typing the variable name at the R prompt; e.g.,
> x=c(0,1,2)
> f=c(.09,.42,.49)
> x
[1] 0 1 2
> f
[1] 0.09 0.42 0.49
The function plot() can be used to plot one variable as a function of
another. Try plot(x,f,type=’h’). The resulting graph should look like
Figure 1.2. There are several types of plots in R including scatter plots, bar
plots, and line plots. The type of plot is specified with the type option. The
option type=’h’ specifies thin vertical lines, which is appropriate for plotting
probability mass functions. Help on any R command is available by typing
help with the command name in parentheses, e.g., help(plot). The points
on top of the lines were added with the command points(x,f).
The random variable X, which denotes the number of butter-side-down
flips, is known as a discrete random variable. The reason is that probability
is assigned to discrete points; for X, it is the discrete points of 0, 1, and
2. There is another type of random variable, a continuous random variable,
in which probability is assigned to intervals rather than points. The differences between discrete and continuous random variables will be discussed in
Chapter 4.
1.1.4
Parameters
Up to now, we have assumed probabilities on some events (those corresponding to outcomes) and used the laws of probability to assign probabilities to
the other events. In experiments, though, we do not assume probabilities; instead, we estimate them from data. We introduce the concept of a parameter
10
0.6
0.4
0.2
0.0
Probability Mass Function
CHAPTER 1. BASIC CONCEPTS
−1
0
1
2
3
Value of X
Figure 1.2: Probability mass functions for random variable X, the number
of butter-side down flips, in Experiment 2.
to avoid assuming probabilities.
Definition 10 (Parameter) Parameters are mathematical
variables on which a random variable may depend. Probabilities
on events are functions of parameters.
For example, let the outcomes of Experiment 1 depend on parameter p
as follows: P r(D) = p. Because the probability of all outcomes must sum
1.0, the probability of event U must be 1 − p. The resulting probability mass
function for X, the number of butter-side down flips, is
1−p x =0
x=1
f (x; p) = p
0
Otherwise
(1.3)
The use of the semicolon in f (x; p) indicates that the function is of one
variable, x, for a given value of the parameter p.
Let’s consider the probabilities in Experiment 2 to be parameters defined
as p1 = P r(D, D), p2 = P r(D, U), p3 = P r(U, D). By the laws of probability,
11
1.2. BINOMIAL DISTRIBUTION
P r(U, U) = 1 − p1 − p2 − p3 . The resulting probability mass function on X is
1 − p1 − p2 − p3
p2 + p3
f (x; p1 , p2 , p3 ) =
p
1
0
x=0
x=1
x=2
Otherwise
(1.4)
This function is still of one variable, x, for given values of p1 , p2 , and p3 .
Although the account of probability and random variables presented here
is incomplete, it is sufficient for the development that follows in the book.
A more complete basic treatment can be found in mathematical statistics
textbooks such as Hogg & Craig (1978) and Rice (1995).
Problem 1.1.1 (Your Turn)
You and a friend are playing a game with a four-sided die. Each of the
sides, labeled A, B, C, and D, has equal probability of landing up on
any given throw.
1. There are two throws left in the game; list all of the possible
outcomes for the last two throws. Hint: these outcomes may be
expressed as ordered pairs.
2. In order to win, you need to throw an A or B on each of the final
two throws. List all the outcomes that are elements in the event
that you win.
3. In this game there are only two possible outcomes: winning and
losing. Given the information in Problem 2, what is the probability mass function for the random variable that maps the event
that you win to 0 and the event that you lose to 1? Plot this
function in R.
1.2
Binomial Distribution
Experiment 1, in which a piece of toast is flipped once, plays an important
role in developing more sophisticated models. Experiment 1 is an example
12
CHAPTER 1. BASIC CONCEPTS
of a Bernoulli trial, defined below.
Definition 11 (Bernoulli Trial) Bernoulli trials are experiments with two mutually exclusive (or dichotomous) outcomes.
Examples include a flip of toast; the sex of a baby (assuming
only male or female outcomes); or, for our purposes, whether
a participant produces a correct response (or not) on a trial in
a psychology experiment. By convention, one of the outcomes
is called a success, the other a failure. A random variable is
distributed as a Bernoulli if it has two possible values: 0 (for
failure) and 1 (for success).
As a matter of notation, we will consider the butter-side-down result in
toast flipping as a success and the butter-side-up result as a failure. Random
variables from Bernoulli trials have a single parameter p and probability mass
function given by Eq. (1.3). In general, the value of p is not known a priori
and must be estimated from the data.
Experiment 2 is a sequence of two Bernoulli trials. We let X1 and X2
denote the outcomes (either success of failure) of these two trials. Let p1 and
p2 be the probability of success parameter for X1 and X2 , respectively. If
p1 = p2 , then random variables X1 and X2 are called identical. Note that
identical does not mean that the results of the flips are the same, e.g., both
flips are successes or both are failures. Instead, it means that the probabilities
of a success are the same. The concept of identical random variables can be
extended: whenever two random variables have the same probability mass
function, they are identical. If the result of a Bernoulli trial is not affected by
the result of the others, then the RVs are called independent. If two random
variables are both independent and identically distributed, then they are
called iid.
In order to understand the concepts of independent and identically distributed, it may help to consider a concrete example. Suppose a basketball
player is shooting free throws. If one throw influences the next, for instance,
if a player gets discouraged because he or she misses a throw, this is a violation of independence, but not necessarily of identical distribution. Although
one throw may effect the next, if on average they are the same, identical
1.2. BINOMIAL DISTRIBUTION
13
distribution is not violated. If a player gets tired and does worse over time,
regardless of the outcome of his or her throws, then identical distribution is
violated. It is possible to violate one and not the other.
Definition 12 (Bernoulli Process) A sequence of independent and identically distributed Bernoulli trials is called a
Bernoulli Process.
In a Bernoulli process, each Xi is a function of the same parameter p.
Furthermore, because each trial is independent, the order of outcomes is
unimportant. Therefore, it makes sense to define a new random variable, Y ,
P
which is the total number of successes, i.e., Y = N
i=1 Xi .
Definition 13 (Binomial Random Variable) The random
variable which denotes the number of successes in a Bernoulli
process is called a binomial. The binomial random variable has
a single parameter, p (probability of success on a single trial). It
is also a function of a constant, N, the number of trials. It can
take any integer value from 0 to N.
Definition 14 (Random Variable Notation) It is common
to use the character “∼” to indicate the distribution of a random variable. A binomial random variable is indicated as
Y ∼ Binomial(p, N). Here, Y is the number of successes in
N trials where the probability of success on each trial is p.
The variable p is considered a parameter but the variable N is not. The
value of N is known exactly and supplied by the experimenter. The true
value of p is generally unknown and must be estimated. The probability
14
CHAPTER 1. BASIC CONCEPTS
mass function of a binomial random variable describes the probability of
observing y successes in N trials:
P r(Y = y) = f (y; p) =
The term
N
y
!
N
y
!
py (1 − p)N −y y = 0, .., N
0
(1.5)
Otherwise
refers to the “choose function,” given by
N
y
!
=
N!
.
y!(N − y)!
Let’s look at the probability mass function in R. Try the following for
N = 20 and p = .7:
y=0:20
#assigns x to 0,1,2,..,20
f=dbinom(y,20,.7) #probability mass function
plot(y,f,type=’h’)
points(y,f)
In the above code, dbinom() is an R function that returns the probability
mass function of a binomial. Variable y is a vector taking on 21 values
(0,1,..,20). Because the first argument of dbinom() is a vector the output is
also a vector with one element for each element of y. Type f to see the 21
values. The first value of f corresponds to the first value of y; the second
value of f to the second value of y; and so on. The resulting plot is shown
in Figure 1.3.
The goal of Experiment 1 and 2 is to learn about p. These experiments,
however, are too small to learn much. Instead, we need a larger experiment
with more flips; for generality, consider the case in which we had N flips. One
common-sense approach is to take the number of successes in a Bernoulli
process, Y and divide by N, the number of trials. A function of random
variables that estimates a parameter is called an estimator. The commonsense estimator of p is p̂ = Y /N. It is conventional to place the caret over
an estimator as in p̂. This distinguishes it from the true, but unknown
parameter p. Note that because p̂ is a function of a random variable, it is
also a random variable. Because estimators are random variables themselves,
studying them requires more background about random variables.
15
0.10
0.00
Probability Mass Function
1.3. EXPECTED VALUES OF RANDOM VARIABLES
0
5
10
15
20
Value of X
Figure 1.3: Probability Mass function for a binomial random variable with
N = 20 and p = .7
1.3
Expected Values of Random Variables
1.3.1
Expected Value
The expected value is the center or theoretical average of a random variable.
For example, the center of the distribution in Figure 1.3 is at 14. Expected
value is defined as:
Definition 15 (Expected Value) The expected value of a
discrete random variable X is given as:
E(X) =
X
xf (x; p).
(1.6)
x
The expected value of a random variable is closely related to the concept of
an average, or mean. Typically, to compute a mean of a set of values, one
adds all the values together and divides by the total number of values. In
the case of an expected value, however, each possible value is weighted by
the probability that it will occur before summing. The expected value of a
random variable is also called its first moment, population mean, or simply
its mean. It is important to differentiate between the expected value of a
random variable and the sample mean, which will be discussed subsequently.
16
CHAPTER 1. BASIC CONCEPTS
Consider the following example. Let X be a random variable denoting the
outcome of a Bernoulli trial with parameter p = .7. Then, the expected value
P
is given by E(X) = x xf (x; p) = (0 × .3) + (1 × .7) = .7. More generally,
the expected value of a Bernoulli trial with parameter p is E(X) = p.
1.3.2
Variance
Whereas the expected value measures the center of a random variable, variance measures its spread.
Definition 16 (Variance) The variance of a discrete random
variable is given as:
V(X) =
X
x
[x − E(X)]2 f (x; p)
(1.7)
Just as the expected value is a weighted sum, so is the variance of a random
variable. The variance is the sum of all possible squared deviations from
the expected value, weighted by their probability of occuring. An equivalent
equation for variance is given as V(X) = E[(X − E(X))2 ]; that is, variance
is the expected squared deviation of a random variable from its mean. The
variance of a random variable is different from the variance of a sample, which
will be discussed subsequently.
Another common measure of the spread of a random variable is the standard deviation. The standard
q deviation of a random variable is square-root
of variance, e.g. SD(X) = V(X). Standard deviation is often used as a
measure of spread rather than variance because it is in the same units as the
random variable. Variance, in contrast, is in squared units, which are more
difficult to interpret. The standard deviation of an estimator has its own
name: standard error.
Definition 17 (Standard Error) The standard deviation of
a parameter estimator is called the standard error of the estimator.
1.3. EXPECTED VALUES OF RANDOM VARIABLES
17
Problem 1.3.1 (Your Turn)
1. It is common in psychology to ask people their opinion of statements, e.g., “I am content with my life.” Responses are often
collected on a Likert scale; e.g., 1=strongly disagree, 2=disagree,
3=neutral, 4=agree, 5=strongly agree. The answer may be considered a random variable. Suppose the probability mass function
for the above question is given as f (x) = (.05, .15, .25, .35, .2) for
x = 1, . . . , 5, respectively. Plot this probability mass function.
Compute the expected value. Does the expected value appear to
be at center of the distribution? Compute the variance.
2. Let Y be a binomial RV with N = 3 and parameter p. Show
E(Y ) = 3p.
1.3.3
Expected Value of Functions of Random Variables
It is often necessary to consider functions of random variables. For example,
the common sense estimator of p, p̂ = Y /N, is a function of random variable
Y . The following two rules are convenient in computing the expected value
of functions of random variables.
18
CHAPTER 1. BASIC CONCEPTS
Definition 18 (Two rules of expected values) Let X, Y ,
and Z all denote random variables, and let Z = g(X). The
following rules apply to the expected values:
1. The expected value of the sum is the sum of the expected
values:
E(X + Y ) = E(X) + E(Y )
, and
2. the expected value of a function of a random variable is
E(Z) =
X
g(x)f (x; p),
x
where f is the probability mass function of X.
The first rule can be used to find the expected value of a binomial random
P
variable. By definition, binomial RV Y is defined as Y = N
1=1 Xi , where the
Xi are iid Bernoulli trials. Hence, by Rule 1,
E(Y ) = E(
X
Xi ) =
X
E(Xi ) =
p = Np
i
i
i
X
The second rule can be used to find the expected value of p̂. The random
variable p̂ = g(Y ) is g(Y ) = Y /N. The expected value of p̂ is given by:
E(p̂) = E(g(Y ))
X
=
(x/N)f (x; p)
x
= (1/N)
X
xf (x; p)
x
= (1/N)E(Y )
= (1/N)(Np)
= p.
While p̂ may vary from experiment to experiment, its average will be p.
1.4. SEQUENCES OF RANDOM VARIABLES
1.4
19
Sequences of Random Variables
1.4.1
Realizations
Consider Experiment 1, the single flip of toast and the random variable,
X, the number of butter-side-down flips. Before the experiment, there are
two possible values that X could take with nonzero probability, 0 and 1.
Afterward, there is one result. The result is called the realization of X.
Definition 19 (Realization) The realization of a RV is the
value it attains in an experiment.
Consider Experiment 2 in which two pieces of toast are flipped. Before
the experiment is conducted, the possible values of X are 0, 1, and 2. Afterward, the realization of X can be only one of these values. The same is
true of estimators. Consider the random variables Y ∼ Binomial(p, N) and
common-sense estimate p̂ = Y /N. After an experiment, these will have realizations denoted y and y/N, respectively. The realization of an estimator is
called an estimate.
It is easy to generate realizations in R. For binomial random variables,
the appropriate function is rbinom(): type rbinom(1, 20, .7). The first
argument is the number of realizations, which is 1. The second is N, the
number of trials. The third is p, the probability of success on a trial. Try
the command a few times. The output of each command is one realization
of an experiment with 20 trials.
1.4.2
Law of Large Numbers
Consider rbinom(5, 20, .7). This should yield five replicates; the five
realizations from five separate experiments. There are two interpretations
of the five realizations. The first, sometimes prominent in undergraduate
introductory texts, is that these five numbers are samples from a common
distribution. The second, which is more common in advanced treatments of
probability, is that the realizations are from different, though independent
and identically distributed, random variables. Replicate experiments can be
20
CHAPTER 1. BASIC CONCEPTS
represented as a sequence of random variables, and in this case, we write:
iid
Yi ∼ Binomial(p = .7, N = 20) i = 1, . . . , 5.
Each Yi is a different random variable, but all Yi are independent and distributed as identical binomials. Each i could repeasent a different trial, a
different person, or a different experimental condition.
Of course, we are not limited to 5 replicates; for example y=rbinom(200,
20, .7) produces 200 replicates and stores them in vector y. To see a histogram, type hist(y, breaks=seq(-.5, 20.5, 1), freq=T). We prefer a
different type of histogram for looking at realizations of discrete random
variables—one in which the y-axis is not the raw counts but the proportion, or relative frequency, of counts. These histograms are called relative
frequency histograms.
iid
Definition 20 (Relative Frequency Histogram) Let Yi ∼
Y be a sequence of M independent and identically distributed
discrete random variables and let y1 , .., yM be a sequence of corresponding realizations. Let hM (j) be the proportion of realizations with value j. The relative frequency histogram is a plot of
hM (j) against j.
Relative frequency histograms may be drawn in R with the following
code:
freqs=table(y) #frequencies of realization values
props=freqs/200 # proportions of realization values
plot(props, xlab=’Value’, ylab=’Relative Frequency’)
The code draws the histogram as a series of lines. The relative histogram
plot looks like a probability mass function. Figure 1.4A shows that this is
no coincidence. The lines are the relative frequency histogram; the points
are the probability mass function for a binomial with N = 20 and p = .7
(The points were produced with the points() function. The specific form is
points(0:21,dbinom(0:21,20,.7),pch=21)).
21
0
5
10
15
20
0.10
0.20
B
0.00
0.10
0.20
Relative Frequency
A
0.00
Relative Frequency
1.4. SEQUENCES OF RANDOM VARIABLES
0
Outcome
5
10
15
Outcome
Figure 1.4: A. Relative Frequency histogram and probability mass function
roughly match with 200 realizations. B. The match is near perfect with
100,000 realizations.
The match between the relative histogram and the pmf is not exact. The
problem is that there are only 200 realizations. Figure 1.4B shows the match
between probability mass function and the relative frequency histogram when
there are 10,000 realizations. Here, the match is nearly perfect. This match
indicates that as the number of realizations grows, the relative frequency
histogram converges to the probability mass function. The convergence is a
consequence of the Law of Large Numbers. The Law of Large says, informally,
that the proportion of realizations attaining a particular value will converge
to the true probability of that realization. More formally,
lim hM (j) = f (j; p),
M →∞
where f is the probability mass function of Y .
The fact that the relative frequency histogram of samples converges to
the probability mass function is immensely helpful in understanding random
variables. Often, it is difficult to write down the probability mass function
of a random variable but easy to generate samples of realizations. By generating a sequence of realizations from independent and identically distributed
random variables, it is possible to see how the probability mass functions behaves. This approach is called the simulation approach and we use it liberally
as a teaching tool.
We can use the simulation approach to approximate the probability mass
function for the common-sense estimator p̂ = Y /N with the following R code:
20
22
0.20
0.10
0.00
Probability Mass Function
CHAPTER 1. BASIC CONCEPTS
0.25
0.4
0.55
0.7
0.85
1
Value of estimator
Figure 1.5: Simulated probability mass function for a common-sense estimator of p for a binomial with N = 20 and p = .7.
y=rbinom(10000,20,.7)
p.hat=y/20 #10,000 iid replicates of p-hat
freq=table(p.hat)
plot(freq/10000,type=’h’)
The resulting plot is shown in Figure 1.5. The plot shows the approximate
probability mass function for the p̂ estimator. The distribution of an estimator is so often of interest that it has a special name: a sampling distribution.
Definition 21 (Sampling Distribution) A sampling distribution is the probability mass function of an estimator.
1.5
Estimators
Estimators are random variables that are used to estimate parameters from
data. We have seen one estimator, the common-sense estimator of p in a
23
1.5. ESTIMATORS
binomial: p̂ = Y /N. Two others are the sample mean and sample variance defined below, which are used as estimators for the expected value and
variance of an RV, respectively.
Definition 22 (Sample Mean and Sample Variance)
Let Y1 , Y2, ..., YM be a collection of M random variables. The
sample mean and sample variance are defined as
Ȳ
s2Y
Yi
and
M
P
2
i (Yi − Ȳ )
=
,
M −1
=
P
i
(1.8)
(1.9)
respectively.
How good are these estimators? To answer this question, we first discuss
properties of estimators.
1.5.1
Properties of estimators
To evaluate the usefulness of estimators, statisticians usually discuss three
basic properties: bias, efficiency, and consistency. Bias and efficiency are
illustrated in Table 1.7. The data are the results of weighing a hypothetical
person of 170 lbs on two hypothetical scales four separate times. Bias refers
to the mean of repeated estimates. Scale A is unbiased because the mean of
the estimates equals the true value of 170 lbs. Scale B is biased. The mean
is 172 lbs which is 2 lbs. greater than true value of 170 lbs. Examining the
values for scale B, however, reveals that scale B has a smaller degree of error
than scale A. Scale B is called more efficient than Scale A. High efficiency
means that expected error is low. Bias and efficiency have the same meaning
for estimators as they do for scales. Bias refers to the difference between
the average value of an estimator and a true value. Efficiency refers to the
amount of spread in an estimator around the true value.
The bias and efficiency of any estimator depends on the sample size. For
P
example, the common-sense estimator p̂ is p̂ = ( N
i=1 Yi /N) provides a better
24
CHAPTER 1. BASIC CONCEPTS
Table 1.7: Two Hypothetical Scales
Scale A
180
160
175
165
Mean 170
Bias
0
RMSE 7.91
Scale B
174
170
173
171
172
2.0
2.55
estimate with increasing N. Let θ̂N denote an estimator which estimates
parameter θ, for a sample size of N.
Definition 23 (Bias) The bias of an estimator is given by BN :
BN = E(θˆN ) − θ
Bias refers to the expected value of an estimator. We have already proven
that estimator p̂ is unbiased (Section 1.3.3). Both sample mean and sample
variance are also unbiased. Other common estimators, however, are biased.
One example is the sample correlation. Fortunately, this bias reduces toward
zero with increasing N. Unbiasedness is certainly desirable, but not critical.
Many of the estimators discussed in this book will have some degree of bias.
Problem 1.5.1 (Your Turn)
Let Yi ; i = 1..N be a sequence of N independent and identically distributed random variables. Show that the sample mean is unbiased for
all N (hint: use the rules of expected value in Definition 18).
25
1.5. ESTIMATORS
Definition 24 (Efficiency) Efficiency refers to the expected
degree of error in estimation. We use root-mean-squared error
(RMSE) as a measure of efficiency:
RMSE =
q
E[(θ̂N − θ)2 ]
(1.10)
More efficient efficient estimators have less error, on average, than less efficient estimators. Sample mean and sample variance are the most efficient
unbiased estimators of expected value and variance, respectively. One of the
main issues is estimation is the trade-off between bias and efficiency. Often, the most efficient estimator of a parameter is biased, and this facet is
explored in the following section.
The final property of estimators is consistency.
Definition 25 (Consistency) An estimator is consistent if
lim RMSE(θ̂N ) = 0
N →∞
Consistency means that as the sample sizes gets larger and larger, the estimator converges to the true value of the parameter. If an estimator is consistent, then one can estimate the parameter to arbitrary accuracy. To get
more accurate estimates, one simply increases the sample size. Conversely, if
an estimator is inconsistent, then there is a limit to how accurately the parameter can be estimated, even with infinitely large samples. Most common
estimators in psychology, including the sample mean, sample variance, and
sample correlation, are consistent.
Because sample means and sample variances converge to expected value
and variances, respectively, they can be used to estimate these properties.
For example, let’s approximate the expected value, variance, and standard
error of p̂ with the sample statistics in R. We first generate a sequence of
iid
realizations y1 , .., yM for binomial random variables Yi ∼ Y i = 1, .., M. For
each realization, we compute an estimate pi = yi /N. The sample mean,
sample variance, and sample standard deviation approximate the expected
value, variance, and standard error. To see this, run the following R code:
26
CHAPTER 1. BASIC CONCEPTS
y=rbinom(10000,20,.7)
p.hat=y/20
mean(p.hat)
#sample mean
var(p.hat)
#sample variance (N-1 in denominator)
sd(p.hat)
#sample std. deviation (N-1 in denominator)
Problem 1.5.2 (Your Turn)
How does the standard error of p̂ depend on the number of trials N?
Let’s use the simulation method to further study the common-sense estimator of the expected value of the binomial, the sample mean. Suppose in
an experiment, we had ten binomial RVs, each the result of 20 toast flips.
Here is a formal definition of the problem:
Yi
Ȳ
iid.
∼ Binomial(p, 20), i = 1...10,
P
i Yi
=
.
10
The following code generates 10 replicates from a binomial, each of 20
flips. Here we have defined a custom function called bsms() (bsms stands
for “binomial sample mean sampler”). Try it a few times. This is analogous
to having 10 people each flip 20 coins, then returning the mean number of
heads across people.
#define function
bsms=function(m,n,p)
{
z=rbinom(m,n,p)
mean(z)
}
#call function
bsms(10,20,.7)
27
0.06
0.04
0.02
0.00
Relative Frequency
1.5. ESTIMATORS
12
13
14
15
16
Sample Mean of Binomials
Figure 1.6: Relative Frequency plot of 10,000 calls to the function bsms().
for this plot, bsms() computed the mean of 10 realizations from binomials
with N = 20 and p = .7
The above code returns a single number as output: the sample mean of 10
binomials. Since the sample mean is an estimator, it has a sampling distribution. The bsms() function returns one realization of the sample mean. If we
are interested in the sampling distribution of the sample mean, we need to
sample it many times and plot the results in a relative frequency histogram.
This can be done by repeatedly calling bsms(). Here is the code for 10,000
replicates of bsms():
M=10000
bsms.realization=1:M #define the vector ppes.realization
for(m in 1:M) bsms.realization[m]=bsms(10,20,.7)
bsms.props=table(bsms.realization)/M
plot(ppes.props, xlab="Estimate of Expected Value (Sample Mean)",
ylab="Relative Frequency", type=’h’)
The resulting histogram is shown in Figure 1.6. The new programming
element is the for loop. Within it, function bsms() is called M times, each
result being stored to a different element of bsms.realization. However,
we cannot reference elements in a vector without first reserving space. The
line bsms.realization=1:M defines the vector, and in the process, reserves
space for it.
28
CHAPTER 1. BASIC CONCEPTS
Problem 1.5.3 (Your Turn)
1. What is the expected value of the sample mean of ten binomial
random variables with N = 20 and p = .5? What is the approximate value from the above simulation? Are the values close?
What is the simulation approximation for the standard error?
2. Manipulate the number of trials, N, in each binomial RV through
a few levels: 5 trials, 20 trials, 80 trials. What is the effect on the
sampling distribution of Ȳ ?
3. Manipulate the number of random variables in the sample mean
though a few levels: e.g., a mean of 4, 10, or 50 binomials. What
is the effect on the sampling distribution Ȳ ?
4. What is the effect of raising or lower the number of replicates M?
1.6
Three Binomial Probability Estimators
Consider the three following estimators for p: p̂0 , p̂1 , and p̂2 .
Y
,
N
Y + .5
=
,
N +1
Y +1
.
=
N +2
p̂0 =
(1.11)
p̂1
(1.12)
p̂2
(1.13)
Let’s use R to examine the properties of these three estimators for 10
flips with p = .7. The following code uses the simulation method. It draws
10,000 replicates from a binomial distribution and computes the value for
each estimator for each replicate.
1.6. THREE BINOMIAL PROBABILITY ESTIMATORS
29
p=.7
N=10
z=rbinom(10000,N,p)
est.p0=z/N
est.p1=(z+.5)/(N+1)
est.p2=(z+1)/(N+2)
bias.p0=mean(est.p0)-p
rmse.p0=sqrt(mean((est.p0-p)^2))
bias.p1=mean(est.p1)-p
rmse.p1=sqrt(mean((est.p1-p)^2))
bias.p2=mean(est.p2)-p
rmse.p2=sqrt(mean((est.p2-p)^2))
Figure 1.7 shows the sampling distributions for the three estimators.
These sampling distributions tend to be roughly centered around the true
value of the parameter, p = .7. Estimator p̂2 is the least spread out, followed
by pˆ1 and pˆ0 . Bias and efficiency of the estimators are indicated. Although
estimator pˆ0 is unbiased, it is also the least efficient! Figure 1.8 shows bias
and efficiency for all three estimators for the full range of p. The conventional
estimator p̂0 is unbiased for all true values of p, but the other two estimators
are biased for extreme probabilities. None of the estimators are always more
efficient than the others. For intermediate probabilities, estimator p̂2 is most
efficient; for extreme probabilities, estimator p̂0 is most efficient. Typically,
researchers have some idea of what types of probabilities of success to expect
in their experiments. This knowledge can therefore be used to help pick the
best estimator for a particular situation. We recommend p̂1 as a versatile alternative to p̂0 for many applications even though it is not the common-sense
estimator.
30
CHAPTER 1. BASIC CONCEPTS
0.30
Probablity
0.25
0.20
p0
Bias=0
RMSE=.145
0.15
0.10
0.05
0.00
Probablity
0.25
0.20
0.15
p1
Bias=−.018
RMSE=.133
0.10
0.05
0.00
Probablity
0.25
0.20
0.15
p2
Bias=−.033
RMSE=.125
0.10
0.05
0.00
0.0
0.2
0.4
0.6
0.8
1.0
Estimated Probability of Success
Figure 1.7: Sampling distribution of p̂0 , p̂1 , and p̂2 . Bias and root-meansquared-error (RMSE) are included. This figure depicts the case that there
are N = 10 trials with a p = .7.
31
1.6. THREE BINOMIAL PROBABILITY ESTIMATORS
Bias
0.05
p1
0.00
p2
p0
−0.05
0.0
0.2
0.025
0.4
p0
0.8
1.0
0.8
1.0
p1
0.020
RMSE
0.6
0.015
p2
0.010
0.005
0.000
0.0
0.2
0.4
0.6
Probability of Success
Figure 1.8: Bias and root-mean-squared-error (RMSE) for the three estimators as a function of true probability. Solid, dashed, and dashed-dotted lines
denote the characteristics of p̂0 , p̂1 , and p̂2 , respectively.
32
CHAPTER 1. BASIC CONCEPTS
Problem 1.6.1 (Your Turn)
The estimators of the binomial probability parameter discussed above
all have the form (Y + a)/(N + 2a). We have advocated using the
estimator p̂1 = (Y + .5)/(N + 1), but there are many other possible
estimators besides a = .5.. Examine what happens to the efficiency of
an estimator as a gets large. Why would we choose a = .5 over, say,
a = 20?
Chapter 2
The Likelihood Approach
Throughout this book, we use a general set of techniques for analysis that are
based on the likelihood function. In this chapter we present these techniques
within the context of three examples involving the binomial distribution.
At the end of the chapter, we provide an overview of the theoretical justification for the likelihood approach. In the following chapters, we use this
approach to analyze pertinent models in cognitive and perceptual psychology.
Throughout this book, analysis is based on the following four steps:
1. Define a hierarchy of models.
2. Express the likelihood functions for the models.
3. Find the parameters that maximize the likelihood functions.
4. Compare the values of likelihood to decide which model is best.
2.1
Estimating a Probability
We illustrate the likelihood approach within the context of an example. As
previously discussed, the binomial describes the number of successes in a
set of Bernoulli trials. Let’s consider the toast-flipping experiment discussed
previously. The goal is to estimate the true probability, p, that a piece of
toast lands butter-side down. As before, let N denote the number of pieces
flipped and let random variable Y denote the number of butter-side down
flips and y denote the datum, a realization of Y .
33
34
CHAPTER 2. THE LIKELIHOOD APPROACH
2.1.1
A hierarchy of models
In this example, we define a single model,
Y ∼ Binomial(N, p).
Clearly, there is no hierarchy with a single model; subsequent examples will
include multiple models arranged in a hierarchy.
2.1.2
Express the likelihood function
The next step in a likelihood analysis is to express the likelihood function.
We first define the function for this example and then give a more general definition. Likelihood functions are closely related probability mass functions.
For the binomial, the probability mass function, P r(Y = y), is
f (y; p) =
N
y
!
py (1 − p)N −y y = 0, .., N
0
(2.1)
Otherwise
We can rewrite the probability mass function as a function of the parameter given realization y:
L(p; y) =
N
y
!
py (1 − p)N −y
(2.2)
The right-hand side of the equation is the same as the probability mass
function; the difference is on the left-hand side. Here, we have switched the
arguments to reflect the fact that the likelihood function, denoted by L, is a
function of the parameter p.
Definition 26 (Likelihood Function) For a discrete random
variable, the likelihood function is the probability mass function
expressed as a function of the parameters.
Let’s examine the likelihood function for the binomial in R. First, we
define a function called likelihood().
2.1. ESTIMATING A PROBABILITY
35
likelihood=function(p,y,N)
return(dbinom(y,N,p))
Now, let’s examine the likelihood, a function of p, for the case in which 5
successes were observed in 10 flips.
p=seq(0,1,.01)
like=likelihood(p,5,10)
plot(p,like,type=’l’)
The seq() function in the first line assigns p to the vector (0, .01, .02, ..., 1).
The second line computes the value of the likelihood for each of these values of p. Figure 2.1 shows the resulting plot (Panel A). The likelihood is
unimodal and is centered over .5. Also shown in the figure are likelihoods
for 50 successes out of 100 flips, 7 successes out of 10 flips, and 70 successes
out of 100 flips. Two patterns are evident. First, the maximum value of the
likelihood is at y/N. Second, the width of the likelihood is a function of N.
The larger N, the smaller the range of parameter values that are likely for
the observation y.
It is a reasonable question to ask what a particular value of likelihood
means. For example, in Panel A, the maximum of the likelihood (about
.25) is far smaller than that in Panel B. In most applications, the actual
value of likelihood is not important. For the binomial, likelihood depends
on the observation, the parameter p and the number of Bernoulli trials, N.
For estimation, it is the shape of the function and the location of the peak
that are important, as discussed subsequently. For model comparison, the
difference in likelihood values among models is important.
0.08
CHAPTER 2. THE LIKELIHOOD APPROACH
0.00
0.00
0.0
0.2
0.4
0.6
0.8
1.0
0.0
0.08
Likelihood
B
0.04
A
0.10
0.20
36
0.4
0.6
0.8
1.0
0.2
0.4
0.6
0.8
1.0
D
0.00
0.00
0.10
0.04
0.20
C
0.2
0.0
0.2
0.4
0.6
p
0.8
1.0
0.0
p
Figure 2.1: A plot of likelihood for (A) 5 successes from 10 flips, (B) 50
successes from 100 flips, (C) 7 successes from 10 flips, and (D) 70 successes
from 100 flips
37
2.1. ESTIMATING A PROBABILITY
Problem 2.1.1 (Your turn)
The Poisson random variable describes the number of rare events in
an interval. For example, it may be used to model the number of car
accidents during rush hour; the number of earthquakes in a year in a
certain region; or the number of deer in a area of land. The probability
mass function of a Poisson random variable is given by
f (y; λ) =
λy e−λ
y!
y = 0, 1, 2, ...; λ > 0
0
Otherwise
(2.3)
where λ is the parameter of interest. Using R, draw two probability
mass functions, one for λ = .5, the other for λ = 10. Draw a graph of
the likelihood for y = 4.
2.1.3
Find the parameters that maximize the likelihood
The basic premise of maximum likelihood estimation is given in the following
definition:
Definition 27 (Maximum Likelihood Estimate) A maximum likelihood (ML) estimate is the parameter value that maximizes the likelihood function for a given set of data.
There are two basic methods of deriving maximum likelihood estimators
(MLEs): (1) use calculus to derive solutions, (2) use a numerical method to
find the value of the parameter that maximizes the likelihood. This second
method is easily implemented in R.
The first step in maximizing a likelihood is to use the natural logarithm
of the likelihood (the log likelihood, denoted by l) as the main function of interest. This transformation is helpful whether one uses calculus or numerical
38
CHAPTER 2. THE LIKELIHOOD APPROACH
methods. Figure 2.2 shows log likelihood functions for the binomial distribution. To find the log likelihood, one takes the logarithm of the likelihood
function, e.g., for the binomial:
l(p; y) = log[L(p; y)]
#
"
!
N
y
N −y
= log
p (1 − p)
y
= log
N
y
!
+ log(py ) + log((1 − p)N −y )
= log
N
y
!
+ y log p + (N − y) log(1 − p).
(2.4)
Definition 28 (log likelihood Function) The log likelihood
function is the natural logarithm of the likelihood function.
There are two types of methods to find maximum likelihood estimates.
The first is based on calculus and is discussed below. The calculus methods
are limited in their application and many problems must be solved with
numerical methods. Throughout the book, we will the second type of method,
numerical methods, and their implementation in R.
2.1.4
Calculus Methods to find MLEs
In this section, we briefly show how calculus may be used to solve the MLE
for the binomial example. Calculus is not necessary to understand the vast
majority of the material in this book. We provide this section as a service to
those students with calculus. Those students without it can skip this section
without loss. This section is therefore both advanced and optional.
Our goal is to find the value of p that maximizes the log likelihood function, l(p; y). To do so we take the derivative of l(p; y) and set it equal to
zero. From Eq. (2.4):
l(p; y) = log
N
y
!
+ y log p + (N − y) log(1 − p).
39
2.1. ESTIMATING A PROBABILITY
−30
−50
−50
0.0
0.2
0.4
0.6
0.8
1.0
0.0
0.2
0.4
0.6
0.8
1.0
0.2
0.4
0.6
0.8
1.0
−30
−50
−30
−10
D
−10
C
−50
Log−Likelihood
−30
−10
B
−10
A
0.0
0.2
0.4
0.6
p
0.8
1.0
0.0
p
Figure 2.2: Plots of log likelihood for (A) 5 successes from 10 flips, (B) 50
successes from 100 flips, (C) 7 successes from 10 flips, and (D) 70 successes
from 100 flips
40
CHAPTER 2. THE LIKELIHOOD APPROACH
Step 1: Differentiate l(p, y) with respect to p.
"
!
#
∂l(p; y)
∂
N
log
+ y log p + (N − y) log(1 − p) ,
=
y
∂p
∂p
"
!#
∂
∂
∂
N
log
+
[y log p] +
[(N − y) log(1 − p)] ,
=
y
∂p
∂p
∂p
y N −y
= 0+ −
,
p
1−p
y N −y
−
.
=
p
1−p
Step 2: Set the derivative to zero and solve for p.
y N −y
−
p
1−p
y
p
(1 − p)y
y − py
y
= 0,
=
=
=
=
p̂ =
N −y
,
1−p
(N − y)p,
Np − yp,
Np,
y
,
N
(2.5)
where y is the number of observed successes in a particular experiment. For
a binomial, the proportion of successes is the maximum likelihood estimator
of parameter p.
Problem 2.1.2 (Your Turn)
Using calculus methods, derive the maximum likelihood estimator of
parameter λ in the Poisson distribution (see Eq. 2.3).
2.1. ESTIMATING A PROBABILITY
2.1.5
41
Numerical Optimization to find MLEs
There are three steps in using R to find MLEs. The first is to define a log
likelihood function; the second is to enter the data; the third is to maximize
the log likelihood function.
• Step 1: Define a log likelihood function for the binomial:
loglike=function(p,y,N)
return(dbinom(y,N,p,log=T))
Note the log=T option in dbinom(). With this option, dbinom() returns the log of the probability mass function.
• Step 2: Enter the data. Suppose we observed five successes on ten flips.
N=10
y=5
• Step 3: Find the maximum likelihood estimate. There are a few different numerical methods implemented in R for optimization. For models
with one parameter, the function optimize() is an appropriate choice
(Brent, 1973). Here is an example.
optimize(loglike,interval=c(0,1),maximum=T,y=y,N=N)
The first argument is the function to be maximized; the second argument is the interval on which the parameter may range, the third
argument indicates that the function is to be maximized (the other
alternative is that it be minimized); the other arguments are passed to
the function loglike(). Here is the output from R.
$maximum
[1] 0.5
$objective
[1] -1.402043
42
CHAPTER 2. THE LIKELIHOOD APPROACH
The data were 5 successes in 10 flips. The maximum is what we expect,
p̂ = .5. The objective is the value of the log likelihood function at the
maximum.
Problem 2.1.3 (Your Turn)
Use optimize() to find the ML estimate of λ of a Poisson distribution
for y = 4 counts.
2.1.6
Select the best model
In this case, there is one model and it is best by default. In all subsequent
examples, we will have more than one model to choose from.
2.2
Is buttered toast fair?
Coins are considered fair; that is, the probability of the coin landing on either
side is p = .5. In our exploration of Murphy’s Law, we can ask whether
buttered toast is fair. We define two models: one, the general model, has p
free to be any value. The second model, the restricted model, has p fixed at
.5. The restricted model instantiates the proposition that buttered toast is
fair.
2.2.1
Define a Hierarchy of models
General Model:
Restricted Model:
Y ∼ Binomial(p, N).
Y ∼ Binomial(.5, N).
It is clear here that there is a hierarchical relationship between the general
and restricted model. The restricted model may be obtained by limiting the
parameter value of the general model. In this sense, the restricted model is
nested within the general one.
43
2.2. IS BUTTERED TOAST FAIR?
Definition 29 (Nested Models) Model B is considered
nested within Model A if there exists a restriction on the
parameters of Model A that yield Model B.
2.2.2
Express the likelihoods
The likelihood functions for the general model (denoted by Lg ) and restricted
model (denoted by Lr ) are:
Lg (p; y) =
N
y
!
N
y
Lr (.5; y) =
py (1 − p)N −y
!
.5y .5N −y
(2.6)
(2.7)
Note that the likelihood for the restriction is not quite a function of parameters. In all other applications, the likelihoods will be proper functions of
parameters.
2.2.3
Maximize the likelihoods
We can use the calculus-based results we found above to maximize the likelihood functions. Estimates are:
y
,
N
= .5,
pˆg =
pˆr
where y is the number of observed successes.
2.2.4
Select the best model
Model selection is done through comparing maximized likelihoods of nested
models. Let’s suppose we observed 13 successes out of 20 trials. Is this
number of successes extreme enough to reject the restricted model in favor
of the general model? Using R, we can find the following log likelihood values
for 13 successes of 20 trials.
44
CHAPTER 2. THE LIKELIHOOD APPROACH
0
General
Model
Log−likelihood
−1
−2
−3
−4
Restricted
Model
−5
0
5
10
15
20
Number of Successes
Figure 2.3: Maximized log likelihoods as a function of the number of successes
for the general and restricted models.
N=20
y=13
mle.general=y/N
mle.restricted= .5 #by definition
log.like.general=dbinom(y,N,mle.general,log=T)
log.like.restricted=dbinom(y,N,mle.restricted,log=T)
log.like.general
[1] -1.690642
log.like.restricted
[1] -2.604652
The log likelihood is greater for the general model than the restricted
model (-1.69 vs. -2.60). This fact, in itself, is not informative. Figure 2.3
shows maximized log likelihood for the restricted and general models for all
possible outcomes. Maximized likelihood for the general model is as great or
greater than that of the restricted model for all outcomes. This trend always
holds: general models always have higher log likelihood than their nested
restrictions.
45
2.2. IS BUTTERED TOAST FAIR?
This leads to the question of whether a restricted model is ever appropriate. The answer is overwhelmingly positive. The restricted model, if it is
true, is a simpler and more parsimonious account of flipping toast. Having
evidence for its plausibility or implausibility represents the accumulation of
knowledge.
We will fail to reject the restricted model if the difference in log likelihood
between the restricted and the general model is not too great. If there is
a great difference, however, then we may reject the restricted model. For
example, if the number of successes is 4, there is a large difference between the
general model and the restricted model indicating that the restriction is illsuited. The likelihood ratio statistic (G2 ) is used to formally assess whether
the difference in log likelihood is sufficiently large to reject a restricted model
in favor of a more general one.
Definition 30 (Likelihood ratio statistic) Consider likelihoods functions for a general and restricted (properly nested)
model, Lg and Lr with parameters pg and pr , respectively. Then
Lr (pˆr ; y)
G = −2 log
Lg (pˆg ; y)
2
!
Statistic G2 is conveniently expressed with log likelihoods. In this case:
G2 = −2[lr (pˆr ; y) − lg (pˆg ; y)],
(2.8)
where lg and lr are the respective log likelihood functions.
If the restricted model holds, then G2 follows a chi-square distribution.
The chi-square distribution has a single parameter, the degrees of freedom
(df). For a likelihood ratio test of nested models, the degrees of freedom is
the difference in the number of parameters between the general and restricted
model. In this case, the general model has one parameter while the restricted
model has none; hence, the difference is 1. The critical .05 value of a chisquare distribution with 1 degree of freedom is about 3.84. Hence for our
case, if G2 < 3.84 then we fail to reject the restriction; otherwise, we reject
the restriction.
Here is G2 for 13 successes of 20 trials:
46
Log−Likelihood
Ratio Statistic
CHAPTER 2. THE LIKELIHOOD APPROACH
25
20
15
10
5
0
0
5
10
15
20
Number of Successes
Figure 2.4: Likelihood ratio statistics (G2 ) as a function of outcome. The
horizontal line denotes the critical value of 3.84. We fail to reject the restricted model (fair toast) when the number of successes is in between 6 and
14, inclusive. If the number of successes is more extreme, then we can reject
the fair-toast model
G2=-2*(log.like.restricted-log.like.general)
G2
[1] 1.828022
The value is about 1.83, which is less than 3.84. Hence, for 13 successes in
20 trials, we cannot reject the restriction that buttered toast is fair.
Figure 2.4 shows which outcomes will lead to a rejection of the fair-toast
restriction: all of those outcomes with likelihood ratio statistics above 3.84.
The horizontal line denotes the value of 3.84. From the figure, it is clear that
observing fewer than 6 or more than 14 successes out of twenty flips would
lead to a G2 greater than 3.84 and, therefore, to a rejection of the statement
that toast is fair.
Testing the applicability of a restricted model against a more general one
is a form of null hypothesis testing. In this case, the restricted model serves
as the null hypothesis. When G2 is high, the restricted model is implausible,
leading to us reject the restriction and accept the more general alternative.
2.3. COMPARING CONDITIONS
2.3
47
Comparing Conditions
Suppose we have two different conditions in a memory experiment. For
example, we are investigating the effects of word frequency on recall performance. In our experiment, participants study two types of words: high
and low frequency. High frequency words are those used often in everyday
speech (e.g., horse, table); low frequency words are used rarely (e.g., lynx, ottoman). We wish to know whether recall performance is significantly better
in one condition than the other. Although there are several ways of tackling
this problem, we use the likelihood framework here. Let’s suppose that each
condition consists of a number of trials. On each trial, performance may either be a success (e.g., a word was recalled) or a failure (e.g., a word was not
recalled). We let Yl and Yh be the number of successes in the low-frequency
and high-frequency, conditions. Likewise, we let Nl and Nh be the number
of trials in these conditions, respectively. Suppose the data are as follows:
yl = 6 of Nl = 20 low-frequency words were recalled and yh = 7 of Nh = 13
high-frequency words were recalled.
2.3.1
Define a hierarchy of models
General Model:
Yl ∼ Binomial(pl , Nl )
Yh ∼ Binomial(ph , Nh )
(2.9)
(2.10)
Yl ∼ Binomial(p, Nl )
Yh ∼ Binomial(p, Nh )
(2.11)
(2.12)
Restricted Model:
The difference in the above models is in the probability of a success. In
the general model, there are two such probabilities, pl and ph , with one for
each condition. This notation is used to indicate that these two probabilities
need not be the same value. The restricted model has one parameter p,
indicating that the probability of success in both conditions must be the
same. If the restricted model does not fit well when compared to the general
model, then we may conclude that performance depends on word frequency.
The next step is expressing the likelihoods. The likelihood functions for
these models are more complicated than for previous models. The complication comes about because the models involve two random variables, Yl and
48
CHAPTER 2. THE LIKELIHOOD APPROACH
Yh . These complications are overcome by introducing a new concept: joint
probability mass function. The probability mass function was introduced earlier; it describes how probability is distributed for a single random variable.
The joint probability mass function describes how probability is distributed
across all combinations of outcomes for two (or more) random variables.
2.3.2
Joint Probability Mass Functions
Definition 31 (Joint Probability Mass Function) Let X
and Y be two discrete random variables, then the joint probability mass function is given by
f (x, y) = P r(X = x and Y = y).
The following is an example of a joint probability mass function. Suppose
X is a random variable that has equal mass on 1, 2, and 3 and zero mass
otherwise. The probability mass function for X is given by:
1/3
1/3
fX (x) =
1/3
0
x=1
x=2
x=3
Otherwise
Because we will be considering more than one random variable, we subscript
probability mass functions, i.e., fX for clarity.
Suppose Y = X + 1 with probability .5 and Y = X − 1 with probability
.5. The joint probability mass function for X and Y is denote fX,Y and given
by:
1/6 x = 1, y = 0
1/6 x = 1, y = 2
1/6 x = 2, y = 1
fX,Y (x, y) = 1/6 x = 2, y = 3
1/6 x = 3, y = 2
1/6 x = 3, y = 4
0
Otherwise
2.3. COMPARING CONDITIONS
49
The likelihood of the parameters is the joint probability mass function
expressed as a function of the parameters. For the general model:
L(pl , ph ; l, h) = fYl ,Yh (l, h; pl , ph ).
(2.13)
We introduced independence earlier in the previous chapter; if two random variables are independent, then the outcome of one does not influence
the outcome of the other. Under this condition the joint probability mass
function is the product of the individual mass functions. These individual
probability mass functions are called marginal probability mass functions.
Definition 32 (Independent Random Variables) Let X
and Y be random variables with probability mass functions fX
and fY , respectively. If these random variables are independent,
fX,Y (x, y) = fX (x)fY (y).
Is independence justified for either the general or restricted model? It
would seem, at first consideration, that independence cannot be used, especially for the restricted model in which both random variables are functions
of the same parameter p. If p is high, both Yl and Yh tend to be high in value;
if p is low, both Yl and Yh tend to be low in value. Hence, knowledge of Yl
would imply knowledge of p which would imply knowledge of Yh , seemingly
violating independence.
This reasoning, fortunately, is not fully accurate. Models, such as the
restricted model, may be specified dependencies among random variables
through common parameters. The critical issue is whether there are dependencies outside this specification. In other words, given a common value of
p, does knowing the value of Yh provide any more information about Yl ?
Implicit in the definition of a model is that there are no other dependencies. Therefore, the likelihood for a model may be made with recourse to
independence. For the general model,
fYl ,Yh (l, h; pl , ph ) = fYl (l; pl , ph ) × fYh (h; pl , ph )
(2.14)
fYl ,Yh (l, h; p) = fYl (l; p) × fYh (h; p)
(2.15)
For the restricted model,
50
CHAPTER 2. THE LIKELIHOOD APPROACH
2.3.3
Expressing the Likelihoods
With this digression on conditional independence finished, we return to the
problem of expressing the likelihood for the models. For the general model,
we start with Eq. (2.14). Because for the general model Yh does not depend
on pl and Yl does not depend on ph , the joint likelihood could be simplified:
fYl ,Yh (l, h; pl , ph ) = fYl (l; pl ) × fYh (h; ph ).
Substituting the probability mass function of a binomial yields:
fYl ,Yh (l, h; pl , ph )(l, h, pl ; ph ) =
Nl
l
!
pll (1
− pl )
Nl −l
Nh
h
!
phh (1 − ph )Nh −h l = 0, .., N,
h = 0, .., N
0
Otherwise
The likelihood function for pl and ph in the general model is therefore
LG (pl , ph , yl , yh ) =
Nl
yl
!
pyl l (1
− pl )
Nl −yl
Nh
yh
!
pyhh (1 − ph )Nh −yh (2.16)
The restricted model has a single parameter p. The likelihood function
is obtained by setting ph = p and pl = p:
LR (p, yl , yh ) =
Nl
yl
!
yl
p (1 − p)
Nl −yl
Nh
yh
!
pyh (1 − p)Nh −yh
(2.17)
log likelihood functions are given by:
!
!
Nl
Nh
lG (pl , ph , yl , yh ) = log
+ log
+ yl log(pl ) +
yl
yh
(Nl − yl ) log(1 − pl ) + yh log(ph ) +
(Nh − yh ) log(1 − ph )
!
!
Nl
Nh
lR (p, yl , yh ) = log
+ log
+ yl log(p) +
yl
yh
(Nl − yl ) log(1 − p) + yh log(p) +
(Nh − yh ) log(1 − p)
(2.18)
(2.19)
Eq. (2.19) can be simplified further:
"
!
!#
Nl
Nh
= log
+ (yl + yh ) log(p) +
yl
yh
[(Nl + Nh ) − (yl + yh )] log(1 − p)
(2.20)
2.3. COMPARING CONDITIONS
2.3.4
51
Maximize the Likelihoods
General Model
There are a few approaches we can take to maximizing likelihoods. The most
intuitive is to note that Yh and Yl are independent samples with no common
parameters. In this case, we can maximize them separately. So, we use our
standard binomial MLE result:
yl
p̂l =
,
(2.21)
Nl
yh
pˆh =
.
(2.22)
Nh
Applying these to the data yields p̂l = 6/20 = .3 and pˆh = 7/13 = .538.
If we failed to see this intuitive approach, we can use R to maximize
likelihood numerically as follows.
The log likelihood function in Eq. (2.18) may be maximized as written,
but it is not optimal. One property of likelihood is that the maximum and the
shape do not change when the function is multiplied by a constant. Likewise,
the maximum of log likelihood does not change with addition of a constant.
This conservation is shown in Figure 2.5. Because of this property, likelihood
is often defined only up to a constant of multiplication. Therefore, if l is a
log likelihood, then l + c is also a log likelihood, where c is a constant that
does not involve the parameters.
This fact helps facilitate the time and precision of finding the parameters
!
Nl
that maximize log likelihood for the general model. The terms log
yl
!
Nh
and log
do not depend on parameters. An equally valid log likelihood
yh
for lG is:
lG = yl log(pl ) + (Nl − yl ) log(1 − pl ) + yh log(ph ) + (Nh − yh ) log(1 − ph ).
Precision and speed are gained by not having to evaluate the constant terms
involving the log of the choose function.
The code below uses numerical methods to maximize the log likelihood
function for the general model. In the code, pL, pH, yL, yH, NL, and
NH correspond to pl , ph , yl , yh , Nl , and Nh respectively. The function
nll.general() returns the negative of the log likelihood. The reason for
this choice is discussed below.
52
−20
B
−100
−60
Log likelihood
−60
−20
A
−100
Log likelihood
CHAPTER 2. THE LIKELIHOOD APPROACH
0.0
0.2
0.4
0.6
0.8
1.0
p
0.0
0.2
0.4
0.6
0.8
1.0
p
Figure 2.5: The maximum of the log likelihood function does not change with
the addition of a constant. Panel A shows the log likelihood function for a
binomial with N=100 and 70 observed successes. The vertical line denotes
the maximum at .7. Panel B shows the log likelihood with constant log Ny
subtracted. The function has the same shape and maximum as the function
in Panel A.
# negative log likelihood of the general model
nll.general=function(par,y,N)
{
pL=par[1]
pH=par[2]
yL=y[1]
yH=y[2]
NL=N[1]
NH=N[2]
ll=yL*log(pL)+(NL-yL)*log(1-pL)+yH*log(pH)+(NH-yH)*log(1-pH)
return(-ll)
}
Function nll.general() is a function of 2 parameters. The function
optimize() In R, finds the maximum of a function of one parameter. The
function optim() is used when there are more than one parameter. By
default, the optim() command minimizes instead of maximizing functions.
This is the reason we use the negative log likelihood. The parameter values
that minimize the negative log likelihood maximize the log likelihood.
par=c(.5,.5) #starting values
2.3. COMPARING CONDITIONS
53
N=c(20,13) #number of trials in each condition
y=c(6,7) #number of successes
#use maximization of two or more variables
optim(par,nll.general,y=y,N=N)
Here is the output from the optim()1 call above:
$par
[1] 0.2999797 0.5384823
$value
[1] 21.1897
$counts
function gradient
69
NA
$convergence
[1] 0
$message
NULL
The numerically-maximized parameter values match those from the intuitive
method above.
Function optim() can use a few different algorithms; the default is based
on the simplex algorithm of Nelder & Mead (1965) which, while slow, is
often successful in finding local minima (Press, Flannery, Teukolsky, and
Vetterling, 1992). We discuss minimization in more depth in Chapter 5.
Restricted Model
The restricted model can also be solved intuitively. Under the model, performance in the low- and high-frequency conditions reflects a single Bernoulli
process. Hence, we can pool the data across conditions. Pooling the number
1
For more information about what each of these values mean, try ?optim in R to get
help on optim().
54
CHAPTER 2. THE LIKELIHOOD APPROACH
of successes across both conditions yields y = 6 + 7 = 13. Doing the same
for the number of trials yields N = 20 + 13 = 33. This pooling is evident in
Eq. (2.20); when we group terms, the restricted log likelihood is the log likelihood function of a binomial with N = Nl +Nh trials and y = yl +yh successes.
If we fail to see this intuitive approach, R may be used to solve the problem
numerically. Because the restricted model has a single parameter, it is best
to use optimize() instead of optim(). The function ll.restricted() is
the log likelihood for the restricted model:
#log likelihood of restricted model
nll.restricted=function(p,y,N)
{
yL=y[1]
yH=y[2]
NL=N[1]
NH=N[2]
ll=yL*log(p)+(NL-yL)*log(1-p)+yH*log(p)+(NH-yH)*log(1-p)
return(-ll)
}
N=c(20,13)
y=c(6,7)
optimize(nll.restricted,interval=c(0,1),y=y,N=N,maximum=F)
Here is the output from the optimize() call above:
$maximum
[1] 0.393931
$objective
[1] 22.12576
2.3.5
Select the best model
We use the likelihood ratio statistic, G2 , to select the best model. To do so
in R, we first assign the output of the numerical minimization to variables:
gen=optim(par,nll.general,y=y,N=N)
res=optimize(nll.restricted,interval=c(0,1),y=y,N=N,maximum=F)
2.4. STANDARD ERRORS FOR MAXIMUM LIKELIHOOD ESTIMATORS55
The results can be seen by typing gen or res. Unfortunately, outputs of
optim() and optimize() are not consistent. The value of the minimized
function is called value in optim() and objective in optimize(). The
value of log likelihood is given by:
gen.ll=-gen$value
res.ll=-res$objective
Statistic G2 is obtained by G2=-2*(res.ll-gen.ll); for the example, G2 is
about 1.87. In this case we retain the restricted model because G2 < 3.84.
Problem 2.3.1 (Your turn)
Let’s revisit the Poisson model. You are a traffic engineer and there
have been too many accidents at rush hour at one particular intersection. To see if the accidents are caused by excessive speed, you place a
police car 100 meters before the intersection. The day before you place
the police car, there are 12 accidents; the day after, 4 accidents. Use a
likelihood ratio test to determine if placing the police car was effective
in lowering the accident rate λ.
2.4
Standard Errors for Maximum Likelihood
Estimators
Maximum likelihood estimators are random variables. Consequently, they
have sampling distributions and standard errors. We provide here a brief description of how to derive standard errors for maximum likelihood estimates.
We present here the implementation without the theoretical underpinning.
Unfortunately, the mathematics in this underpinning is beyond the scope of
this book. A formal treatment may be found in several standard advanced
statistics texts including Lehman (1991).
The standard error of a maximum likelihood estimator is related to the
curvature of the log likelihood function at the maximum. In Figure 2.2, the
likelihood function in Panel B has more curvature at the maximum than
56
CHAPTER 2. THE LIKELIHOOD APPROACH
that in Panel A. This curvature reflects how much variability there is in
the sampling distribution of the MLE with more curvature corresponding to
smaller variability. Not surprisingly, log likelihood curvature and variability
are both functions of the number of observations.
To compute an index of curvature at the maximum in R, we add the
option hessian=TRUE to the optim() call. The following code returns an
index of curvature:
par=c(.5,.5)
gen=optim(par,nll.general,y=y,N=N,hessian=TRUE)
Type gen and notice the additional field $hessian, which is a matrix. The
diagonal elements are of interest and standard errors for the parameters are
calculated by genSE=sqrt(diag(solve(gen$hessian))). The elements of
the vector genSE correspond to the standard errors for the parameters in the
vector par.
Many researchers report standard errors along with estimates. Standard
errors, in our opinion, give a rough guide to the amount of variability in
the estimate as well as calibrate the eye as to the magnitude of significant
effects (Rouder & Morey, 2005). We discuss the related concept of confidence
intervals in Chapter 7.
The following code plots standard errors in R. For convenience, we define
a new function errbar(), which we can use for adding error bars to any plot.
errbar=function(x,y,height,width,lty=1){
arrows(x,y,x,y+height,angle=90,length=width,lty=lty)
arrows(x,y,x,y-height,angle=90,length=width,lty=lty)
}
We plot the estimates in bar plot format:
xpos=barplot(gen$par,names.arg=c(’Low Frequency’,
’High Frequency’),col=’white’,ylim=c(0,1),space=.5,
ylab=’Probability Estimate’)
The x-axis in a bar plot is labeled by condition. R assigns numeric values to x-axis so that you may add lines, annotations, or error bars. These are
stored in variable xpos. To add error bars, use errbar(xpos,gen$par,genSE,.3).
57
0.8
0.6
0.4
0.2
0.0
Probability Estimate
1.0
2.5. WHY MAXIMUM LIKELIHOOD?
Low Frequency
High Frequency
Condition
Figure 2.6: Standard errors and confidence intervals on maximum likelihood
estimates from the word frequency example. Bars represent point estimates,
and error bars represent standard errors of the estimator.
The last value is simply the width of the error bar. The resulting bar plot
with standard error bars is shown in Figure 2.6. The overlap of the error bars
indicates that any effect of word frequency is quite small given the variability of the estimate. The likelihood ratio tests confirms the lack of statistical
significance.
Problem 2.4.1 (Your Turn)
Consider the previous problem with the Poisson distribution model of
traffic accidents. Plot your estimates of λ for the general model with
standard error bars.
2.5
Why Maximum Likelihood?
The likelihood approach is used throughout this book for analysis. In this
section, we provide an overview of the theoretical justification for this choice.
There are several other good alternatives to maximum likelihood including
58
CHAPTER 2. THE LIKELIHOOD APPROACH
minimizing the mean squared error between model predictions and observed
data. Likelihood has the following advantages under mild technical conditions2 :
1. Maximum likelihood estimates, while often biased in small sample sizes,
are consistent; i.e., in the limit of a large number of samples, the estimates of a parameter converge to its true value.
2. Maximum likelihood estimates are asymptotically maximally efficient—
in the limit of a large number of samples, no other estimate has a
smaller error. This lower bound is known as the Cramer-Rao lower
limit in the statistics literature, and it can be shown that maximum
likelihood estimators approach the limit with large sample sizes.
3. Maximum likelihood estimates are asymptotically normally distributed.
As the sample size becomes larger, the sampling distribution converges
to a normal distribution. This licenses the use of normal-model based
statistics in analyzing maximum likelihood estimates. A discussion of
normal-model based statistics is provided in Chapter 7.
4. The likelihood approach is tractable for the simple, nonlinear models.
These models are more realistic accounts of psychological processes
than standard normal-based models. Because they are more realistic,
they provide more detailed tests of psychological theory than standard
ANOVA or regression models. We join others who recommend likelihood as a viable alternative for the types of models well-suited for
cognitive and perceptual psychology (see also Glover & Dixon, 2004;
Myung, 2001.
5. Likelihood is a stepping stone in understanding Bayesian techniques.
We have argued that these techniques will become increasingly valuable
in psychological contexts (e.g., Rouder & Lu, in press; Rouder, Lu,
Speckman, Sun, and Jiang, in press; Rouder, Sun, Speckman, Lu &
Dzhou, 2003). Knowledge gained in this book about analysis with
likelihoods transfers to the Bayesian framework more so than that of
other estimation methods.
2
For these advantages to hold, the model must be regular. Regularity is a set of
conditions that guarantee that the likelihood function is sufficiently smooth. A formal
definition may be found in Lehmann, 1991.
2.5. WHY MAXIMUM LIKELIHOOD?
59
Given these advantages, it is worth considering the drawbacks of maximum likelihood. There are three related drawbacks:
1. Maximum likelihood estimates are often biased in finite samples. Researchers may simply feel uncomfortable with biased estimators. Consequently, there is much development in maximally efficient unbiased
estimators. We feel comfortable recommending ML and are not overly
concerned with bias. We do recommend that researchers understand
the magnitude of bias for their application and the simulation method
is useful for this purpose.
2. Although maximum likelihood estimates have excellent asymptotic properties, they may not be optimally efficient for small sample sizes. Sometimes, other methods are just as good asymptotically and better in
finite samples (see Heathcote, Brown, and Mewhort, 2002, Brown &
Heathcote, 2004, and Speckman & Rouder, 2004, for an exchange about
a different estimation appears to outperform likelihood in finite samples).
3. Likelihood tests of composite hypotheses are not necessarily the most
powerful. Composite hypotheses are the type that we consider in this
book and correspond to the case in which one model is a restriction of
another. For some model classes, there are more powerful tests (a good
example is the t-test). Of course, developing most-powerful tests for
nonlinear models is a difficult problem for many applications whereas
the application of the likelihood method is often straightforward and
tractable.
In sum, the likelihood method is not necessarily ideal for every situation,
but it is straightforward and tractable with many nonlinear models. With
it, psychologists can test theories to a greater level of detail than is possible
with standard linear models.
More advanced, in-depth treatments of maximum likelihood techniques
can be found in mathematical statistics texts. While these in-depth treatments do require some calculus knowledge, the advanced student can benefit from learning about properties of maximum likelihood estimators. Some
good texts to consider for further reading are Hogg & Craig (1978), Lehmann
(1991), and Rice (1998).
60
CHAPTER 2. THE LIKELIHOOD APPROACH
Chapter 3
The High-Threshold Model
Experimental psychologists learn about cognition by measuring how people
react to various stimuli. In many cases, these reactions indicate how well
people process stimuli; e.g., how well people can detect a faint tone or how
well they can remember words. In most investigations, it is essential to have
a measure of how well people perform on a task.
3.1
The Signal-Detection Experiment
Consider the problem of assessing how well a participant can hear a tone
of a given frequency and volume when it is embedded in noise. A simple
experiment may consist of two types of trials: one in which the target tone
is presented embedded in noise, and a second in which the noise is presented
without the tone. Trials with an embedded tone are called signal trials; trials
without the tone are called noise trials. Participants listen to a sequence of
signal and noise trials in which both types are intermixed. Their task is
to indicate whether the target tone is present or absent. Let’s consider an
experiment in which the experimenter presents 100 signal trials and 50 noise
trials. Table 3.1 shows a sample set of data.
Experiments with data in the form of Table 3.1 are called signal-detection
experiments. Psychologists express the results of such experiments in terms
of four events:
1. Hit: Participant responds “tone present” on a signal trial.
61
62
CHAPTER 3. THE HIGH-THRESHOLD MODEL
Stimulus
Signal
Noise
Total
Response
Tone Present Tone Absent
75
25
30
20
105
45
Total
100
50
150
Table 3.1: Sample data for a signal-detection experiment.
2. Miss Participant responds “tone absent” on a signal trial.
3. False Alarm: Participant responds “tone present” on a noise trial.
4. Correct Rejection: Participant responds “tone absent” on a noise trial.
Hit and correct rejection events are correct responses while false alarm and
miss events are error responses.
Signal detection experiments are used in many domains besides the detection of tones. One prominent example is in the study of memory. In
many memory experiments, participants study a set of items, and then at
a later time, are tested on them. In the recognition memory paradigm both
previously studied items and unstudied novel items are presented at the test
phase. The participant indicates whether the item was previously studied or
is novel. In this case, a studied item is analogous to the signal stimulus and
the novel item is analogous to a noise stimulus. Consequently, the miss error
occurs when a participant is presented a studied item and indicates that it is
novel. The false alarm error occurs when a participant is presented a novel
item and indicates it was studied.
It is reasonable to ask why there are two different types of correct and
error events. Why not just measure overall accuracy? One of the most
dramatic example of the importance of differentiating the errors comes from
the repressed-memory literature. The controversy stems from the question of
whether it is possible to recall events that did not happen, especially those
regarding sexual abuse. According to some, child sexual abuse is such a
shocking event that memory for it may be repressed (Herman & Schatzow,
1987). This repressed memory may then be “recovered” at some point later
in life. The memory, when repressed is a miss; but when recovered, is a
hit. Other researchers question the veracity of these recovered memories,
claiming it is doubtful that a memory of sexual abuse can be repressed and
then recovered (Loftus, 1993). The counter claim is that the sexual abuse
3.1. THE SIGNAL-DETECTION EXPERIMENT
63
may not have occurred. The “recovered memory” is actually a false alarm.
In this case, differentiating between misses and false alarms is critical in
understanding how to evaluate claims of recovered memories.
The results of a signal detection experiment are commonly expressed as
the following rates:
• Hit Rate: The proportion of tone-present responses on signal trials .
The hit rate in Table 3.1 is .75.
• Miss Rate: The proportion of tone-absent responses on signal trials.
The miss rate in Table 3.1 is .25
• False-Alarm Rate: The proportion of tone-present responses on noise
trials. The false-alarm rate in Table 3.1 is .6.
• Correct-Rejection Rate: The proportion of tone-absent responses on
noise trials. The correct-rejection rate in Table 3.1 is .4.
To perform analysis on data from signal-detection experiments, let’s use
random variables to denote counts of events. Let RVs Yh , Ym , Yf , and Yc
denote the number of hits, misses, false alarms, and correct rejections, respectively. For example, Yh denote the number of hits. Data, such as the
entries in Table 3.1, are denoted with yh , ym , yf , and yc respectively. Let
Ns and Nn refer to the number of signal and noise trials, respectively. The
hit rate is yh /Ns ; the miss rate is ym /Ns ; the false-alarm rate is yf /Nn ; the
correct-rejection rate is yc /Nn .
Each signal trial results in either a hit or miss event; likewise, each noise
trial results in either a false-alarm or correct-rejection event. Hence,
Ns = yh + ym ,
Nn = yf + yc
Whereas Ns and Nn are known rather than estimated, it is only necessary
to record the numbers of hits and false alarms. From these numbers, the
numbers of misses and correct rejections can be calculated. Therefore, there
are only two independent pieces of data in the signal detection experiment.
64
3.1.1
CHAPTER 3. THE HIGH-THRESHOLD MODEL
A Simple Binomial Model of Hits and False Alarms
The simplest model of the signal-detection experiment is given as
Yh ∼ B(ph , Ns ),
Yf ∼ B(pf , Nn ),
(3.1)
(3.2)
where ph and pf refer to the true probabilities of hits and false alarms, respectively. The other two probabilities, the probability of a miss and the probability of a correct rejection are denoted pm and pc , respectively. The outcome
of a signal trial may only be a hit or a miss. Consequently, ph + pm = 1.
Likewise, pf + pc = 1. Hence, there are only two free parameters (ph , pf ) of
concern. Once these two are estimated, estimates of (pm , pc ) can be obtained
by subtraction.
The model may be analyzed by treating each component independently.
Hence, by the results with the binomial distribution, maximum likelihood
estimates are given by
p̂h = yh /Ns
p̂f = yf /Nn
The terms p̂h and p̂f are the hit and false alarm rates, respectively.
3.2
The High-Threshold Model
The problem with the binomial model above is that it yields two different
measures of performance: the hit rate and the correct-rejection rate. In most
applications, researchers are interested in a single measure of performance.
The high-threshold model provides this. It posits that perception is all-ornone. A participant is either in one of two mental states on a signal trial.
They either have detected the target tone, with probability d, or failed to do
so, with probability 1 − d. When participants fail to detect the tone, they
still may guess that it had been presented with probability g. Figure 3.1
provides a graphical representation of the model. There are two ways for a
hit event: the first is through successful detection. A hit may also occur when
detection fails yet the participant still guesses that the stimulus is present.
This route to a hit occurs with probability (1 − d)g. Summing these yields
the probability of a hit: ph = d + (1 − d)g. For noise trials, there is no target
65
3.2. THE HIGH-THRESHOLD MODEL
Hit
High Threshold Model
d
False Alarm
Hit
g
g
1−d
1−g
1−g
Correct Rejection
Miss
Signal Trials
Noise Trials
Figure 3.1: The high-threshold model.
to detect. False alarms are produced only by guessing: pf = g. Substituting
these relations into the binomial model on hits and false alarms (Eqs. 3.1 &
3.2) yields:
Yh ∼ Binomial(d + (1 − d)g, Ns )
Yf ∼ Binomial(g, Nn ),
(3.3)
(3.4)
where 0 < d, g < 1. The goal is to estimate parameters d and g. We use the
four-step likelihood approach. The first step, defining a hierarchy of models
has been done. The above model is the only one.
3.2.1
Express the likelihood
The likelihood is derived from the joint probability mass function f (yh , yf ).
As discussed in Chapter 2, given common parameters, we treat yh and yf as
independent. Therefore the joint probability mass function may be obtained
by multiplying the marginal probability mass functions. These marginals are
given by:
f (yh ; d, g) =
Ns
yh
!
(d + (1 − d)g)yh (1 − (d + (1 − d)g))Ns−yh ,
and
f (yf ; d, g) =
Nn
yf
!
(g)yf (1 − g))Nn −yf .
66
CHAPTER 3. THE HIGH-THRESHOLD MODEL
Theses equations may be simplified by substituting ym for Ns − yh , yc for
Nn − yf and (1 − d)(1 − g) for 1 − (d + (1 − d)g). Making these substitutions
and multiplying these marginal probability mass functions yields
Ns
yh
f (yh , yf ; d, g) =
×
!
(d + (1 − d)g)yh ((1 − d)(1 − g))ym
Nn
yf
!
(g)yf (1 − g))yc .
The log likelihood may be obtained by rewriting this equation as a function of d and g and then taking logarithms:
l(d, g; yh, yf ) = log
+ log
Ns
yh
!
Nn
yf
+ yh log(d + (1 − d)g) + (ym ) log((1 − d)(1 − g))
!
+ yf log(g) + (yc ) log(1 − g).
Some of the terms are not functions of parameters and may be omitted:
l(d, g; yh, yf ) = − (yh log(d + (1 − d)g) + (ym ) log((1 − d)(1 − g)) + yf log(g) + (yc ) log(1 − g
3.2.2
Maximize the Likelihood
Either calculus methods or numerical methods may be used to provide maximum likelihood estimates. The calculus methods provide the following solutions:
dˆ =
yh
Ns
y
− Nfn
,
y
1 − Nfn
ĝ = yf /Nn .
(3.5)
(3.6)
Typically, these are written in terms of the hit and false alarm rates, p̂h and
p̂f , respectively:
p̂h − p̂f
dˆ =
,
1 − pˆf
ĝ = p̂f .
3.2. THE HIGH-THRESHOLD MODEL
67
These equations are used to generate estimates from the sample data in
Table 3.1:
75
100
30
− 50
30 = .375
1 − 50
ĝ = yf /Nn = .6.
dˆ =
The following is an implementation in R. We present it for two reasons:
(1) the programming techniques implemented here are useful in analyzing
subsequent models; and (2) estimates of standard errors are readily obtained
in the numerical approach. In the program y is a vector of data. It has
four elements, (yh , ym , yf , yc ) and may be assigned values in Table 3.1 with
y=c(100,50,30,20). Vector par is the vector of parameters (d, g)
The first step is to compute the negative log likelihood:
#negative log likelihood of high-threshold model
nll.ht=function(par,y)
{
d=par[1]
g=par[2]
ll=y[1]*log(d+(1-d)*g)+y[2]*log((1-d)*(1-g))+
y[3]*log(g)+y[4]*log(1-g)
return(-ll)
}
Although the above code is valid, we may rewrite it to make it easier to
read and modify. The change is motivated by noting that the log likelihood
can be put in following form:
l = C + yh log(ph ) + ym log(pm ) + yf log(pf ) + yc log(pc ),
where C is the log of the choose terms that do not depend on parameters.
This log likelihood can be rewritten as
l=
X
yi log(pi ),
i
where i ranges over the four events. Let p denote a vector of probabilities
(ph , pm , pf , pc ). The function is rewritten as:
68
CHAPTER 3. THE HIGH-THRESHOLD MODEL
#negative log likelihood of high-threshold model
nll.ht=function(par,y)
{
d=par[1]
g=par[2]
p=1:4 # reserve space
p[1]=d+(1-d)*g #probability of a hit
p[2]=1-p[1] # probability of a miss
p[3]=g # probability of a false alarm
p[4] = 1-p[3] #probability of a correct rejection
return(-sum(y*log(p)))
}
The next step is to maximize the function:
y=c(75,25,30,20)
par=c(.5,.5)
optim(par,nll.ht,y=y)
Execution of this code yields estimates of dˆ = .3751 and ĝ = .5999, which
are acceptably close to the closed-form answers of dˆ = .375 and ĝ = .6.
3.3
Selective Influence in the High Threshold
Model
ˆ it
Although the high-threshold model provides estimates of performance (d),
does so by assuming that perception is all-or-none. Is this aspect correct?
One way of testing it is to perform a selective influence experiment. In
a selective influence experiment of the high-threshold model, the researcher
designates a manipulation that should affect one parameter and not the other.
Parameter d is a bottom-up strength parameter. In a tone experiment, it
would reflect factors that determine the strength of perception of the tone
including its volume and frequency. Parameter g is a top-down parameter.
To influence g and not d, a researcher may use differing payoffs. In one
condition, Condition 1, the researcher may pay 10c for each hit and 1c for
each correct rejection. In a second condition, Condition 2, the researcher may
3.3. SELECTIVE INFLUENCE IN THE HIGH THRESHOLD MODEL69
Frequencies of Responses
Hit Miss False Alarm
Condition 1 40
10
30
Condition 2 15
35
2
Correct Rejection
20
48
Table 3.2: Hypothetical data to test selective influence in the High Threshold
model.
pay the reverse (1c for each hit and 10c for each correct rejection). Condition
1 favors tone-present responses; condition 2 favors a tone-absent responses.
Parameter g should therefore be higher in Condition 1 than Condition 2.
Parameter d does not reflect these payoffs and should be invariant to the
manipulation. Suppose the experiment was run with 50 signal and 50 noise
trials in each condition. Hypothetical data is given in Table 3.2.
There are two parts to the selective influence test: The first is whether
the manipulation affected g as hypothesized. The second part is whether the
the manipulation had no effect on d. This second test is as least as important
as the first; the invariant of d, if it occurs, is necessary for support of the
model. We follow the four steps in answering this question.
3.3.1
Hierarchy of Models
We form three models. Let di and gi be the sensitivity and guessing rate in
the ith condition, where i is 1 or 2. Likewise, let Yh,i, Yf,i denote the number
of hits and false alarms in the ith condition, respectively. Let Ns,i , Nn,i denote
the number of signal and noise trials in the ith condition, respectively. Model
1, the most general model, is constructed by allowing separate sensitivity and
guessing rates.
Model 1
Yh,i ∼ B(di + (1 − di )gi, Ns,i ),
Yf,i ∼ B(gi , Nn,i ).
Model 1 has four parameters: d1 , g1 , d2 , g2 .
(3.7)
(3.8)
(3.9)
70
CHAPTER 3. THE HIGH-THRESHOLD MODEL
Model 2 is constructed as a restriction on Model 1—assume that sensitivity is equal in both conditions:d = d1 = d2 .
Model 2
Yh,i ∼ B(d + (1 − d)gi , Ns,i),
Yf,i ∼ B(gi , Nn,i).
(3.10)
(3.11)
(3.12)
Model 2 has three parameters (d, g1, g2 )
Model 3 is the other restriction of Model 1. Although sensitivity may vary
across conditions, the guessing rate is assumed to be equal: g = g1 = g2 .
Model 3:
Yh,i ∼ B(di + (1 − di )g, Ns,i),
Yf,i ∼ B(g, Nn,i).
(3.13)
(3.14)
The three models form a hierarchy with Model 1 being the most general
and Models 2 and 3 being proper restrictions. This hierarchy allows us to
test the selective influence hypotheses. Accordingly, the expected variation
of g can be tested by comparing Model 3 to Model 1. Likewise, the expected
invariance of d can be tested by comparing Model 2 to Model 1.
3.3.2
Express the Likelihoods
We express the likelihoods within R. Our approach is to specify a general log
likelihood function for any one condition. It takes as input the four hit, miss,
false alarm, and correct rejection counts and two parameters. We will use this
function repeatedly, even when fitting the restricted models. The comments
indicate how par and y should be assigned when calling the function.
#negative log likelihood for high-threshold
#assign par=c(d,c)
#assign y=c(h,m,f,c)
nll.condition=function(par,y)
{
p=1:4
d=par[1]
g=par[2]
3.3. SELECTIVE INFLUENCE IN THE HIGH THRESHOLD MODEL71
p[1]=d+(1-d)*g
p[2]=1-p[1]
p[3]=g
p[4]=1-p[3]
return(-sum(y*log(p)))
}
Given common parameters, data from the different conditions are independent. The joint likelihood across conditions is the product of likelihoods for
each condition, and the joint log likelihood across conditions is the sum of
the log likelihoods for each condition. The following function, nll.1() computes the negative log likelihood for Model 1. It does so by calling individual
condition log likelihood function nll.condition() twice and adding the results. Because Model 1 specifies different parameters for each condition, each
call to nll.condition() has different parameters.
#negative log likelihood for Model 1:
#assign par4=d1,g1,d2,g2
#assign y8=(h1,m1,f1,c1,h2,m2,f2,c2)
nll.1=function(par4,y8)
{
nll.condition(par4[1:2],y8[1:4])+ #condition 1
nll.condition(par4[3:4],y8[5:8])
#condition 2
}
The input to the function are the vector of four parameters (d1 , g1, d2 , g3 )
and the vector of eight data points from the two conditions.
In Model 2, there is a single detection parameter d. The log likelihood
for this model is evaluated similarly to that in Model 1. The difference is
that when nll.condition() is called for each condition, it is done with a
common detection parameter. The input is the vector of three parameters
and eight data points:
#negative log likelihood for Model 2:
#assign par3=d,g1,g2
#assign y8=(h1,m1,f1,c1,h2,m2,f2,c2)
nll.2=function(par3,y8)
{
72
CHAPTER 3. THE HIGH-THRESHOLD MODEL
nll.condition(par3[1:2],y8[1:4])+
nll.condition(par3[c(1,3)],y8[5:8])
}
The negative log likelihood function for Model 3 is given by
#negative log likelihood for Model 3:
#par3=d1,d2,g
#y8=(h1,m1,f1,c1,h2,m2,f2,c2)
nll.3=function(par3,y8)
{
nll.condition(par3[c(1,3)],y8[1:4])+
nll.condition(par3[2:3],y8[5:8])
}
3.3.3
Maximize the Likelihoods
Maximization may be done with the optim call:
dat=c(40,10,30,20,15,35,2,48)
#Model 1
par=c(.5,.5,.5,.5) #starting values
mod1=optim(par,nll.1,y8=dat,hessian=T)
#Model 2
par=c(.5,.5,.5) #starting values
mod2=optim(par,nll.2,y8=dat,hessian=T)
#Model 3
par=c(.5,.5,.5) #starting values
mod3=optim(par,nll.3,y8=dat,hessian=T)
The above code produces a number of warnings: “NaNs produced in
log(x).” These warnings are inconsequential for this application. They come
about because optim does not know that sensitivity and guess parameters
may only be between 0 and 1. In Section ?? we will discuss a solution to this
problem.
0.2
0.4
0.6
0.8
Condition 1
Condition 2
0.0
Parameter Estimates
1.0
3.3. SELECTIVE INFLUENCE IN THE HIGH THRESHOLD MODEL73
Detection (d)
Guessing (g)
Parameter
Figure 3.2: Parameter estimates and standard errors from Models 1 and 3.
Bars are parameter estimates from the general model and points are estimates
from the restricted model.
The output is in variables mod1, mod2, and mod3. There is one element
of the analysis that is of concern. The estimate of dˆ2 for Model 3, given in
mod3 is -.03. This estimate is invalid and we discuss a solution in Section ??.
For now the value of dˆ2 may be set to 0. Figure 3.2 provides an appropriate
graphical representation of the results. It was constructed with barplot and
errbar as discussed in Chapter 2. The bar plots are from Model 1, the
general model. The point between the two bar-plotted detection estimates
is the common detection estimate from Model 2. The point between the two
bar-plotted guessing estimates is the common guessing estimate from Model
3. From these plots, it would seem that the manipulation certainly affected
g. The case for d is more ambiguous, but it seems plausible that d depends on
the payoff, which would violate selective influence and question the veracity
of the model.
3.3.4
Testing Selective Influence
Although Figure 3.2 is informative, it is no substitute for formal hypothesis
tests. The first part of the selective influence test is whether g depended on
condition. The test is performed with a likelihood ratio test. The value of G2
may be computed in R as 2*(mod3$value-mod1$value). The value is 41.28
74
CHAPTER 3. THE HIGH-THRESHOLD MODEL
(with dˆ2 set to 0 the value is 41.37). Under the null-hypothesis that g1 = g2 ,
this value should be distributed as a chi-square. As mentioned previously, the
degrees of freedom for the test is the difference in the number of parameters
in the models, which is 1. The criterial value of the chi-square statistic with
1 degree of freedom is 3.84. Hence, Model 3 can be rejected in favor of Model
1. The payoff manipulation did indeed influence g as hypothesized.
The second part of selective influence is the invariance of d. From Figure 3.2, it is evident that there is a large disparity of sensitivity across the
conditions (.50 vs. .27). This difference appears relatively large given the
standard errors. Yet, the value of G2 (2*(mod2$value-mod1$value)) is 1.26,
which is less than the criterial value of 3.84. Therefore, the invariance cannot
be rejected.
This later finding is somewhat surprising given the relatively large size of
the effect in Figure 3.2. As a quick check of this obtained invariance, it helps
to inspect model predictions. The model predictions for a condition can be
obtained by:
ˆ
p̂h = dˆ + (1 − d)ĝ
p̂f = ĝ.
(3.15)
(3.16)
With these equations, the predictions from Model 1 and Model 2 are
shown in Table 3.3. As can be seen, Model 2 does a fair job at predicting
the data, even though the parameter estimate of d is different than d1 and
d2 in Model 1. This result is evidence for the invariance of d. When there
is a common detection parameter, the ability to predict the data is almost
as good as with condition-specific detection parameters. The lesson learned
is that it may be difficult with nonlinear models to inspect parameter values
with standard errors and decide if they differ significantly.
3.3. SELECTIVE INFLUENCE IN THE HIGH THRESHOLD MODEL75
Condition 1
Data
Model 1 Prediction
Model 2 Prediction
Condition 2
Data
Model 1 Prediction
Model 2 Prediction
Hit Rate
False-Alarm Rate
.800
.800
.750
.600
.600
.640
.300
.300
.322
.040
.040
.038
Table 3.3: Predictions derived from the High Threshold model.
Problem 3.3.1 (Your Turn)
You are testing the validity of the high-threshold model for the perception of faint audio tones with a selective influence test. You manipulate
the volume of the tone through two levels: low and very low. The manipulation is hypothesized to affect d and not g. The obtained data are
given below. Use R to test for selective influence.
Hits Misses
Low
33
17
Very Low 40
10
False Alarms Correct Rejections
42
9
30
20
76
CHAPTER 3. THE HIGH-THRESHOLD MODEL
Problem 3.3.2 (Your Turn)
Let’s gain some insight into why the detection estimates across conditions seem quite different in Figure 3.2 even though they are not
statistically so. One reason for the appearance of difference is that the
standard error bars are drawn symmetrically around the parameter estimate. Let’s see if this is accurate. We ask whether there is skew to the
sampling distribution of the common d estimator in Model 2. Use the
simulation method to construct the sampling distribution of d in Model
2 for true values (d = .3, g1 = .64, g2 = .04) with 50 noise and signal
trials in each of the two conditions. Plot the sampling distribution as
a relative-frequency histogram. Is it skewed? If so, in which direction.
For the purposes of this problem, set negative estimates of g2 to zero.
3.4
Receiver Operating Characteristic
There is a common graphical approach to assessing models in signal detection
experiments. Psychologists typically graph the hit rate as a function of the
false-alarm rate. The resulting plot is called a receiver operating characteristic or ROC plot. Table 3.4 shows data from a hypothetical experiment which
is a test of the selective of payoffs. There are 500 observations per condition.
The resulting hit rates are .81, .70, .57, .50, .30 for Conditions A through E,
respectively. The resulting false-alarm rates are .60, .47, .37, .20, .04 for Conditions A through E, respectively. The ROC plot for these data is shown in
Figure 3.3. There is a point for each condition. The x-axis value is the false
alarm rate, the y-axis value is the hit-rate.
The lines in Figure 3.3 are predictions from the high-threshold model.
Each line corresponds to a particular value of d. The points on the line are
obtained by varying g. The line is the prediction for the case of invariance
of sensitivity and it is called the isosensitivity curve (Luce, 1963). The highthreshold model predicts straight line isosensitivity curves with a slope of
(1 − d) and an intercept of d. The following is the derivation of the result:
77
3.4. RECEIVER OPERATING CHARACTERISTIC
Reward for Correct Response
Signal Trial
Noise Trial
Condition A
10c
1c
Condition B
7c
3c
Condition C
5c
5c
Condition D
3c
7c
Condition E
1c
10c
Data
Hit Miss
404 96
348 152
287 213
251 249
148 352
FA
301
235
183
102
20
CR
199
265
317
398
480
0.8
1.0
Table 3.4: Hypothetical data for a signal detection experiment with payoffs.
A
0.6
B
Hit Rate
d=.6
C
0.4
D
d=.35
0.2
E
0.0
d=.1
0.0
0.2
0.4
0.6
0.8
1.0
False Alarm Rate
Figure 3.3: ROC plot. The points are the data from Table 3.4. Lines denote
predictions of the high-threshold model.
78
CHAPTER 3. THE HIGH-THRESHOLD MODEL
Let i index condition. By the high-threshold model
ph,i = d + (1 − d)gi , pf,i = gi .
Substituting the later equation into the former yields
ph,i = d + (1 − d)pf,i ,
which is a straight-line relationship. Straight-line ROCs are characteristic of
models with all-or-none mental processes.
ROC plots can be made in R with the plot() command. Straight lines,
denoting isosensitivity curves, may be added with the lines() command.
The details are left as part of the exercise below.
Problem 3.4.1 (Your Turn)
Fit the high-threshold model to the data in Table 3.4. Fit a general
model with separate parameters for each condition. The 10 parameters
in this model are (dA , gA , dB , gB , dC , gC , dD , gD , dE , gE ). Fit a common
detection model; the six parameters are (d, gA , gB , gC , gD , gE ).
1. Estimate parameters for both models. Make a graph showing the
detection parameters for each condition (from the general model)
with standard errors.
2. Plot the data as an ROC and add a line denoting the commondetection model.
3. Perform a likelihood ratio test to see if detection varies across
conditions.
3.5
3.5.1
The Double High-Threshold Model
Basic Model
The high-threshold model is useful because it provides separate estimates of
detection and guessing probabilities. It is, however, not the only model to do
79
3.5. THE DOUBLE HIGH-THRESHOLD MODEL
Double High−Threshold Model
Hit
d
Correct Rejection
d
g
Hit
1−d
g
False Alarm
1−d
1−g
Miss
Signal Trials
1−g
Correct Rejection
Noise Trials
Figure 3.4: The double high-threshold model.
so. A closely related alternative is the double high-threshold model. Like the
high-threshold model, the double high-threshold model is also predicated
on all-or-none mental processes. In contrast to the high-threshold model,
however, the double high-threshold model posits that participants may enter
a noise-detection state in which they are sure no signal has been presented.
The model is shown graphically in Figure 3.4. The model is the same as
the high-threshold model for signal trials: either the signal is detected, with
probability d, or not. If the signal is not detected, the participant guesses as
before. On noise trials, participants either detect that the target is absent
(with probability d) or enter a guessing state. Model equations are given by
Yh ∼ Binomial(d + (1 − d)g, Ns )
Yf ∼ Binomial((1 − d)g, Nn),
(3.17)
(3.18)
where 0 < d, g < 1.
Analysis of this model is analogous to the high-threshold model. The log
likelihood is given by:
l(d, g; yh, yf ) = yh log(d + (1 − d)g) + (ym ) log((1 − d)(1 − g)),
+yf log((1 − d)g) + (yc ) log(d + (1 − d)(1 − g)).
Either calculus methods or numerical methods may be used to provide
maximum likelihood estimates. The calculus methods provide the following
estimates:
yh
yf
dˆ =
−
(3.19)
Ns Nn
yf /Nn
.
(3.20)
ĝ =
1 − dˆ
80
CHAPTER 3. THE HIGH-THRESHOLD MODEL
These can be rewritten in terms of hit and false alarm rates:
dˆ = p̂h − p̂f
p̂f
ĝ =
.
1 − dˆ
(3.21)
(3.22)
The estimate dˆ in this case is simply the hit rate minus the false alarm
rate. This estimate is often used as a measure of performance in memory
experiments (e.g., Anderson, Craik, & Naveh-Benjamin, 1998). Equation
(3.19) shows that this measure is may be derived from a double high-threshold
model. Moreover, the validity of the measure may be assessed in a particular
domain via a suitable selective influence test.
Implementation in R is straightforward. The log likelihood is computed
by
#negative log likelihood for double high-threshold model
nll.dht=function(par,y)
{
d=par[1]
g=par[2]
p=1:4 # reserve space
p[1]=d+(1-d)*g #probability of a hit
p[2]=1-p[1] # probability of a miss
p[3]= (1-d)*g# probability of a false alarm
p[4] = 1-p[3] #probability of a correct rejection
return(-sum(y*log(p)))
}
The ROC of the double high-threshold model can be derived by observing
that ph = d + pf . Hence, the ROC is a straight line with y-intercept of d and
a slope of 1.0. Plots of ROCs for a few values of d are shown in Figure 3.5.
Problem 3.5.1 (Your Turn)
Do the analyses in Your Turn 3.4 for the double high-threshold model.
81
3.5. THE DOUBLE HIGH-THRESHOLD MODEL
1.0
0.8
0.6
Hit Rate
d=0.6
0.4
d=0.35
0.2
d=0.1
0.0
0.0
0.2
0.4
0.6
0.8
1.0
False Alarm Rate
Figure 3.5: ROC lines of the double high-threshold model for several values
of d.
3.5.2
Double High-Threshold Model and Overall Accuracy
In the preceding sections, we stressed the importance of differentiating miss
errors from false alarm errors. Some researchers prefer to use overall accuracy
as a measure of performance. Overall accuracy, denoted by c, is
c=
Yh + Yc
.
Ns + Nn
ĉ =
yh + yc
.
Ns + Nn
It may be estimated by
In many experiments, the number of signal and noise trials is equal (N =
Nn = Ns ). In this case, accuracy is given by
c = (Yh + Yc )/2N.
It may be surprising that this measure may be theoretically motivated
by the double high-threshold model. For signal trials, the number of correct
82
CHAPTER 3. THE HIGH-THRESHOLD MODEL
responses is Yh = Ns (d + (1 − d)g); for noise trials, the number of correct
responses is Yc = Nn (d + (1 − d)(1 − g)). Overall accuracy is
c=
Ns (d + (1 − d)g) + Nn (d + (1 − d)(1 − g))
.
Ns + Nn
If Ns = Nn , then the above equation may be reduced to
c=
d 1
+ .
2 2
This relationship indicates that overall accuracy is a simple linear transform
of d for when Nn = Ns . Overall accuracy for this case may be derived from
a double-high threshold model; its validity as a measure is tested by testing
the selective influence of the double-high threshold model.
3.5.3
Process Dissociation Procedure
The most influential double high-threshold model is Jacoby’s process dissociation procedure for the study of human memory (Jacoby, 1991). Currently,
human memory is often conceptualized as consisting of several separate systems or components. Perhaps the most fundamental piece of evidence for
separate components comes from the study of anterograde amnesics. These
patients typically suffer a stroke or a head injury. They have fairly-well preserved memories from before the stroke or injury, but have impairment in
forming new memories. They do poorly in direct tests for the memory of
recent events such as a recognition memory test. They make many more
miss and false alarm errors than appropriate control participants.
Although amnesics are greatly impaired in direct memory tasks such as
recognition memory, they are far less impaired on indirect memory tasks.
One such task is stem completion. In this task participants are shown a list
of words at study as before. At test, they are given a word stem, such as
br
. Their task is to complete the stem with the first word that comes
to mind. Control participants without amnesia typically show a tendency to
complete stems with studied words. For example, if no word starting with
stem br is studied, typical completions are words like bread and brother but
not bromide. If bromide is studied, however, it is far more likely to be used as
the stem completion than otherwise. This type of test is considered indirect
because the experimenter does not ask whether an item was studied or not.
3.5. THE DOUBLE HIGH-THRESHOLD MODEL
83
Instead, the presence of memory is inferred from its ability to indirectly affect
a mental action such as completing the stem with the first word that comes to
mind. Most surprisingly, amnesics have somewhat preserved performance on
indirect tests. While an amnesic may not recall studying a specific word, that
word still has an elevated chance of being used by the amnesic to complete
the stem at test (Graf & Schacter,1985).
This finding, as well as many related ones, have led to the current conceptualization of memory as consisting of two systems or components. One
of these components reflects conscious recollection, which is willful and produces the feeling of explicitly remembering an event. This type of memory is
primarily used in direct tasks. The other component is the automatic, unconscious, residual activation of previously processed or encountered material.
Automatic activation corresponds to the feeling of familiarity but without
explicit memorization. Within this conceptualization, amnesics’ deficit is in
the conscious recollective component but not in the automatic component.
Hence, they tend to have less impairment in tasks that do not require a
conscious recollection. There are a few variations of this dichotomy (e.g.,
Jacoby, 1991; Schacter, 1990; Squire, 1994), but the conscious-automatic one
is influential.
The goal of the process dissociation procedure is to measure the degree
of conscious and automatic processing and we describe its application to the
stem completion task. The task starts with a study phase in which participants are presented a sequence of items. There are two test conditions in
process dissociation: an include condition and an exclude condition. In the
include condition, participants are instructed to complete the stem with a
previously studied word. In the exclude condition, participants are instructed
to complete the stem with any word other than the one studied. In the include condition, stem completion of studied word can occur either through
successful conscious recollection or automatic activation. In the exclude condition, successful conscious recollection does not lead to stem completion
with the studied item, instead it leads to stem completion with a different
item.
The following notation is used to implement the model. Let Ni and
Ne denote the number of words in the include and exclude test conditions,
respectively. Let random variables Yi,s and Yi,n be the frequency of stems
completed with a studied word and a word not studied, respectively, in the
include condition. Let random variables Ye,s and Ye,n denote the same for
words in the exclude condition. It is assumed that recollection is all-or-none
84
CHAPTER 3. THE HIGH-THRESHOLD MODEL
Process Dissociation Procedure
studied word
r
word not studied
r
a
studied word
1−r
a
studied word
1−r
1−a
1−a
word not studied
word not studied
Exclude Trials
Include Trials
Figure 3.6: The process dissociation procedure model.
and occurs with a probability of r. Automatic activation is also all-or-none
and occurs with probability a. Figure 3.6 depict the model. Resulting data
are modeled as:
Yi,s ∼ Binomial(Ni , r + (1 − r)a)
Ye,s ∼ Binomial(Ne , (1 − r)a).
(3.23)
(3.24)
The model is a double-high threshold model where conscious recollection
plays the role of detection and automatic activation plays the role of guessing
(Buchner, Erdfelder, Vaterrodt-Plunneck, 1995). Because process dissociation is a double high-threshold model, all of the previous development may
be used in analysis. Estimators for R and A are given by:
yi,s ye,s
−
Ni
Ne
ye,s /Ne
â =
.
1 − r̂
r̂ =
(3.25)
(3.26)
3.5. THE DOUBLE HIGH-THRESHOLD MODEL
Problem 3.5.2 (Your Turn)
As people age, their memory declines. Let’s use the process dissociation procedure to assess the locus of the decline. The table, below,
shows hypothetical data for younger and elderly adults. The columns
“Studied” and “Not Studied” shows the number of stems completed by
a studied word and a word not studied, respectively. The total number
of stems is the sum of these two numbers. Given these data, test for
the effects of aging on r and a.
Younger
Elderly
Condition
Include
Exclude
Studied Not Studied Studied Not Studied
69
81
37
113
36
64
12
88
85
86
CHAPTER 3. THE HIGH-THRESHOLD MODEL
Chapter 4
The Theory of Signal Detection
The theory of signal detection (Green & Swets, 1966) is the dominant modelbased method of assessing performance in perceptual and cognitive psychology. We describe the model for the tone-in-noise signal detection experiment.
The model, however, is applied more broadly to all sorts of two-choice tasks
in the literature including those in assessing memory. McMillan and Creelman (1991) provide an extensive review of the variations and uses of this
flexible model. It is important to distinguish between the theory of signal
detection and signal-detection experiments. The former is a specific model
like the high-threshold model; the latter is an experimental design with two
stimuli and two responses.
The presentation of the model relies on continuous random variables,
which were not covered in Chapter 1. We first discuss this type of random
variable as well as density, cumulative distribution, and quantile functions.
Then we introduce the normal distribution, upon which the theory of signal
detection is based. After covering this background material in the first half
of the chapter, we present the theory itself in the second half.
4.1
Continuous Random variables
In previous chapters, we proposed models based on the binomial distribution.
One feature of the binomial is that there is only mass on a select number
outcomes. Figure 4.1, top-left, shows the probability mass function for a
binomial with 50 trials (p = .3). Only the points 0,1,...,50 have nonzero
mass. Because mass falls on select discrete points, the binomial is a called
87
CHAPTER 4. THE THEORY OF SIGNAL DETECTION
0.10
B
0.00
0.04
Probability
0.08
0.20
A
0.00
Probability
0.12
88
0
10
20
30
40
50
0
5
15
D
Density
0.005
0.0
0.4
0.8
C
0.010
0.015
10
Number of Events
0.000
Density (1/pounds)
Number of Successes
100
150
200
Weight in Pounds
250
−0.5
0.0
0.5
1.0
Outcome
Figure 4.1: Top: Probability mass functions for discrete random variables.
Bottom: Density functions for continuous random variables.
a discrete distribution. Another example of a discrete distribution is the
Poisson distribution. The top-right panel shows the Poisson probability mass
function. For the Poisson, mass occurs on all positive integers and zero.
There are gaps between points where there is no mass.
In contrast to discrete distributions, continuous distributions have mass
on intervals rather than on discrete points. The best-known continuous distribution is the normal. The bottom-left panel (Figure 4.1) shows a density function of a normal distribution. Here there is density on every single
point. The bottom-right panel shows another example of a continuous random variable—the uniform distribution. Notice that where there is density,
it is on an interval rather than on single points separated by gaps. The density function serves a similar purpose to the probability mass function and is
discussed in detail below.
1.5
4.1. CONTINUOUS RANDOM VARIABLES
4.1.1
89
The Density Function
In discrete distributions, probability mass functions describe the probability
that a random variable takes a specific realization. The density function
is different. At first consideration, it is tempting to interpret density as
a probability. This interpretation, however, is flawed. To see this flaw,
consider density as probability for Figure 4.1D. If density were probability,
then we could ask, ”What is the probability of observing either 1/3 or 2/3?”
The density at each point is 1.0, hence, the sum density for two points is
2.0. Probability for any event can never exceed 1; hence, density cannot be
probability.
Density is interpretable on intervals rather than on individual points. To
compute the probability that a random variable takes a realization within
an interval, we compute the area under the density function on the interval.
Consider again the uniform distribution of Figure 4.1D. Suppose we wish to
know the probability that an observation is between 1/4 and 3/4. The area
under the density function on this interval, which is the probability, is p =
1/2. The uniform is a particularly easy distribution to compute the area on
an interval because the area is that of a rectangle. The relationship between
the density function, area, and probability is formalized in the following
definition:
Definition 33 (Density Function) The density function of a
continuous random variable X is a function such that the area
under the function between a and b corresponds to P r(a < X ≤
b)
.
What is the probability that a continuous random variable takes any single value? It is the area under a single point, which is zero. This condition
makes sense. For example, we can ask what the probability that a person
weighs 170lbs. There are many people who report their weight at 170lbs,
but this report is only an approximation. In fact, very few people worldwide weigh between 169.99lbs and 170.01lbs. Surely almost nobody weighs
between 169.999999lbs and 170.000001lbs. As we decrease the size of the
interval around 170lbs, the probability that anybody’s weight falls in the
90
CHAPTER 4. THE THEORY OF SIGNAL DETECTION
interval becomes smaller. In the limit, nobody can possibly weigh exactly
170lbs. Hence the probability of someone weighing exactly some weight, to
arbitrary precision, is zero.
In order to more fully understand the density function, it is useful to
consider the units of the axes. The units of the x-axis of a density function is
straightforward—it is the units of the measurement of the random variable.
For example, the x-axis of the normal density in Figure 4.1 is in units of
pounds. The units of the y-axis is more subtle. It is found by considering
the units of area. On any graph, the units of area under the curve is given
by:
Units of Area = x-axis units × y-axis units .
(4.1)
The area under the density function corresponds to probability. Probability
is a pure number without any physical unit such as pounds or inches. For
the normal density over weight in Figure 4.1 in which the x-axis units is
pounds, the y-axis unit must be 1/pounds in order for Equation 4.1 to hold.
In general, then, the units of density on the y-axis is the reciprocal of that
of the random variable.
One of the more useful properties of densities is that they describe the
convergence of histograms with increasingly large samples. In Chapter 1 with
discrete RVs, we advocated a relative-frequency histograms because they
converge to the appropriate probability mass function. For continuous RVs,
the appropriate histogram is termed a relative-area histogram. In relativearea histograms, the area of a bin corresponds to the proportion of responses
that fall within the bin. Figure 4.2 (left) shows a relative-area histogram
of 100 realizations from a normal distribution. The bin between 150lbs and
160lbs has a height value .015 (in units of 1/lbs) and an area of .15. Fifteen
percent of the realizations fell in this bin. The advantage of relative-area
histograms is that they converge1 to the density function. The right panel
shows and example of this convergence. There are 20,000 realizations in the
histogram.
1
Convergence involves shrinking the bin size. As the number of realizations grows,
the bins should become smaller, and, in the limit, the bins should become infinitesimally
small.
91
0.015
0.010
0.005
Density (1/pounds)
0.000
0.010
0.000
Density (1/pounds)
4.1. CONTINUOUS RANDOM VARIABLES
100
120
140
160
180
200
220
100
Weight in pounds
150
200
250
Weight in pounds
Figure 4.2: Convergence of relative-area histograms to a density function.
Left: Histogram of 100 observations. Right: Histogram of 20,000 observations.
4.1.2
Cumulative Distribution Functions
The area under the density function for an interval describes the probability
that an outcome will be in that interval. The cumulative distribution function
describes the probability than an outcome will be less than or equal to a point.
Definition 34 (Cumulative Distribution Function (CDF))
Let F denote the cumulative distribution function of random
variable X. Then, F (x) = P r(X ≤ x).
Figure 4.3 shows the relationship between density and cumulative distribution functions for a uniform between 0 and 2. There are two dotted
vertical lines, labeled a and b, are for a = .5 and b = 1.3. The values of
density and CDF are shown in left and right panels, respectively. The area
under the density function to the left of a is .25, and this is the value of the
CDF in the right panel. Likewise, the area under the density function to the
left of b is .65, and this is also graphed in Panel B. Cumulative distribution
functions are limited to the [0, 1] interval and are always increasing.
The cumulative distribution function can be used to compute the probability an observation occurs on the interval [a, b]:
P r(a < X ≤ b) = F (b) − F (a).
(4.2)
92
0.8
a
b
0.0
0.2
0.4
b
0.4
Cumulative Probability
a
0.0
Density
0.6
CHAPTER 4. THE THEORY OF SIGNAL DETECTION
0.0
0.5
1.0
1.5
0.0
2.0
Outcome
0.5
1.0
1.5
2.0
Outcome
Figure 4.3: Density (left) and cumulative distribution function (right) for a
uniform random variable. The cumulative distribution function is the area
under the density function to the left of a value.
The relationship between density and cumulative distribution functions
for continuous variables may be expressed with calculus. We provide them
for students with knowledge of calculus:
F (x) =
Z
τ =x
τ =−∞
f (τ )dτ,
d
F (x).
dx
Cumulative distribution functions are also defined for discrete random
variables. For the binomial model of toast-flipping, the CDF describes the
probability of obtaining x or fewer butter-side-down flips. Figure 4.4 shows
the CDF for 10 flips (with p = .5). A few points deserve comment. First,
some points are open (not filled-in) while others are closed. The associated
value of the function at these points is the closed point. For example, the
CDF at x = 4 is .38 and is not .17. Second, the CDF is defined easily for
fractions, like 3.5, which do not correspond to outcomes. The probability of
observing 3.5 or fewer butter-side-down flips is the same as observing 3 or
fewer butter-side-down flips. This facet accounts for the stair-step characteristic in the graph.
f (x) =
4.1.3
Quantile Function
Quantiles play a major role in some of the psychological models we consider.
The easiest way to explain quantiles is to consider percentiles. The 75th
93
0.8
0.6
0.4
0.2
0.0
Cumulative Probability
1.0
4.1. CONTINUOUS RANDOM VARIABLES
0
2
4
6
8
10
Outcome
Figure 4.4: Cumulative distribution function for a binomial with N = 10
and p = .5.
percentile is the value below which 75% of the distribution lies. For example,
for the uniform in Figure 4.3, the value 1.5 is the 75th percentile because
75% percent of the area is below this point. Quantiles are percentiles for
distributions, except they are indexed by fractions rather than by percentage
points. The .75 quantile corresponds to the 75th percentile.
Definition 35 (Quantile) The pth quantile of a distribution is
the value qp such that P r(X ≤ qp ) = p.
The quantile function takes a probability p and returns the associated pth
quantile for a distribution. The quantile function is the inverse of the cumulative distribution function. Whereas the CDF returns the proportion of mass
below a given point, the quantile function returns the point below which a
given proportion of the mass lies. Examples of density functions, cumulative
distribution functions and quantile functions for three different continuous
distributions are shown in Figure 4.5. The top row is for a uniform between
0 and 2; the middle row is for a normal distribution; the bottom row is for
an exponential distribution. The exponential is a skewed distribution used
to model the time between events such as earthquakes, light bulb failures,
94
CHAPTER 4. THE THEORY OF SIGNAL DETECTION
Quantile Function
Cumulative Distribution Function
2.0
Density Function
0.0
0.5
1.0
1.5
0.0
1.0
Outcome
0.8
0.4
Probability
0.0
0.2
0.0
Density
0.4
Uniform
2.0
0.0
0.5
Outcome
1.0
1.5
2.0
0.0
0.2
Outcome
0.4
0.6
0.8
1.0
0.8
1.0
0.8
1.0
Probability
150
200
200
120
0.0
100
160
Outcome
0.8
0.4
Probability
0.010
0.000
Density
Normal
250
100
Outcome
150
200
250
0.0
0.2
Outcome
0.4
0.6
Probability
0
2
4
6
Outcome
8
10
8
6
0
0.0
0.0
2
4
Outcome
0.8
0.4
Probability
0.2
Density
0.4
Exponential
0
2
4
6
Outcome
8
10
0.0
0.2
0.4
0.6
Probability
Figure 4.5: Density, cumulative probability, and quantile functions for uniform, normal, and an exponential distributions.
or action potential spikes in neurons. The inverse relationship between CDF
and quantile functions is evident in the graph.
Several quantiles have special names. The .25, .50 and .75 quantiles of a
distribution are known as the first, second, and third quartiles respectively,
because they divide the distribution into quarters. The .50 quantile is also
known as the median.
4.1.4
Expected Value of Continuous Random Variables
The following section is based on calculus. It is not critical for understanding
the remaining topics in this book. The expected value of a distribution is its
center, and is also often called the mean of the distribution. In Chapter 1
we defined the expected value of discrete random variables in terms of their
4.1. CONTINUOUS RANDOM VARIABLES
95
probability mass functions, E(X) = x xf (x). This definition is not appropriate for continuous random variables. Instead, expected value is defined
in terms of integrals. The following hold for expected value and variance of
continuous random variables:
P
E(X) =
V(x) =
Z
∞
−∞
Z ∞
−∞
xf (x)dx,
(x − E[X])2 f (x)dx.
Likewise, the expected of a function of a random variable g(X) is given by:
E[g(X)] =
Z
∞
−∞
g(x)f (x)dx.
The other properties of expected value discussed in Chapter 1 hold for
continuous random variables. Most importantly, the expected value can typically be estimated by the sample mean. Therefore, instead of calculating
expected values by evaluating integrals, we can use simulation instead. As
before, we simply draw an appropriately large number of random samples
and then calculate the sample mean.
4.1.5
The Normal Distribution
The normal distribution is the basis of many common inferential tests including t-tests, ANOVA, and regression. The normal has two parameters, µ
and σ 2 . These are called the mean and variance, respectively, because if X is
a normal random variable, it can be shown that E(X) = µ and V(X) = σ 2 .
A concept essential for understanding signal detection is the standard
normal distribution:
Definition 36 (Standard Normal) A standard normal random variable is distributed as a normal with µ = 0 and σ 2 = 1.
The cumulative distribution function and quantile function of the standard
normal is denoted by Φ(x) and Φ−1 (p), respectively.
96
4.1.6
CHAPTER 4. THE THEORY OF SIGNAL DETECTION
Random variables in R
The R package has built-in density, cumulative distribution, and quantile
functions for a large number of distributions. Functions dnorm(), pnorm(),
and qnorm() are the density, cumulative distribution function, and quantile
function of the normal, respectively. Likewise, functions dunif(), punif(),
qunif() are the corresponding R functions for the uniform distribution. The
syntax generalizes: a ’d’ before a random variable name refers to either the
density or a probability mass function (depending on whether the random
variable is continuous or discrete). A ’p’ and a ’q’ before the name refer to
the CDF and quantile functions, respectively. An ’r’ before the name, as in
rnorm() or rbinom(), produces realizations from the random variable. Syntax for these functions can be obtained through help(), e.g.; help(dunif).
Quantile functions are useful in finding criterial values for test statistics.
The chi-squared distribution, for example, describes the distribution of G2
under the null hypothesis. In conventional hypothesis testing, the goal is to
specify the probability of mistakenly rejecting the null hypothesis when it
is true. The probability is called the Type I error rate and is often denoted
as α. In psychology the convention is to set α = .05. This setting directly
determines the criterion of the test statistic. For G2 , we wish to set a criterion
so that when the null is true, we reject it 5% of the time. The situation is
depicted in the left column of Figure ??. The criterion is therefore the
value of a chi-squared distribution below which 95% of the mass lie; i.e.,
the .95 quantile. This value is given by qchisq(.95,df), where df is the
degrees of freedom. Criterial bounds for t-tests, F-tests, and z-tests can be
found with qt(), qf(), and qnorm() , respectively. With the t-tests and
z-tests, researchers are often interested in two-tail alternative hypotheses.
These bounds are provided with .025 and .975 quantiles. An example for
a t-distribution with four degrees of freedom is shown in the right panel of
Figure 4.6.
97
4
4.1. CONTINUOUS RANDOM VARIABLES
qt(.025,4)
0.2
Density
3
95%
0.1
2
1
Density
qt(.975,4)
0.3
qchisq(.95,1)
95%
0
1
2
3
4
0.0
0
5%
5
2.5%
−4
2.5%
−2
0
2
4
Figure 4.6: Criterial bounds for the chi-square distribution with 1 df and the
t distribution with 4 df.
Problem 4.1.1 (Your turn)
1. Use R to plot the density function of a standard normal from -3
to 3.
2. Use R to plot the cumulative distribution function of a standard
normal from -3 to 3.
3. Use the cumulative distribution function to find the median of
the standard normal. Is your answer what you’d expect? Where
is this point on the density function?
4. Use R to plot the quantile function of a standard normal from
.001 to .999. Use abline(v=c(.025,.975)) to put vertical lines
on your plot. What are the y-values where these lines cross the
quantile function? How much probability mass is on the interval
between these lines?
5. Use R to plot a histogram of 10,000 realizations drawn from a
standard normal. How does this compare with your plot from
Problem #1 above?
6. Use these 10,000 realizations to estimate the expected value, median, and variance of the standard normal.
98
CHAPTER 4. THE THEORY OF SIGNAL DETECTION
Tone Absent
Tone Present
0.2
0.0
0.1
Density
0.3
0.4
d’
c
−4
−2
0
2
4
6
Sensory Strength
Figure 4.7: The signal detection model.
4.2
Theory of Signal Detection
It is important to distinguish between a signal detection experiment and the
theory of signal detection: one is a type of experiment the other is a model.
There are many theories applicable signal detection experiment besides the
theory of signal detection including the threshold theories of the previous
chapter.
We describe the theory of signal detection for the tone detection experiment. Participants monitor the input for a tone. The resulting sensation
is assumed to be a random variable called strength. Tone-absent trials tend
to have low strength while tone-present trials tend to higher strength. Hypothetical distributions of strength are shown in Figure 4.7; in the figure
the distribution for tone-present trials has greater strength on average than
that for tone-absent trials. These distributions are modeled as a normal
distributions:
S∼
(
Normal(µ = 0, σ 2 = 1), for tone-absent trials,
Normal(µ = d′ , σ 2 = 1), for tone-present trials.
(4.3)
To make a response, the participant sets a criterial bound on strength. This
bound is denoted by c and is presented as a vertical line in Figure 4.7. If the
strength of a stimulus is larger than c, then the participant responds “tone
present;” otherwise the participant responds “tone absent.”
4.2. THEORY OF SIGNAL DETECTION
99
Analysis begins with model predictions about hit, false-alarm, miss, and
correct-rejection probabilities. Correct rejection probability is the easiest to
derive. Correct rejection events occur when strengths from the tone-absent
distribution are below the criterial bound c. This probability is the CDF of
a standard normal at c, which is denoted as Φ(c).
pc = Φ(c).
(4.4)
The probability of a false alarm is 1 minus probability of a correct rejection.
Hence,
pf = 1 − Φ(c).
(4.5)
The equations for hits and misses are only a bit more complicated. The
probability of a miss is the probability that an observation from a normal
with mean of d′ and variance of 1 is less than c. This can be written as
pm = F (c; µ = d′ , σ 2 = 1). It is more standard, however, to express these
probabilities in relation to the standard normal. This is done by noting that
the probability that an observation from N(d′ , 1) is less than c is the same
as an observation from N(0, 1) is less than c − d′ :
Because ph = 1 − pm ,
pm = Φ(c − d′ ).
(4.6)
ph = 1 − Φ(c − d′ ).
(4.7)
Equations 4.5 through 4.7 describe underlying probabilities, and not data.
The resulting data is distributed as a binomials, e.g.,
Yh ∼ Binomial(ph , Ns )
Yf ∼ Binomial(pf , Nn ).
Substituting in for ph and pm provides the complete specification of the signal
detection model:
Yh ∼ Binomial(1 − Φ(c − d′ ), Ns )
Yf ∼ Binomial(1 − Φ(c), Nn ).
4.2.1
(4.8)
(4.9)
Analysis
In this section, we provide analysis for a single condition. Data are the
numbers of hit, miss, false alarm and correct rejects and denoted with the
100
CHAPTER 4. THE THEORY OF SIGNAL DETECTION
vector y = (yh , ym , yf , yc ). The most common method of deriving estimators
is as follows: Equation 4.5 can be rewritten as c = Φ−1 (1 − pf ). Equation 4.7
can be rewritten as c−d′ = Φ−1 (1−ph ). The standard normal is a symmetric
distribution. Hence, the area below a point x is the same as the area above
point −x. This fact implies that Φ−1 (p) = −Φ−1 (1 − p). Using this fact and
a little algebraic rearrangement, the following hold:
c = −Φ−1 (pf )
d′ = Φ−1 (ph ) − Φ−1 (pf )
Conventional estimators are obtained by using empirically observed hit and
false-alarm rates:
ĉ = −Φ−1 (p̂f )
dˆ′ = Φ−1 (p̂h ) − Φ−1 (p̂f )
(4.10)
(4.11)
Fortunately, these conventional estimators are the maximum likelihood
estimators. Hence for a single condition, it is easiest to simply use the above
estimators rather than numerically maximizing likelihood. These estimators
may be computed in R:
dprime.est=qnorm(hit.rate)-qnorm(fa.rate)
c.est=-qnorm(fa.rate)
Although these estimators are useful in single-condition cases, they are
not useful for testing the invariance of parameters across conditions. For
example, the above method may not be used to estimate a common sensitivity
parameter across two conditions with different bounds. These more realistic
cases may be analyzed with a likelihood approach.
The first step is writing down the log likelihood. The log likelihood for a
binomial serves as a suitable starting point:
l(ph , pf ; yh , yf ) = yh log(ph ) + ym log(pm ) + yf log(pf ) + yc log(pc )
(4.12)
Substituting expressions for the probabilities yields:
l(d′ , c; yh , yf ) = yh log(1−Φ(c−d′ ))+ym log(Φ(c−d′ ))+yf log(1−Φ(c))+yc log(Φ(c))
(4.13)
This log likelihood may be computed in R:
4.2. THEORY OF SIGNAL DETECTION
101
#log likelihood for signal detection
#par=c(d’,c)
#y=c(hit,miss,fa,cr)
ll.sd=function(par,y)
{
p=1:4
p[1]=1-pnorm(par[2],par[1],1)
p[2]=1-p[1]
p[3]=1-pnorm(par[2],0,1)
p[4]=1-p[3]
sum(y*log(p))
}
Let’s estimate d′ and c for data y = (40, 10, 30, 20):
y=c(40,10,30,20)
par=c(1,0) #starting values
optim(par,ll.sd,y=y,maximum=T)
The results are d̂′ = .588, and ĉ = −.253. These estimates match those
obtained from Equations 4.10 and 4.11.
4.2.2
ROC Curves for Signal Detection
In Chapter 3, we described the receiver operating characteristic (ROC) plots.
The points in Figure 4.8A shows the ROC for the hit and false-alarm rate data
provided in Table 3.4. The ROC is useful when an experimental manipulation
is assumed to not affect sensitivity. For this case, different models make
different predictions about the isosensitivity curve. The isosensitivity curve
for the signal detection model for a few values of d′ are shown in Figure 4.8A.
These isosensitivity curves are different than those from the high-threshold
model and double-high threshold model; they are curved rather than straight
lines.
There is an alternative to the ROC plot specifically suited for the signal detection model. The alternative, called a zROC plot, is shown in Figure 4.8B. In this plot, standard normal quantiles of hit and false-alarm rates
(i.e., Φ−1 (p̂h ) and Φ−1 (p̂f )) are plotted rather than the hit and false-alarm
102
2
d=1.5
d=0.75
0.2
−1
0.4
d=0.1
d=0.75
0
z(Hit Rate)
0.6
1
d=1.5
d=0.1
0.0
A
0.0
0.2
0.4
0.6
0.8
B
−2
Hit Rate
0.8
1.0
CHAPTER 4. THE THEORY OF SIGNAL DETECTION
1.0
−2
False Alarm Rate
−1
0
1
z(False Alarm Rate)
Figure 4.8: ROC and zROC plots for the data from Table 3.4. Signal detection model predictions are overlaid as lines.
rates themselves. The main motivation for doing so is that isosensitivity
curves on this plot are straight lines with a slope of 1.0. To see why this is
true, note that Eq. (4.11) can be generalized for i conditions as follows:
d′i = Φ−1 (p̂h,i) − Φ−1 (p̂f,i ).
To derive an isosensitivity curve, we assume each condition has the same
sensitivity; therefore, d′i may be replaced by d′ . Rearranging yields
Φ−1 (p̂h,i) = Φ−1 (p̂f,i ) + d′ .
For notational convenience, let yi = Φ−1 (p̂h,i) and xi = Φ−1 (p̂f,i ). Then,
yi = xi + d′ ,
which is the equation for a straight line with a slope of 1.0 and an intercept
of d′ . If the signal detection model holds and the conditions each have the
same sensitivity, then the zROC points should fall on a straight line with
slope 1.0. The function Φ−1 is also called a z-transform and z-transformed
proportions are also called z-scores.
To draw a zROC, we use qnorm(p), where p is either the hit or false-alarm
rate. The following code plots the zROC for the data in Table 3.4.
2
4.2. THEORY OF SIGNAL DETECTION
103
hit.rate=c(.81,.7,.57,.5,.30)
fa.rate=c(.6,.47,.37,.2,.04)
plot(qnorm(fa.rate),qnorm(hit.rate),ylab="z(Hit Rate)",
xlab="z(False-Alarm Rate)",ylim=c(-2,2),xlim=c(-2,2))
Signal-detection isosensitivity lines can be overlaid on the plot by drawing lines with the abline() function. The syntax for drawing lines with
specified slope and intercept is abline(a=intercept,b=slope). For example, the diagonal with slope of 1 and an intercept of 1.5 is drawn with
abline(a=1.5,b=1) or simply abline(1.5,1).
Problem 4.2.1 (Your Turn)
Fit the signal-detection model to the data in Table 3.4. Fit a general
model with separate parameters for each condition. The 10 parameters
in this model are (d′A , cA , d′B , cB , d′C , cC , d′D , cD , d′E , cE ). Fit a common
sensitivity model; the six parameters are (d′ , cA , cB , cC , cD , cE ).
1. Estimate parameters for both models. Make a graph showing
the sensitivity parameters for each condition (from the general
model) with standard errors.
2. Plot the data as a zROC; add a line denoting the commonsensitivity model.
3. Perform a likelihood ratio test for the common-sensitivity hypothesis.
4. Estimate the common-detection high-threshold model fit for the
data (see Your Turn 3.4). Plot this model’s prediction in the
zROC plot.
104
4.2.3
CHAPTER 4. THE THEORY OF SIGNAL DETECTION
Null Counts
One vexing problem in the analysis of the signal detection model is the
occasional absence of either miss or false-alarm events. When there are no
misses, the hit-rate is p̂h = 1.0. Recall that d̂′ = Φ−1 (p̂h ) − Φ−1 (p̂f ). When
p̂h = 1.0, the term Φ−1 (p̂h ) is infinite (try qnorm(1)), leading to an estimate
of d̂′ = ∞. Likewise, when there are no false alarms, the term Φ−1 (p̂f ) is
negatively infinite (try qnorm(0)), leading again to an estimate of d̂′ = ∞.
An infinite sensitivity estimate is indeed problematic.
The presence of null counts is therefore a problem in need of redress. The
problem occurs in the estimates of ph and pf when there are no misses or no
false alarms. It is reasonable to assume that the underlying true values ph and
pf are never 1.0 and 0.0, respectively. As the number of trials is increased, it is
expected that participants eventually make both false alarm and miss errors.
Accordingly, estimates of p̂h and p̂f should not be 1 and 0, respectively, even
when there are no misses and false alarms. Snodgrass and Corwin (1988)
recommend using the p̂1 estimator we have previously introduced in Chapter
1 (Equation 1.12): p̂1 = (y + .5)/(N + 1). This estimator keeps the estimates
from extremes of 0 or 1. For signal detection, estimates of hit and false-alarm
rates are given by:
yh + .5
pˆh =
,
Ns + 1
yf + .5
.
pˆf =
Nn + 1
This correction can be implemented within the formal likelihood approach by
adding .5 to each observed cell count. For moderate values of probabilities,
it will lead to more efficient estimation (see Figure 1.8).
There is a second correction used in the literature to keep estimates from
being too extreme, called the 1/2N rule (Berkson, 1953):
1
,
2N
y
,
N
y = 0,
0 < y < N,
p̂ =
1
1 − 2N , y = N.
This correction is implemented in R as follows:
p=y/N
p[p==0]=1/(2*N)
p[p==1]=1-1/(2*N)
4.2. THEORY OF SIGNAL DETECTION
105
The above code introduces some new programming elements. Let’s work
through an example with three conditions. Suppose for each condition, there
are 20 observations (N = 20) and the number of successes (hits or false
alarms) is y = (2, 0, 20). The code works as follows: From the first line, p
is a vector with values (.1, 0, 1). The second line of code is more complex.
Consider first the term p==0. The symbol == tests each term for equality
with 0, and so this line returns a vector of true and false values. In this case,
the second element is true, because p[2] does equal 0. The left-hand side,
p[p==0] refers to all of those elements in which the term within the brackets
is true, i.e., all those in which p does indeed equal zero. These elements
are replaced with the value of 1/2N. The third line operates analogously–it
replaces all estimated proportions of 1.0 with the value 1−1/2N. Hautus and
Lee (1998) provide further discussion of the properties of these estimators.
106
CHAPTER 4. THE THEORY OF SIGNAL DETECTION
Problem 4.2.2 (Your Turn)
In application, it matters little whether a Snodgrass-Corwin or a Berkson correction is used. Let’s compare them using R.
1. An experimenter runs a signal detection experiment with 20 signal and noise trials. The true value of d′ and c are 1 and .5,
respectively. Simulate the sampling distributions of d̂′ and ĉ for
both correction methods using 100,000 replicate experiments (if
your computer is old and slow, then perhaps 10,000 replicates is
more appropriate). Plot these as histograms (there are four separate histograms obtained by the combining the 2 parameters (d′ ,
c) with the two correction methods).
2. Estimate the bias and RMSE for sensitivity of each correction
method. Which of these is more efficient? One convenient method
of comparing the two corrections is to compute a ratio of RMSE
for the two methods. Let the numerator be the RMSE from the
Snodgrass-Corwin correction and the denominator be the RMSE
from the Berkson correction. With this convention numbers less
than 1.0 indicate better efficiency for the Snodgrass-Corwin correction; numbers greater than 1.0 indicate the reverse.
3. Of course, true values are not limited to (d′ = 1, c = .5). Try
your code for true values (d′ = 1.5, c = .2).
4. Let’s explore the corrections for a range of true values. Let
true d′ = (.2, .4, .., 2.8). For each of these values, let c =
(0, .1d′ , .2d′, .., .9d′ , d′). Compute the efficiency ratio of the two
methods. There should be 154 different RMSE ratios. Use
the contour function to plot these. This method provides as
assessment of the relative efficiency between the two methods
for a wide range of parameter values. As alternative use the
filled.contour function. Also try these plots for logarithm of
the efficiency ratio. This quantity has the advantage of being
positive when efficiency favors one of the correction methods and
negative when it favors the other. Hint: Use loops through true
values in your R code.
107
4.3. ERROR BARS FOR ROC PLOTS
Condition A
Condition B
Condition C
Reward for Correct Response
Signal Trial
Noise Trial
10c
1c
5c
5c
1c
10c
Data
Hit Miss
82
18
68
36
48
52
FA
62
44
28
CR
36
56
72
Table 4.1: Hypothetical data for a signal-detection experiment with payoffs.
4.3
Error bars for ROC plots
The previously-drawn ROC plots lack error bars. As mentioned previously,
standard errors are a rough guide to the variability in parameter estimates.
In this section, we consider methods of using error bars for ROC plots.
Data points in ROC graphs are composed of hit and false-alarm rates.
These rates are estimates of the true hit and false-alarm probabilities. The
following equation may be used for computing standard errors for probability
estimates from data distributed as a binomial:
SE(p̂) =
s
p(1 − p)
.
N
Figure 4.9A shows an example of an ROC plot with standard errors. For
each point, there is one error bar in the vertical direction indicating the
standard error of the hit-rate estimate and one in the horizontal direction
indicating the standard error of the false-alarm rate estimate. The ROC plot
comes from the hypothetical data in Table 4.1, which serves as a convenient
example in demonstrating how to draw these error bars in R.
The novel element in these plots are horizontal error bars which are drawn
with the following code:
horiz.errbar=function(x,y,height,width,lty=1)
{
arrows(x,y,x+width,y,angle=90,length=height,lty=lty)
arrows(x,y,x-width,y,angle=90,length=height,lty=lty)
}
The following code uses horiz.errorbar to draw the standard errors.
1.0
CHAPTER 4. THE THEORY OF SIGNAL DETECTION
A
0.5
B
0.0
d’=0.56
−1.0
−0.5
z(Hit Rate)
0.6
0.4
0.2
0.0
Hit Rate
0.8
1.0
108
0.0
0.2
0.4
0.6
0.8
1.0
False Alarm Rate
−1.0
−0.5
0.0
0.5
z(False Alarm Rate)
Figure 4.9: A: ROC plot for the data from Table 4.1 with standard errors on
hit and false-alarm rates. The black line represents the isosensitivity curve
for the value of d′ obtained by maximum likelihood. B: zROC plot for the
same data with standard errors on sensitivity estimate. The solid line is the
isosensitivity curve for the best-fitting signal detection model. The dotted
lines are standard errors on sensitivity.
hit=c(82,68,48)
fa=c(62,44,28)
N=100
hit.rate=hit/N
fa.rate=fa/N
std.err.hits=sqrt(hit.rate*(1-hit.rate)/N)
std.err.fa=sqrt(fa.rate*(1-fa.rate)/N)
#plot ROC
plot(fa.rate,hit.rate,xlim=c(0,1),ylim=c(0,1))
errbar(fa.rate,hit.rate,height=std.err.hits,width=.05)
horiz.errbar(fa.rate,hit.rate,height=.05,width=std.err.fa)
An alternative approach is to draw standard errors on more substantive
parameters. Figure 4.9B provides an example for the zROC curve. The
fitted model is a signal detection model with a common sensitivity estimate
1.0
4.3. ERROR BARS FOR ROC PLOTS
109
for all three conditions. The resulting isosensitivity curve is the solid line.
Standard errors were derived from optim with the option hessian=T. For the
data in Table 4.1, the estimate of sensitivity is 0.56 and its standard error is
0.077. These standard errors are plotted as parallel isosensitivity curves and
are denoted with dotted lines. The code for drawing these lines is
dprime.est= .56
# as derived from an optim call
std.err= .077
#as derived from an optim call
plot(qnorm(fa.rate),qnorm(hit.rate))
abline(a=dprime.est,b=1) #isosensitivity curve
abline(a=dprime.est+std.err,b=1,lty=2) #plus standard error
abline(a=dprime.est-std.err,b=1,lty=2) #minus standard error
These two approaches to placing standard errors should not be used on
the same plot. Placing standard errors on both hits and false alarms and on
substantive parameters is often confusing and we recommend using only one
of these approaches. A good rule-of-thumb is that standard errors should be
placed on hit and false-alarm rates when several different models are being
compared, whereas standard errors should be placed on model parameters
when a specific hypothesis is being tested.
Problem 4.3.1 (Your Turn)
Using the hypothetical data in Table 3.4, create two plots:
1. Create an ROC plot with standard errors on hit and false alarms.
2. Create a zROC plot with standard errors on d′ .
These plots should resemble Figure 4.9.
110
CHAPTER 4. THE THEORY OF SIGNAL DETECTION
Chapter 5
Advanced Threshold and
Signal-Detection Models
In the previous two chapters we discussed basic models of task performance.
Unfortunately, there are many examples in the literature in which these basic
models fail to account for empirically observed ROC functions (e.g., Luce,
1963; Ratcliff, Sheu, & Grondlund, 1993). In this chapter, we expand our coverage to more flexible and powerful models. The first model we discuss is the
general high-threshold which is a generalization of both the high-threshold
and double high-threshold model. The second model is a generalization of
the signal detection model. The final model, Luce’s low-threshold model,
introduces the idea that people can perceive stimuli that are not present.
The models introduced in the previous chapter have two basic parameters:
one for detection or sensitivity and another for guessing or response bias.
The models introduced in this chapter have three basic parameters: two for
sensitivity and a third for response bias. Three-parameter models cannot
be fit to a single condition of a signal-detection experiment because each
condition provides only two independent observations: the numbers of hits
and false alarms. Instead, these models are estimated with observations from
many conditions. In order to demonstrate analysis, we consider the payoff
experiment example of Table 3.4. In this hypothetical experiment, the stimuli
are constant across conditions, hence parameters describing sensitivity should
not vary. Models that are consistent with invariance of sensitivity parameters
are appropriate whereas those that are inconsistent are not.
Although these new models are more flexible, they present two problems
in analysis. First, the methods we have used for numerically minimizing
111
112CHAPTER 5. ADVANCED THRESHOLD AND SIGNAL-DETECTION MODELS
the likelihood fail for these models. They do not return parameters that
maximize likelihood. Second, the previously introduced techniques for model
comparison are insufficient for the three new models. None of the new models
is a restriction of another; e.g., the models are not nested. The likelihood
ratio statistic, while appropriate for nested models, is not appropriate for
non-nested ones. We discuss an alternative method, based on the Akaike
Information Criterion (AIC, Akaike, 1974), to make these comparisons. The
remainder of the chapter is divided into three sections: a discussion of more
advanced numerical techniques for improving optimization, a discussion of
the three new models, and a discussion of nonnested model comparisons.
5.1
5.1.1
Improving Optimization
The Problem
The likelihoods of the models we consider in this chapter cannot be maximized with the methods of the previous chapters without modification. The
problem is that with default settings, the optim() function often fails to
find the maximum in problems with many parameters. To demonstrate this
failure , we start with a simple optimization problem. Suppose we have a set
of observations y1 , ..., yN and a set of parameters θ1 , ..., θN . We wish to find
the best values of θ1 ...θN that minimize the following function h:
h=
n
X
i=1
(yi − θi )2 .
The function is a sum-of-squared-differences formula. It is at a minimum
when all the differences are zero; i.e., when θ1 = y1 , θ2 = y2 , ..., θn = yn . If
θi = yi, then the sum of squared differences is h = 0. Function optim() does
a good job of finding this minimum for a handful of parameters. The following
code serves as an example. It minimizes h with respect to parameters θ1 , .., θ4
for four pieces of data: y1 = 3, y2 = 8, y3 = 13, y4 = 18.
h=function(theta,y) return(sum((theta-y)^2))
y=c(3,8,13,18)
par=rep(10,4) #starting values
optim(par,h,y=y)
5.1. IMPROVING OPTIMIZATION
113
The results are:
$par
[1] 3.000866 8.000700 13.000482 17.999283
$value
[1] 1.986393e-06
As expected, the minimum of h is very close to h = 0 and parameter values
are nearly equal to their respective data points.
The results are not so good for twenty parameters. Consider the following
code:
y=1:20 #integers from 1 to 20
par=rep(10,20) #starting values
optim(par,h,y=y)
Results are:
$par
[1] 0.3535526 1.7091429 4.1538558 3.0865141 5.8710791 5.2499118
[7] 7.8665955 8.8864199 7.8365200 9.2342382 11.6387186 13.8285510
[13] 13.6197244 13.8850712 12.5633528 15.3360254 16.7935952 16.6734207
[19] 19.3004116 19.8297374
$value
[1] 19.91514
These results are troubling as the estimates are surprisingly far from their
true values, and the function minimizes to a value of 19.9 instead of 0.
This example demonstrates that the optim() function with default settings is unable to handle problems with more than a few parameters. The
above case demonstrates that optim() is not foolproof. In the following sections, we explore a few strategies to increase the accuracy of optimization.
5.1.2
Nested Optimization
One good approach to increasing the accuracy of optimization is to frame
the analysis so that it involves several separate optimizations with each being
over a smaller number of parameters. Consider the above example with the
function h and twenty parameters. Notice that the value of θ1 that minimizes
114CHAPTER 5. ADVANCED THRESHOLD AND SIGNAL-DETECTION MODELS
h is a function of only y1 and not of the other nineteen data points. This fact
holds analogously for the other parameters as well—the appropriate value of
each parameter depends on a single data point. Consider the following code
for minimization that takes advantage of this fact:
h=function(theta,obs) (theta-obs)^2 #to-be-minimized function
y=1:20
par.est=rep(0,20) #reserve space
min=0
for (i in 1:20)
{
est=optimize(h,interval=c(-100,100),obs=y[i])
min=min+est$objective
par.est[i]=est$minimum #store the estimates
}
In this code, we call optimize twenty times from within the loop. Each pass
through the loop optimizes a single parameter. The resulting parameters
values are stored in vector par.est. The minimum of the function is stored
in min. The results are that the estimated values (par.est) are nearly the
true values and the function minimizes to nearly 0. For this case, performing
20 one-parameter optimizations is far more accurate than performing a single
twenty-parameter optimization.
This strategy of framing analysis so that it involves multiple optimizations
with smaller numbers of parameters is often natural in psychological contexts.
Consider the analysis of the high-threshold model for payoffs data (Table 3.4)
as an example. The model reflecting selective influence has six parameters:
(d, gA , gB , gC , gD , gE ). We start by assuming the true value of d. Of course,
this assumption is unwarranted. We only use the assumption to get started
and will soon dispense of it during estimation. If the true value of d is known,
then estimation of gA only depends on data in Condition A, estimation of gB
only depends on the data of Condition B and so on. This fact leads naturally
to multiple optimization calls. The first step is to compute the likelihood for
g in a single condition given a fixed value of d:
#negative log likelihood for high-threshold model
#one condition, function of g for given d
#y=c(hit,mis,fa,cr) for one condition
5.1. IMPROVING OPTIMIZATION
115
nll.ht.given.d=function(g,y,d)
{
p=1:4
# reserve space
p[1]=d+(1-d)*g #probability of a hit
p[2]=1-p[1]
# probability of a miss
p[3]=g
# probability of a false alarm
p[4] = 1-p[3]
#probability of a correct rejection
return(-sum(y*log(p)))
}
The body of the function is identical to that in Section 3.2.3; the difference
is how the parameters are passed. The function in Section 3.2.3 is minimized
with respect to two parameters. The current function will be minimized with
respect to g alone. The maximum log likelihood for all five conditions for a
known value of d is:
#d is detection parameter
#dat is hA,mA,faA,cA,...hE,mE,faE,cE
nll.ht=function(d,dat)
{
return(
optimize(nll.ht.given.d,interval=c(0,1),y=dat[1:4],d=d)$objective+
optimize(nll.ht.given.d,interval=c(0,1),y=dat[5:8],d=d)$objective+
optimize(nll.ht.given.d,interval=c(0,1),y=dat[9:12],d=d)$objective+
optimize(nll.ht.given.d,interval=c(0,1),y=dat[13:16],d=d)$objective+
optimize(nll.ht.given.d,interval=c(0,1),y=dat[17:20],d=d)$objective
)
}
The function nll.ht can be called for any value of d, and when it is called, it
performs five one-parameter optimizations. Of course, we wish to estimate d
rather than assume it. This can be done by optimizing nll.ht with respect
to d. Here is the code for the data in Table 3.4:
dat=c(404,96,301,199,348,152,235,265,
287,213,183,317,251,249,102,398,148,352,20,480)
g=optimize(nll.ht,interval=c(0,1),dat=dat)
116CHAPTER 5. ADVANCED THRESHOLD AND SIGNAL-DETECTION MODELS
The result of this last optimization provides the minimum of the negative log
likelihood as well as the ML estimate of d. The minimum here is 2901.941,
which is probably lower than the negative log likelihood you found for the
restricted model in Your Turn 3.4.1 (depending on the starting values you
chose). To find the ML estimates of the guessing parameters, each of the
optimization statements in nll.ht may be called with the ML estimate of d.
We call this approach nested optimization. Optimization of parameters
specific to conditions (gA , gB , .., gE ) is nested within parameters common
across all conditions (d). Overall nested optimization often provides for more
accurate results than a large single optimization. The disadvantage of nested
optimization is that it does not immediately provide standard error estimates
for parameters. One strategy is to do two separate optimizations. The first
of these is with nested optimizations. Afterward, the obtained ML estimates
can be used as starting values for a single optimization of all parameters with
a single optim() call. In this case, optim() should return these starting values as parameter estimates. In addition, it will return the Hessian which can
be used to estimate standard errors as discussed in Chapter 2.
Problem 5.1.1 (Your Turn)
1. Use nested optimization to estimate parameters of the highthreshold model for multiple conditions with a common detection
parameter. The code in this section provides a partial solution–it
returns common detection parameter d. It does not return guessing parameters (gA , gB , ..., gE ). You will need to modify the code
to return these estimates. Test your code with the data of Table 3.4 and compare the results with those found in Your Turn
4.2.1. Estimate standard errors for the six parameters.
2. Use nested optimization to estimate the signal detection model
with common d′ . Estimate all six parameters and their standard
errors.
117
5.1. IMPROVING OPTIMIZATION
5.1.3
Parameter Transformations
When optimizing log likelihood for the high-threshold and double high-threshold
models in Chapter 3, you may have noticed that R reported some warnings.
The warnings reflect a mismatch between the models and the minimization.
In the models, parameters are probabilities and must be constrained between zero and one. The default algorithm in optim(), however, assumes
that parameters can take on any real value. In the course of optimization,
simplex may try to evaluate the log likelihood for an invalid value, for example, d = −.5. When this happens, the logarithms are undefined and optim()
reports the condition as a warning and goes on optimizing.
There are two disadvantages to having this mismatch. The first is that
optim() may return an invalid parameter value. For example, in Chapter
3 we presented a high-threshold model with a common guessing parameter
(Model 3, page 68). Unfortunately, the obtained ML estimate of d2 was
negative. We previously ignored this transgression, but that is not an ideal
solution. The second disadvantage is that having a mismatch often results in
longer and less accurate optimization. It takes time to evaluate functions with
invalid parameter values and many such calls will lead to inaccurate results.
Parameter transformations are a general and easy-to-implement solution.
The basic idea is to construct a mapping from all real values into valid
ones. Figure 5.1 provides an example for the high-threshold model. The
x-axis shows all real values; the y-axis shows the valid values between zero
and one. We allow optim() algorithm to choose any value of the x axis, and
term this value z. For example, in optimizing the high-threshold model, the
algorithm might choose to try a value z = −1. Instead of trying to evaluate
the log likelihood with this value, we evaluate the log likelihood with the
value on the y axis, in this case, .27.
The function in Figure 5.1 is
1
.
(5.1)
1 + e−z
This particular transform is the logit or log-odds transform of a probability
parameter. The inverse mapping from p to z is given by:
p=
!
p
z = log
.
1−p
(5.2)
These two equations are built into R. Equation 5.1 may be evaluated with
the plogis() function; Equation 5.2 may be evaluated with the qlogis()
0.0
0.2
0.4
p
0.6
0.8
1.0
118CHAPTER 5. ADVANCED THRESHOLD AND SIGNAL-DETECTION MODELS
−6
−4
−2
0
2
4
6
z
Figure 5.1: The logit transform.
function. Here is an example of using this transform for the high-threshold
model. We pass the vector z, the unconstrained parameter values to function
and then transform them to valid values with plogis().
# negative log likelihood, high-threshold model
#par ranges across all reals
nll=function(z,y)
{
par=plogis(z) #transform to [0,1]
d=par[1]
g=par[2]
p=1:4 # reserve space
p[1]=d+(1-d)*g #probability of a hit
p[2]=1-p[1] # probability of a miss
p[3]=g # probability of a false alarm
p[4] = 1-p[3] #probability of a correct rejection
return(-sum(y*log(p)))
}
This function may be called as usual:
y=c(75,25,30,20)
z=c(qlogis(.5),qlogis(.5)) #ranges from -infty to infty
5.1. IMPROVING OPTIMIZATION
119
results=optim(z,snll,y=y)
plogis(results$par)
Function optim() evaluates the log likelihood function with various values
of z that are free to vary across the reals. The results are for z, which ranges
across all reals. To interpret these values, they should also be transformed
to probabilities; this is done with the plogis(result$par) statement.
Rerun the code from Chapter 3 and then run the above code for comparison. The parameter estimates are almost identical. Look at the the $counts
returned by optim(). It is lower for the current code (45 evaluations) than
for that in Chapter 3 (129 evaluations). When we transformed parameters, R
required only one-third the evaluation calls, saving time in the optimization
process.
Problem 5.1.2 (Your Turn)
In our discussion of the high-threshold model (Chapter 3), we presented
a model we called “Model 3” with a common guessing parameter. Unfortunately, the obtained ML estimate of d2 was negative. Use the
transformed-parameters strategy to re-estimate this model. Compare
the results with those presented in Chapter 3.
5.1.4
Convergence
Optimization routines work by repeatedly evaluating the function until a
minimum is found. By default optim() continues to search for better parameter values until one of two conditions is met: 1. new iterations do not
lower the value of the to-be-minimized function much or 2. a maximum
number of iterations occurs. If the optim() reaches this maximum number
of iterations, then it is said to have not converged and optim() returns a
value of 1 for the $convergence field. If the algorithm stops before this
maximum number of iterations because new iterations do not lower the function value much, then the algorithm is said to have converged and a value of
0 is returned in the convergence field. The maximum number of iterations
defaults to 500 for the default algorithm in optim().
120CHAPTER 5. ADVANCED THRESHOLD AND SIGNAL-DETECTION MODELS
Convergence should be monitored. If convergence is not obtained, it is
possible that the parameter estimates found are not the maximum likelihood estimates. Sometimes the brute force method of raising the maximum
number of iterations is effective. The maxit option in optim() controls this
maximum. Let’s consider the previous example in which a single optim()
P
2
call is used to minimize the function h = 20
i=1 (yi − θi ) . To increase the
maximum number of iterations, the following call is made:
y=1:20
par=rep(10,20)
optim(par,h,control=list(maxit=10000),y=y) #takes a while
We achieve convergence and the estimates are much better than for 500 iterations (although the function minimizes to .08 instead of zero). A related
approach for convergence is to run the algorithm repeatedly. For each repetition, the previous parameter values serve as the new starting values. For
example,
par=rep(10,20)
a1=optim(par,h,control=list(maxit=10000),y=y)
a2=optim(a1$par,h,control=list(maxit=10000),y=y)
The results of the second call are reasonable (the function minimizes to .006
instead of zero).
The advantage of these brute-force approaches of raising the number of
function evaluations is that they are trivially easy to implement. The disadvantage is that they often are not as effective as nesting optimization and
transforming parameters.
5.1.5
An alternative algorithm
The default algorithm in optim() is simplex (Mead and Nelder, 1967), which
is known for its versatility and robustness (e.g., Press et al., 1992). The simplex algorithm, however, is one of many algorithms for function optimization1
1
The scope of this book precludes a discussion on the theory behind and comparisons
between optimization methods, The interested reader is referred to Press et al., 1992 and
Nocedal and Wright (1999).
5.1. IMPROVING OPTIMIZATION
121
We have used simplex within optim() because it is known to work moderately well for problems with constraints, such as those in which parameters
are constrained to be between zero and one. There are other choices in R
and the function nlm is often useful. Suppose we wish to minimize function
P
h = i (yi − θi )2 for 200 parameters rather than 4 or 20. This is impossible
with a single simplex call, yet it works quickly with a single nlm() call:
y=1:200
par=rep(100,200)
optim(par,h,y=y)
nlm(h,par,y=y)
The outputted estimates are accurate and the function h is minimized to
zero. The syntax of nlm() closely resembles that in optimize() and may be
seen with the statement help(nlm). Function nlm returns fields $value and
$estimates, the minimized value and estimates, respectively. Field $code
provides diagnostics which are covered in help(nlm). Function nlm() also
returns the Hessian for standard errors.
Function nlm() has some drawbacks. It often fails when parameters are
constrained. It cannot be used, for example, with the high-threshold model
without parameter transformation. We have found that it sometimes fails
even after parameters are transformed. When nlm() fails, it tends to return
implausible parameter estimates. This is an advantage as the poor quality of
estimation is easily detected. One effective optimization strategy is to combine optim() (with simplex) and nlm(). First run optim() to get somewhat
close to ML values, and then run nlm() to find the true minimum.
5.1.6
Caveats
Optimization is far from foolproof. The lack of an all-purpose, sure-fire numerical optimizer is perhaps the most significant drawback to the numerical
approach we advocate. As a result, it is incumbent on the researcher to
use numerical optimization with care and wisdom. We recommend that a
researcher consider additional safeguards to understand the quality of their
optimizations. Here are a few:
• Repeat optimization with different starting points. It is a good
idea to repeat optimization from a number of different starting values.
Hopefully, many starting values lead to the same minimum.
122CHAPTER 5. ADVANCED THRESHOLD AND SIGNAL-DETECTION MODELS
• Fix some parameters. Once a minimization is found, it is often easy
to fix some of the parameters and re-fit the remaining ones as free.
For example, in the high-threshold model, we can fix detection to the
estimated value of d and re-fit a model in which guessing parameters
are free to vary. If the original optimization is good, the resulting
parameter estimates for guessing should be identical in the original fit
and in the re-fit.
• Examine simulated data. Once a minimization has been found, the
parameter estimates can then be used to generate a large data set. This
new, artificial data set can then be minimized. The difference between
the parameter estimates from the empirical data and those from the
artificial data should not only be small, but should decrease as the
sample size of the artificial data is increased.
5.2
General High-Threshold Model
With the discussion of optimization complete, we return to models of psychological process. The first of the three models presented in this chapter is
the general high-threshold model (Figure 5.2). The three parameters are the
sensitivity to signal (ds ), the sensitivity to noise (dn ), and the guessing bias
(g). The equations for hits and false alarms are
ph = ds + (1 − ds )g,
pf = (1 − dn )g
(5.3)
(5.4)
This model is a generalization of both the high-threshold model and
the double high-threshold model. It reduces to the high-threshold model
if dn = 0, and it reduces to a double high-threshold model if ds = dn . The
isosensitivity curve on the ROC plot is obtained by varying g while keeping
ds and dn constant. The predicted curve is a straight line with y-intercept
at ds and slope of (1 − ds )/(1 − dn ). Two examples of isosensitivity curves
are drawn in Figure 5.3A. In this chapter, we fit all of the models to the
hypothetical data from the payoff experiment in Table 3.4. The model we fit
has selective invariance of the detection parameters dn and ds . There are a
total of seven parameters (ds , dn , gA , gB , gC , gD , gE ) across the five conditions.
It is helpful to fit the model with the nested optimization strategy. First,
log likelihood of g is expressed for known detection parameters (ds , dn ).
123
5.2. GENERAL HIGH-THRESHOLD MODEL
Double High−Threshold Model
Hit
ds
g
Correct Rejection
dn
Hit
False Alarm
g
1−d s
1−d n
1−g
1−g
Miss
Signal Trials
Correct Rejection
Noise Trials
0.0
0.4
0.8
False Alarms
0.8
2
0.4
1
0.0
Hits
0.4
2
1
Hits
0.8
2
0.0
Hits
1
0.0
0.4
0.8
Figure 5.2: The general high-threshold model.
0.0
0.4
0.8
False Alarms
0.0
0.4
0.8
False Alarms
Figure 5.3: Isosensitivity curves for three models. A: General high-threshold
model. Lines 1 and 2 have detection parameters (ds = .5, dn = .3) and (ds =
.7, dn = .15), respectively. The former is closer to a double high-threshold
model while the later is closer to a high-threshold model. B: Unequal-variance
signal detection model. Lines 1 and 2 have parameters (d′ = 1, σ = 1.4) and
(d′ = .25, σ = .8), respectively. C: Low-threshold model. Lines 1 and 2 have
detection parameters (ds = .6, dn = .3) and (ds = .8, dn = .15), respectively.
124CHAPTER 5. ADVANCED THRESHOLD AND SIGNAL-DETECTION MODELS
#negative log likelihood for general threshold model, 1 condition
#det=c(ds,dn)
#y=c(hit,miss,fa,cr)
#g=guessing
nll.ght.1=function(g,y,det)
{
ds=det[1]
dn=det[2]
p=1:4 # reserve space
p[1]=ds+(1-ds)*g #probability of a hit
p[2]=1-p[1] # probability of a miss
p[3]=(1-dn)*g # probability of a false alarm
p[4] = 1-p[3] #probability of a correct rejection
return(-sum(y*log(p)))
}
We find the best value of g for each condition with optimize(). Because
optimize() works well on restricted intervals such as [0, 1], there is no reason
to transform these guessing parameters. The likelihood across all conditions
is obtained by adding the likelihood from each condition. Note the use of
transformed detection parameters.
#log likelihood of general high-threshold model
#dat=c(h1,m1,f1,c1,...,h5,m5,f5,c5)
#zdet=logit-transformed detection parameters
nll.ght=function(zdet,dat)
{
det=plogis(zdet)
return(
optimize(nll.ght.1,interval=c(0,1),y=dat[1:4],det=det)$objective+
optimize(nll.ght.1,interval=c(0,1),y=dat[5:8],det=det)$objective+
optimize(nll.ght.1,interval=c(0,1),y=dat[9:12],det=det)$objective+
optimize(nll.ght.1,interval=c(0,1),y=dat[13:16],det=det)$objective+
optimize(nll.ght.1,interval=c(0,1),y=dat[17:20],det=det)$objective
)
}
5.3. SIGNAL DETECTION WITH UNEQUAL VARIANCE
125
Next, the log likelihood for the model is maximized by finding the appropriate detection parameters (ds , dn ). Both nlm() and optim() work well with
transformed detection parameters in this application.
dat=c(404,96,301,199,348,152,235,265,
287,213,183,317,251,249,102,398,148,352,20,480)
zdet=rep(0,2) #starting values of zdet
est=optim(zdet,nll.ght,dat)
plogis(zdet$par)
We will discuss the results after introducing the remaining models.
5.3
Signal Detection with Unequal Variance
The signal detection model in the previous chapter assumed that the variance
of the tone-present strength distribution was the same as the tone-absent
strength distribution. A more general model is one in which these variances
are not assumed equal. The variance of the tone-absent distribution is still set
to 1, but the variance of the tone-present distribution is free and denoted as
σ 2 . The model is called the free-variance signal-detection model. Figure 5.4
provides a graphical representation.
The probability of hits and false alarms are:
pf = 1 − Φ(c),
ph = 1 − F (c, d′, σ 2 ),
(5.5)
(5.6)
where F (x, µ, σ 2 ) is the CDF for a normal with parameter (µ, σ 2). It is
conventional to rewrite this equation in terms of the CDF for the standard
normal: F (x, µ, σ 2) = Φ([x − µ]/σ). Therefore,
c − d′
.
ph = 1 − Φ
σ
!
(5.7)
Even though it is more conventional to express the model in terms of the
standard normal, it is more convenient not to do so in the R implementation.
The model reduces to the equal-variance signal detection model if σ 2 = 1.
The isosensitivity ROC curve is obtained by varying c while keeping d′ and
σ 2 constant. The predicted curve is curvilinear with points though the
126CHAPTER 5. ADVANCED THRESHOLD AND SIGNAL-DETECTION MODELS
0.2
σ
0.0
0.1
Density
0.3
0.4
d’
−4
−2
bound
4
6
8
Sensory Strength
Figure 5.4: The free-variance signal detection model.
origin and (1,1); two examples of isosensitivity curves are drawn in Figure 5.3B. For the five conditions in Table 3.4, the model has 7 parameters
(d′ , σ 2 , c1 , c2 , c3 , c4 , c5 ). Evaluation of the log likelihood for known d′ and σ 2
in a single condition may be evaluated in R:
#negative log likelihood of free-variance signal detection model
#par=c(d,sigma)
#y=c(hits,misses,fa’s,cr’s)
nll.fvsd.1=function(c,y,par)
{
d=par[1]
sigma=par[2]
p=1:4 # reserve space
p[1]=1-pnorm(c,d,sigma) #probability of a hit
p[2]=1-p[1] # probability of a miss
p[3]=1-pnorm(c) # probability of a false alarm
p[4] = 1-p[3] #probability of a correct rejection
return(-sum(y*log(p)))
5.4. LOW-THRESHOLD MODEL
127
}
Problem 5.3.1 (Your Turn)
Fit the free variance signal detection model to the data in Table 3.4.
Use a common d′ , a common σ 2 , and a separate c parameter for each
condition. Be sure to use the nested optimization strategy. Do not use
transformed parameters as the parameters of the free-variance signaldetection model are not restricted. Functions optim() and nlm()
should give identical answers.
5.4
Low-Threshold Model
Luce (1963) proposed a qualitatively different model for the detection of tones
than the general high-threshold or signal-detection model. He noted that
empirically obtained ROC curves for auditory detection were not straight
lines, and hence inconsistent with the high-threshold model. Yet, based on
other experiments in which participants needed to both detect tones and
then identify their frequency, Luce concluded that decisions were based on
all-or-none representations of information. Luce proposed the low-threshold
model as a threshold model that does not predict straight-lined ROCs. This
model is similar to the general high-threshold model in that perception is
assumed all-or-none. In the general high-threshold model, it is assumed that
either the correct stimulus is detected or that the participant guesses. In the
low-threshold model, by contrast, it is assumed that people can misperceive
stimuli—that is they can detect a tone’s presence even when it is absent.
When the tone is presented, the participant either detects the tone or
detects its absence. There is no guessing in this model. The probability that
the participant detects the tone is ds and dn for signal and noise stimuli,
respectively. Parameters ds and dn are sensitivity parameters and do not
reflect bias from manipulations such as payoffs. Of courses, responses do
depend on payoffs and an additional specification is needed. The simplest
128CHAPTER 5. ADVANCED THRESHOLD AND SIGNAL-DETECTION MODELS
model is to propose that responses are biased by an amount b:
ph = ds + b,
pf = dn + b.
In this case, there is a bias toward “signal present” responses if b is positive
and one toward “signal absent” responses if b is negative. This model, however, has a logical flaw. To see this, consider the effect of Condition E, in
which participants earn 10c for every correct rejection but only 1c for every
hit. In this case, both hit and false-alarm rates should be quite low and b
should be quite negative. If dn is less than b, then the predicted false alarm
rate will be negative. For example, if b = .3 and dn = .1, then the predicted
false-alarm rate is -.2.
In Luce’s low-threshold model, bias is a relative fraction rather than an
absolute amount. Suppose ds is ds = .7. The largest conceivable positive
bias is .3; any more bias would lead to a hit rate greater than 1. The bias
parameter in Luce’s model indexes the fraction of this largest effect. For
example, if the bias is b = .1, the effect is 10% of the largest conceivable
bias amount. For the case ds =, 7, the hit rate is .7+(.1)(.3)=.73. For the
same example, let’s suppose the dn is .2. The largest conceivable amount of
positive bias is .8; any more would result in false alarm rates above 1.0. If
b = .1, then the false alarm rate is .2+(.1)(.8)=.28. The hit and false-alarm
probabilities are
ph =
(
pf =
(
ds + (1 − ds )b 0 ≤ b ≤ 1
ds + ds b
−1 ≤ b < 0
dn + (1 − dn )b 0 ≤ b ≤ 1
dn + dn b
−1 ≤ b < 0
(5.8)
(5.9)
The relative bias, b, varies between −1 and 1.
Figure 5.3C shows isosensitivity predictions for this model. These are
obtained by keeping dn and ds constant while varying the bias parameter b.
The isosensitivity curve consists of two straight lines: one from the origin to
point (dn , ds ) and the other from point (dn , ds ) to the point (1, 1). The line
starting at the origin is termed the lower limb and results from b < 0. The
line ending at (1, 1) is termed the upper limb and results from b > 0. The
point (dn , ds ) is obtained when b = 0.
To implement the low-threshold model in R, it is convenient to first define
the following function that describes the amount of bias as a function of p
and b.
5.4. LOW-THRESHOLD MODEL
129
amount.of.bias=function(b,p)
ifelse(b>0,(1-p)*b,p*b)
The ifelse function has been introduced earlier (p.xx). If b > 0 then the
second argument ((1-p)*b) is evaluated and returned; otherwise the third
argument (p*b) is evaluated and returned.
With this function, it is straightforward to implement a function that
evaluates the log likelihood for a single condition. The following is the log
likelihood of b for known detection parameters (ds , dn ).
#low-threshold model
#det=c(ds,dn)
#y=c(hits,misses,fa’s,cr’s)
nll.lt.1=function(b,y,det)
{
ds=det[1]
dn=det[2]
p=1:4 # reserve space
p[1]=ds+amount.of.bias(b,ds) #probability of a hit
p[2]=1-p[1] # probability of a miss
p[3]=dn+amount.of.bias(b,dn) # probability of a false alarm
p[4] = 1-p[3] #probability of a correct rejection
return(-sum(y*log(p)))
}
Problem 5.4.1 (Your Turn)
Fit the low-threshold model to the data in Table 3.4. Be sure to use
nested optimization. Also, be sure to use transformed parameters for
ds and dn .
130CHAPTER 5. ADVANCED THRESHOLD AND SIGNAL-DETECTION MODELS
Binomial Model
logL=−32.57
G2=3.99
logL=−34.57
G2=19.66*
High−Threshold
logL=−44.40
AIC=100.80
AIC=85.15
G2=8.25*
G2=8.45*
Low−Threshold
General
High−Threshold
AIC=83.13
logL=−36.70
10 Parameters
AIC=87.40
Free−Variance
Signal Detection
logL=−36.80
G2=3.30
AIC=87.60
G2=22.47*
Double
High−Threshold
logL=−36.21
7 Parameters
AIC=84.43
Equal−Variance
Signal Detection
logL=−48.03
6 Parameters
AIC=108.07
Figure 5.5: A hierarchy of seven models for the payoff experiment in Table 3.4. Included are model comparison statistics G2 , log likelihood, and
AIC.
5.5
Nested-Model and AIC Analyses
We have now discussed seven different models of the payoff experiment of
Table 3.4. These models are expressed as a tree in Figure 5.5. At the top
level is a binomial model with a separate ph and pf probability parameter
for each condition. There are ten free parameters for the five conditions.
This model is the most general and all other models are nested within it. At
the next level are the general high-threshold model, the free-variance signal
detection model, and the low-threshold model. Each of these models has
seven parameters. At the bottom level are six-parameter models: the highthreshold model, the double-high threshold model, and the signal detection
model. The lines between models depict nesting relationships; e.g., the double high-threshold model is nested within the general high-threshold model
but not within the free-variance signal detection model.
The data are plotted as points in an ROC plot in Figure 5.6A. The lines
are the best-fitting isosensitivity curves of the three high threshold models.
Figure 5.6B presents the same data; the lines are best-fitting isosensitivity
curves of the two signal detection models and the low-threshold model. It
is obvious that there are poor fits for the high-threshold model and equal-
131
1.0
0.6
0.0
0.2
0.4
Hits
0.6
0.4
0.2
0.0
Hits
B
0.8
A
0.8
1.0
5.5. NESTED-MODEL AND AIC ANALYSES
0.0
0.2
0.4
0.6
False Alarms
0.8
1.0
0.0
0.2
0.4
0.6
0.8
False Alarms
Figure 5.6: ROC plots of the data and model predictions. Left: Isosensitivity
predictions of the generalized high-threshold, high-threshold, and doublehigh threshold models. Right: Isosensitivity predictions of the low-threshold,
free-variance signal detection, and equal-variance signal detection models.
Standard errors on hit and false alarm rates are never bigger than .022.
variance signal detection model. It is difficult to draw further conclusions
from inspection.
Comparison of nested models may be made with the log likelihood ratio test, as discussed in previous chapters. Values of G2 between nested
models are indicated in the Figure 5.5 for these comparisons. Those values
with asterisks are significant indicating the restricted model is inappropriate. According to this analysis, the only appropriate models are the general
high-threshold model and the double high-threshold model. Neither of these
restrictions can be rejected from their more general model.
For this application, it is not necessary to compare across non-nested
models to decide which is the most parsimonious. In general, however, it is
often helpful to make such comparisons. There are a number of approaches
discussed in the modern statistics literature. We describe the Akaike Information Criteria (AIC) approach (Akaike, 1973) because it has been recommended in psychology (e.g., Ashby and Ells, 2003) and is convenient in a
likelihood framework. There are other approaches to comparing non-nested
1.0
132CHAPTER 5. ADVANCED THRESHOLD AND SIGNAL-DETECTION MODELS
models including Bayesian information criteria (BIC, Schwartz, 1990) and
Bayes factor (Raftery & Kass, 1995; Myung & Pitts, 1997). These other
methods are more complex and are outside the scope of this book.
The AIC measure for a model is:
AIC = −2 log L(θ∗ ) + 2M,
(5.10)
where L is the likelihood of the model, θ∗ are the MLEs of the parameters,
and M is the number of parameters. The lower the AIC, the better the
model fit. The model with the lowest AIC measure is selected as the most
parsimonious. AIC measures for the seven models are shown in Figure 5.5.
The general high-threshold model is the most parsimonious, followed by the
double-high threshold model and the binomial model.
Consider the AIC measure for two models that have the same number
of parameters. In this case, the model with the lower AIC score is the one
with the higher log likelihood. This is a reasonable boundary condition. Log
likelihood values, however, are insufficient when two models have different
numbers of parameters. As discussed in Chapter 2, models with more parameters tend to have higher log likelihoods. For example, the binomial model
with ten parameters will always have higher likelihood than any of its restrictions simply because it has a greater number of parameters. The AIC
measure accounts for the number of parameters by penalizing models with
more parameters. For each additional parameter, the AIC score is raised by
2 points (this value of 2 is not arbitrary; it is derived from statistical theory).
Because of this penalty, the AIC score for the general high-threshold model
is lower than that of the binomial model, even though the latter has greater
log likelihood.
The AIC and likelihood ratio test analyses concord fairly well in that
they both favor the general high-threshold and double-high threshold model
over the other competitors. They appear to disagree, however, on which of
these two is the most appropriate. According to the likelihood ratio test, the
double high-threshold model may not be rejected in favor of the general highthreshold model. In contrast, according to AIC, the general high-threshold
model is preferred over the double high-threshold model. The disagreement
is more apparent than real because the analyses have different logical bases.
The likelihood ratio test is vested in the logic of a null hypothesis testing.
In this case, the double high-threshold restriction (ds = dn ) serves as the
null hypothesis and we do not have sufficient evidence to reject it at the .05
5.5. NESTED-MODEL AND AIC ANALYSES
133
level. We do have some evidence, however, that it is not fitting perfectly
well. The expected value of the chi-square with one degree-of-freedom is
1.0. The obtained value of G2 is 3.3, is reasonably large. While it is not
sufficiently great to reject the restriction at the .05 level, it is at the .1
level (try 1-pchisq(3.3,1)). The AIC measure reflects this information and
accords an advantage to the general high-threshold model. A statement that
reconciles these two approaches is that while the evidence favors the general
high-threshold model, it is not sufficient to reject the double high-threshold
model restriction.
134CHAPTER 5. ADVANCED THRESHOLD AND SIGNAL-DETECTION MODELS
Chapter 6
Multinomial Models
In the previous chapters, we focused on paradigms in which participants were
presented two response options. Here, we consider paradigms in which participants are presented more than two options. One example is a confidenceratings task. In this task, participants indicate their confidence in their
judgments by choosing one of several options such as, “I have low confidence.” The binomial is not the appropriate distribution for this paradigm
because confidence ratings span several values. Instead, we use the multinomial distribution, a generalization of the binomial suitable for more than two
responses. After presenting the multinomial, we present several psychological models: the signal detection model of confidence, Rouder & Batchelder’s
storage-retrieval model (Rouder & Batchelder, 1998), Luce’s similarity choice
model (Luce, 1963b), multidimensional scaling models (Shepard, Romney, &
Nerlove, 1967), and Nosofsky’s generalized context model (Nosofsky, 1986).
Each of these models may be implemented as substantive restrictions on the
general multinomial model.
6.1
Multinomial distribution
The confidence-ratings task provides a suitable paradigm for presenting the
multinomial distribution. In the simplest case, participants are presented
one of two stimuli, such as tones embedded in noise or noise alone. The
participant chooses from a set of options as indicated in Table 6.1. The
table also provides hypothetical data. The multinomial model is applicable
to the results from a particular stimulus. The top row of Table 6.1 shows
135
136
CHAPTER 6. MULTINOMIAL MODELS
Response
Tone Absent
Stimulus High Medium Low
Tone in Noise
3
7
14
Noise Alone
12
16
21
Tone Present
Low Medium High
24
32
12
15
10
6
Total
92
80
Table 6.1: Hypothetical data from confidence ratings task.
data from the tone-in-noise stimuli. The outcomes of N tone-in-noise trials
may be described by values y1 , y2, ..., yI where I is the number of options
and yi is the number trials for which the ith option was chosen. For the top
row of Table 6.1, I = 6, y1 = 3, y2 = 7, y3 = 14, y4 = 24, y5 = 32, and
y6 = 12. The total sum of all counts must equal the number of trials N; i.e.,
P
i yi = N. We let random variable Yi denote the frequency counts for each
category. After data are obtained, values yi are realizations of Yi . Because N
is always known, one of the counts may be calculated from the others: e.g.,
P
yI = N − I−1
i=1 yi . For Table 6.1, there are five independent pieces of data
for each stimulus.
In Chapter 2, we introduced the concept of a joint probability mass function. The joint probability mass function of random variables X and Y was
f (x, y) = P r(X = x and Y = y). The multinomial distribution is a joint
distribution over random variables Y1 , Y2 , ..., YI . It has i probability parameters p1 , .., pI ; where pi denotes the probability that the response on a trial
is the ith option. The multinomial probability mass function is
k
N! Y
f (y1 , y2 , .., yI ) = Qk
pyi i
i=1 yi ! i=1
(6.1)
where denotes the product of the terms, and is analogous to Σ for sums.
P
On each trial, one of the response options must be chosen; hence, i pi = 1.
Consequently, one of the probability parameters may be calculated from the
others. There are, therefore, I − 1 free parameters in the distribution. When
a sequence of random variables Y1 , ..YI are distributed in this manner, we
write,
Y1 , .., YI ∼ Multinomial(p1 , .., pI , N),
(6.2)
Q
where N, as defined above, is the total number of trials.
6.2. SIGNAL DETECTION MODEL OF A CONFIDENCE-RATING TASK137
Problem 6.1.1 (Your Turn)
Show that if I = 2, the pmf of a multinomial distribution is the same
as that of a binomial distribution.
The likelihood of the parameters given the data is obtained by rewriting
the joint pmf as a function of parameters:
Log likelihood is
k
N! Y
pyi i .
L(p1 , .., pI ; y1 , ..., yI ) = Qk
y
!
i=1 i i=1
l(p1 , .., pI ; y1 , ..., yI ) = log N! −
X
i
log yi ! +
X
yi log pi .
(6.3)
i
The terms in log N! and − i log yi are not dependent on the parameters and
P
may be omitted in analysis. The remaining term, i yi log pi , is identical to
the critical term in the log likelihood of the binomial. Maximum likelihood
estimates of pi can be obtained by numerical minimization or by calculus
methods. The calculus methods yield:
P
p̂i =
6.2
yi
.
N
(6.4)
Signal Detection Model of a ConfidenceRating Task
In the previous chapters, we assumed that payoffs affect response parameters
and not perceptual ones. By varying payoff conditions, it was possible to
construct a ROC plot of the data and overlay isosensitivity predictions of
various models. The advantage of the payoff method is that it may be used
to test many different models simultaneously (see Figure 5.5). There is a
second, more common method for achieving the same aim: ask participants
to rate their confidence. This method is somewhat easier and often less costly
to implement.
138
6.2.1
CHAPTER 6. MULTINOMIAL MODELS
The model
It is straightforward to adapt a signal-detection model for confidence-ratings
data. According to the theory of signal detection, stimuli give rise to perceptual strengths. Responses are then determined by a decision bound. For
the two-choice paradigm, there is one decision bound. Strengths below the
bound produce a tone-absent response; strengths above it produce a tonepresent response. Confidence-rating data is accounted for by positing I − 1
bounds. These bounds are denoted c1 , c2 , ..cI−1 with c1 ≤ c2 ≤ ... ≤ cI−1 .
The model with these bounds is shown in Figure 6.1. Strengths below c1
result in a “Tone absent with High Confidence” response; strengths between
c1 and c2 result in a “Tone Absent with Medium Confidence” response, and
so on.
6.2.2
Analysis: General Multinomial Model
In order to analyze this model, we start with a general multinomial model.
We then implement the confidence-ratings signal detection model as a nested
restriction on the multinomial probability parameters. The data from a
confidence-rating experiment can be denoted as Yi,j where i refers to the
response category and j refers to the stimulus. Stimuli are either signal-innoise (j = s) or noise-alone (j = n). Responses are the confidence categories
(i = 1, 2, .., I). The data in Table 6.1 serve as an example.
For the general model, the data are modeled with a pair of multinomial
distributions:
(Y1,s , .., yI,s) ∼ M(p1,s , .., pI,s , Ns )
(Y1,n , .., yI,n) ∼ M(p1,n , .., pI,n , Nn ),
(6.5)
(6.6)
In this model, the Ns and Nn are the number of noise and signal trials,
respectively. Parameters pi,j are the probability of the ith response to the j th
P
P
stimuli and are subject to the restrictions i pi,s = 1 and i pi,n = 1. ML
estimates for pij are analogous to those for the binomial and are p̂i,j = Yi,j /Nj .
Calculation is straightforward and an example for the data of Table 6.1 is:
y.signal=c(3,7,14,24,32,12)
y.noise=c(12,16,21,15,10,6)
NS=sum(dat.signal)
NN=sum(dat.noise)
6.2. SIGNAL DETECTION MODEL OF A CONFIDENCE-RATING TASK139
c3
c2
c5
c4
0.6
c1
High
confidence
Medium
confidence
Tone Present
Low
confidence
Low
confidence
Medium
confidence
High
confidence
0.3
0.2
0.1
0.0
Density
0.4
0.5
Tone Absent
−2
0
2
4
6
Strength
Figure 6.1: The signal detection model for confidence ratings. The left and
right distributions represent the noise and signal-plus-noise distributions, respectively. The five bounds, c1 , .., c5 , divide up the strengths into six intervals
from “Tone absent with high confidence” to “Tone present with high confidence.”
140
CHAPTER 6. MULTINOMIAL MODELS
parest.signal=dat.signal/NS
parest.noise=dat.noise/NN
Although parameter estimates for the general multinomial models are
straightforward, it is useful in analyzing the signal detection model to express
the log likelihood for the multinomial model. The first step is to express the
likelihood for a single stimulus:
#negative log likelihood for one stimulus
nll.mult.1=function(p,y) -sum(y*log(p))
We assume independence holds across stimuli; hence, the overall log likelihood for both stimuli in the general model is
nll.general=nll.mult.1(parest.signal,dat.signal)+
nll.mult.1(parest.noise,dat.noise)
6.2.3
Analysis: The Signal Detection Restriction
Signal detection model for confidence ratings may be implemented as restrictions on multinomial probabilities. We describe here the case for the
free-variance signal-detection model. In this model, the probability of an ith
response is the probability that an observation is in the interval [ci−1 , ci ] (with
c0 defined as −∞ and cI defined as +∞ for convenience). This probability
is simply the difference in cumulative distribution functions (see Eq. 4.2).
Hence:
pi,j = F (ci ; µj , σj2 ) − Fj (ci−1 ; µj , σj2 ),
(6.7)
where F is the CDF of the normal distribution with mean µj and variance σj2 .
The values of µj and σj2 reflect the stimulus. For noise trials, these values are
set to 0 and 1, respectively. For signal trials, however, these values are free
parameters d′ and σ 2 , respectively. The following R function, sdprob.1(),
computes the values of pi,j for any µj , σj2 , and vector of bounds.
sdprob.1=function(mean,sd,bounds)
{
cumulative=c(0,pnorm(bounds,mean,sd),1)
p.ij=diff(cumulative)
6.2. SIGNAL DETECTION MODEL OF A CONFIDENCE-RATING TASK141
return(p.ij)
}
To understand the code, consider the case in which the stimulus is noise
alone (standard normal) and the bounds are (−2, −1, 0, 1, 2). The vector
cumulative is assigned values (0, .05, .16, .5, .84, .95, 1). The middle five values are the areas under the normal density function to the left of each of the
five bounds. Eq. 6.7 describes the area between the bounds, which are the
successive differences between these cumulative values. These differences are
conveniently obtained by the diff(), function.
Now that we can compute model-based probabilities for a single condition,
it is fairly straightforward to estimate d′ , σ, and the bounds. We provide code
that computes the (negative) log likelihood of (d′, σ 2 , c1 , .., c5 ) and leave it to
the reader to optimize it. The first step is to write a function that returns
response probabilities for both stimuli as a function of model parameters:
nll.sigdet=function(par,y)
#negative log likelihood of free-variance signal detection
#for confidence interval paradigm
#par=d,sigma,bounds
#y=y_(1,s),..,y_(I,s),y_(1,n),..,y_(I,n)
{
I=length(y)/2
d=par[1]
sigma=par[2]
bounds=par[3:length(par)]
p.noise=sd.prob.1(0,1,bounds)
p.signal=sd.prob.1(d,sigma,bounds)
nll.signal=nll.mult.1(p.signal,y[1:I])
nll.noise=nll.mult.1(p.noise,y[(I+1):(2*I)
return(-nll.signal-nll.noise)
}
142
CHAPTER 6. MULTINOMIAL MODELS
Problem 6.2.1 (Your Turn)
Decide whether the free-variance signal detection model is appropriate
for the data in Table 6.1 by comparing it with the general multinomial
model with a nested likelihood ratio test.
6.3
Multinomial Process Tree Models
Batchelder and Reifer (1999; Riefer & Batchelder, 1988) have advocated a
class of multinomial models, called multinomial process tree models (MPT),
for many applications in perception and cognition. The formal definition1
of a MPT model is provided by Batchelder and Riefer (1999) and Hu and
Batchelder (1994); we provide a more informal definition and a detailed example. Informally, MPT models are models with the following three properties: (1) latent processes are assumed to be all-or-none, (2) processing at one
stage is contingent on the results of the previous stage, and (3) the models
may be expressed as “tree” diagrams. The high-threshold model meets these
properties: First, the psychological processes of detection and guessing are
all-or-none. Second, processes are contingent—guessing occurs contingent
on a detection failure. Third, the tree-structure of the model is evident in
Figure 3.1.
6.3.1
Storage-retrieval model
Rouder and Batchelder (1998) present a MPT model for separating storage
and retrieval effects in a bizarre-imagery task. Storage is the process of
forming memory traces of to-be-remembered items. Retrieval, in contrast, is
the process of recovering those traces for output. It is well known that bizarre
material, such as a dog riding a bicycle, is easier to recall than comparable
common material, such as a dog chasing a bicycle. Rouder and Batchelder
1
Hu and Batchelder (1994) describe technical restrictions on MPT models that are
often but not always met in practice. The benefit of considering these restrictions is that
if they are met, then a certain statistical algorithm, the expectation-maximization (EM)
algorithm (Dempster, Laird, and Rubin, 1977) is guaranteed to find the true maximum
likelihood estimates (Hu & Batchelder, 1994). A discussion of the restrictions and of the
EM algorithm are outside the scope of this book.
6.3. MULTINOMIAL PROCESS TREE MODELS
Event
E1
E2
E3
E4
E5
E6
Free-Recall Result
both words free recalled
one word free recalled
no words free recalled
two words free recalled
one word free recalled
no words free recalled
143
Cued-Recall Result
correct cued recall
correct cued recall
correct cued recall
incorrect cued recall
incorrect cued recall
incorrect cued recall.
Table 6.2: Possible results for each word pair in the storage-retrieval experiment (Riefer & Rouder, 1992).
asked whether the mnemonic benefit of bizarre material was in storage or in
retrieval processes.
The data we analyze comes from Riefer and Rouder (1992) who presented
participants with sentences such as: “The DOG rode the BICYCLE.” Participants were asked to imagine the event described in the sentence. After a
delay, they were asked to perform two different memory tests. The first test
was a free-recall test in which participants were given a blank piece of paper
and asked to write down as many capitalized words as they can remember.
After the free-recall test, participants were given a cued-recall test. Here, the
first capitalized word of the sentence was given (e.g., DOG) and participants
were asked to write down the other capitalized word (e.g., BICYCLE). For
each sentence, the free-recall test result is scored on the number of words of a
pair recalled (either 0, 1, or 2); the cued-recall test is scored as either correct
or incorrect. The result of both tests is given in Table 6.2
Riefer and Rouder (1992) argued that whereas storage is necessary for
both free recall and cued recall, retrieval is not as needed for cued recall as it
is for free recall. The cue aids retrieval of the second capitalized word allowing
for successful recall even when the participant was unable to retrieve the first
word or association on their own. Rouder and Batchelder (1998) embedded
this argument in a MPT model shown in Figure 6.2. Each cognitive process
is assumed to be all-or-none. The first branch is for associative storage,
which entails the storage of both capitalized words and their association.
Associative storage is sufficient and necessary for cued recall. If associative
storage is successful, then the participant has the opportunity to retrieve the
association and items and does so successfully with probability r. Associative
storage and retrieval are sufficient for free recall of both items and correct
cued recall. If retrieval of the association fails, participants can retrieve each
144
CHAPTER 6. MULTINOMIAL MODELS
E1
r
s
s
1−r
1−s E2
s
a
1−s
1−s
1−a
u
E1
E2
E3
u
E4
1−u
E5
u
E5
1−u
1−u
E6
Figure 6.2: The Rouder and Batchelder storage-retrieval model for bizarre
imagery. See Table 6.2 for description of events E1 , .., E6 .
item independently as a singleton with probability s. If associative storage
fails, the participants still may store and retrieve each item as a singleton
with probability u.
The equations for the model may be derived from Figure 6.2. Probabilities
on a path multiply. For example, the probability of an E3 event is P r(E3 ) =
a(1 − r)(1 − s)2 . When there are two paths to an event, the probabilities of
the paths add. For example, there are two ways to get an E1 event: either
the association is stored and retrieved (with probability ar) or associative
retrieval fails and both items as retrieved as singletons (with probability
a(1 − r)s2 ). Adding these up yields P r(E1 ) = ar + a(1 − r)s2 . The full set
of equations is:
P r(E1)
P r(E2)
P r(E3)
P r(E4)
P r(E5)
P r(E6)
=
=
=
=
=
=
ar + a(1 − r)s2 ,
a(1 − r)[2s(1 − s)],
a(1 − r)(1 − s)2 ,
(1 − a)u2 ,
(1 − a)[2u(1 − u)],
(1 − a)(1 − u)2 .
6.3. MULTINOMIAL PROCESS TREE MODELS
6.3.2
145
Analysis
Estimating parameters of the model is relatively straightforward. The following code computes the log likelihood for the model. Because all of the
parameters are bounded on the (0, 1) interval, we passed logistic transformed
variables.
#Storage Retrieval Model (negative log likelihood)
#y=#E1, #E2,..#E6
#par=c(a,r,s,u) logit transformed
nll.sr.1=function(par,y)
{
a=plogis(par[1]) #transform variables to 0,1
r=plogis(par[2])
s=plogis(par[3])
u=plogis(par[4])
p=1:6
p[1]=a*(r+(1-r)*s^2)
p[2]=2*a*(1-r)*s*(1-s)
p[3]=a*(1-r)*(1-s)^2
p[4]=(1-a)*u^2
p[5]=2*(1-a)*u*(1-u)
p[6]=(1-a)*(1-u)^2
-sum(y*log(p))
}
146
CHAPTER 6. MULTINOMIAL MODELS
Problem 6.3.1 (Your Turn)
Riefer and Rouder (1992) report the following frequencies for bizarre
and common sentences, respectively.
Condition
Bizarre
Common
E1 E2
103 2
80
0
Event
E3 E4
46 0
65 3
E5
7
9
E6
22
2
1. Estimate parameters for the bizarre condition. Be sure to use parameter transformations. Hint: use optim() to get approximate
estimates. Use these estimates as starting values for nlm() to get
true ML estimates.
2. Do the same for the the common condition.
3. Test the hypothesis that the bizarreness effect is a storage advantage. The log likelihood of the general model is simply the sum of
the log likelihoods across the conditions. Use nested optimization
for the restricted model.
4. Test the hypothesis that the bizarreness effect is a retrieval advantage. Use nested optimization for the restricted model.
6.4
6.4.1
Similarity Choice Model
The Choice Axiom
The study of how people choose between competing alternatives is relevant
to diverse fields such as decision-making, marketing, and perceptual psychology. Psychologists seek common decision-making processes when describing
choices as diverse as diagnosing diseases or deciding between ordering apple
and orange juice. Luce’s choice axiom (1957) and similarity choice model
147
6.4. SIMILARITY CHOICE MODEL
Q. Which of the following is brewed and served at
The Flatbranch Brewery in Columbia, Missouri?
FOUR CHOICES
A) Ant Eater Lager
B) Oil Change Stout
C) Tiger Ale
D) Chancellor’s Reserve
BELIEFS
A) .20
B) .19
C) .60
D) .01
TWO CHOICES
B) Oil Change Stout
D) Chancellor’s Reserve
REVISED BELIEFS
B) .95
D) .05
Figure 6.3: An example of the choice axiom.
(1963) have had a large impact in all of these fields.
The choice axiom is explained in the context of the following example (
Figure 6.3). Suppose a contestant in a game show is asked a question about
a local beer in a small Midwestern city. The contestant is unsure of the
correct answer but is able to assign probability values to the four choices
as indicated in the figure. In particular, the contestant believes choice (2)
is nineteen times as likely to be correct as choice (4). The game show host
then eliminates choices (1) and (3). According to the choice axiom, the ratio
between choices (2) and (4) should be retained whether choices (1) and (3)
are available or not. If this 19:1 ratio is preserved, the probabilities in the
two-choice case becomes .95 and .05, respectively.
The choice axiom is also an instantiation of the law of conditional probability. The law of conditional probabilities provides an ideal means of updating
probabilities when conditions change and states that probabilities are nor-
148
CHAPTER 6. MULTINOMIAL MODELS
malized by the available choices. For the example in Figure 6.3, let pi and
p∗i denote the belief in the ith alternative before and after two choices are
eliminated. According to the law of conditional probability:
p∗i =
pi
,
p2 + p4
i = 2, 4.
(6.8)
The correct answer for this question is (2). When probabilities follow the
law of conditional probability, we say the decision maker properly conditions
on events.
6.4.2
Similarity Choice Model
In this section we implement a choice-axiom-derived model, the similarity
choice model (Luce, 1963), for the case of letter identification. Under normal
viewing conditions, we rarely mistakenly identify letters. In order to study
how people identify letters, it is helpful to degrade the viewing conditions
so that people make mistakes. One way of doing this is to present a letter
briefly and follow it with a pattern mask consisting of “###.” If the letter is
presented sufficiently quickly, performance will reflect guessing. As the stimulus duration is increased, performance increases until it reaches near-perfect
levels. Of interest to psychologists is how letters are confused when performance is intermediate. These patterns of confusion provide a means of telling
how similar letters are to each other. As discussed subsequently, measures
of similarity can be used to infer the mental representation of stimuli.
Several authors have proposed that participants use the choice axiom in
decision making (e.g., Clark, 1957; Luce, 1959; and Shepard, 1957). The
choice axiom, however, may fail for the trivial reason of response bias. For
example, a participant may favor the first response option presented (or
the one presented on the left if options are presented simultaneously). The
Similarity Choice Model (SCM) adds response biases to the choice axiom.
The probability of response j to stimulus i is
ηi,j βj
.
k=1 ηj,k βk
pi,j = PJ
(6.9)
In this model, ηi,j describes the similarity between stimuli i and j, and
βj describes the bias toward the jth response. Similarities range between
0 and 1. The similarity between any item and itself is 1. To make the
6.4. SIMILARITY CHOICE MODEL
149
model identifiable it is typically assumed that similarity is symmetric, e.g.,
ηi,j = ηj,i . For example in letter identification, the similarity of a to b is the
same as b to a. The assumption of similarity is not without critics (Tversky,
1979) and is dispensed with in some models (e.g., Keren & Baggen, 1981).
One upshot of these restrictions on similarity is a reduction in the number
of free parameters. To see this reduction, consider the paradigm in which
participants identify the first three letters a, b, c. For this case there are
I = J = 3 stimuli and responses. The full matrix of all ηi,j has 9 elements.
The below matrix shows the effect of imposing the restriction ηi,i = 1 and
ηi,j = ηj,i.
η
η1,2 η1,3 1,1 η1,2 η1,3 η =1, η =η 1
i,j
j,i i,i
(6.10)
η1,2
1 η2,3 7−→
η2,1 η2,2 η2,3 η1,3 η2,3
η3,1 η3,2 η3,3 1
In this case, the restrictions reduce the number of parameters from 9 to 3.
To insure that the SCM predictions for response probabilities are between
0 and 1, response biases are always positive. Although there are J response
bias terms, only J − 1 of them are free. To see this, consider what happens
to pij when all response bias values are doubled. The factor of 2 cancels in
the numerator and denominator of Eq (6.9). Hence, one of the responses can
be set to 1.0 without any loss of generality. As a matter of convenience, we
always set β1 = 1.0 and estimate the remaining J − 1 values.
6.4.3
Analysis
Consider SCM for the identification of the first three letters a, b, c. Table 6.3
depicts the format of data. Random variable Yij is the number of times
stimulus i elicits response j. In the table, the stimuli are denoted by lowercase letters (even though the physical stimuli may be upper-case) and the
responses are denoted by upper-case letters. Table 6.3 is called a confusion
matrix.
The general multinomial model for this confusion matrix is
(Ya,A , Ya,B , Ya,C ) ∼ Multinomial(pa,A , pa,B , pa,C , Na ),
(Yb,A , Yb,B , Yb,C ) ∼ Multinomial(pb,A , pb,B , pb,C , Nb ),
(Yc,A , Yc,B , Yc,C ) ∼ Multinomial(pc,A , pc,B , pc,C , Nc ).
(6.11)
(6.12)
(6.13)
For each of these three component multinomials, the three probability
parameters represent the probability of a particular response given a partic-
150
CHAPTER 6. MULTINOMIAL MODELS
Stimulus
Stimulus a
Stimulus b
Stimulus c
Response
A
B
C
Ya,A Ya,B Ya,C
Yb,A Yb,B Yb,C
Yc,A Yc,B Yc,C
# of trials per stimulus
Na
Nb
Nc
Table 6.3: Confusion matrix for three stimuli (a, b, c) associated with three
responses (A, B, C).
ular stimulus. These three probabilities must sum to 1.0. Hence, for each
component, there are two free parameters. For the three stimuli, there are a
total of six free parameters. The log likelihood is
l=
3 X
3
X
yi,j log pi,j .
(6.14)
i=1 j=1
ML estimates of the probability parameters are the appropriate proportions;
i.e., p̂a,A = ya,A /Na .
The SCM model for the identification of three letters has five free parameters (η1,2 , η1,3 , η2,3 , β2 , β3 ). The other parameters can be derived from
these five. Analysis proceeds by simply expressing the probabilities in Equations (6.11) through (6.13) as functions of the five free parameters. For
example, probability p1,1 may be expressed as
1
η1,1 β1
=
.
p1,1 = P
1 + η1,2 β2 + η1,3 β3
k η1,k βk
Likewise, the multinomial log likelihood function (Eq. 6.14) is expressed as
a function of the five free parameters and maximized with respect to these
parameters. Analysis for a confusion matrix of arbitrary I and J proceeds
analogously.
6.4.4
Implementation in R
In this section, we implement analysis of SCM in R. One element that makes
this job difficult is that the similarities, ηi,j , are most easily conceptualized
as a matrix, as in Eq. 6.10. Yet, all of our previous code relied on passing
parameters as vectors. To complicate matters, not all of the elements of the
similarity matrix are free. Hence, the code must keep careful track of which
6.4. SIMILARITY CHOICE MODEL
151
matrix elements are free and which are derived. Consequently, the code
is belabored. Even so, we present it because these types of problems are
common in programming complex models, and this code serves as a suitable
exemplar. Readers not interested in programming these models can skip this
section without loss.
The first step in implementing the SCM model is to write a function
that yields the log likelihood of the data as a function of all similarity and
response parameters, whether they are free or derived. The following code
does so. It calculates the log likelihood for each stimulus within a loop and
steps through the loop for all I stimuli. Before running the code, be sure to
define I; e.g., I=3.
#eta is an I-by-I matrix of similarities
#beta is an I element array of response biases
#Y is stimulus-by-response matrix of frequencies
nll.scm.first=function(eta,beta,Y)
{
nll=0
for (stim in 1:I)
{
denominator=sum(eta[stim,]*beta) #denominator in Eq 6.8
p=(eta[stim,]*beta)/denominator #all J p[i,] at once
nll=nll-sum(Y[stim,]*log(p)) #add negative log likelihoods
}
return(nll)
}
This function is not directly suitable for minimization because it is a function of both free parameters and derived parameters. The matrix eta has
I 2 elements; yet the model specifies far fewer similarity parameters. In fact,
after accounting for ηi,i = 1 and ηi,j = ηj,i there are only I(I − 1)/2 free similarity parameters. Likewise, there are only I − 1 response bias parameters.
The goal is to maximize likelihood with respect to these free parameters.
The following code partially meets this goal; it maps the free similarity parameters, denoted, par.e, into matrix eta.
#par.e is a I(I-1)/2 element vector of free similarity parameters
#eta is an I-by-I matrix of all similarities
152
CHAPTER 6. MULTINOMIAL MODELS
par2eta=function(par.e)
{
eta=matrix(1,ncol=I,nrow=I)
eta[upper.tri(eta)]=par.e
eta[lower.tri(eta)]=par.e
return(eta)
}
#create an I-by-I matrix of 1s.
The novel elements in this code are the functions upper.tri() and lower.tri().
These functions map the elements of the vector par.e into the appropriate
locations in the matrix eta. To see how these work, try the following lines
sequentially:
x=matrix(nrows=3,1:9,byrow=T) #type x to see the matrix
upper.tri(x)
lower.tri(x)
x[upper.tri(x)]
x[lower.tri(x)]
x[upper.tri(x)]=c(-1,-2,-3) #type x to see the matrix
The following function, par2beta, returns all beta as a function of the
I − 1 free response bias parameters.
#par.b is I-1 free response bias parameters
#code returns all I response bias parameters
par2beta=function(par.b)
{
return(c(1,par.b))
}
In SCM, there are I(I − 1)/2 free similarity parameters and I − 1 free response bias parameters. The following function returns the log likelihood as a
function of these I(I −1)/2+I −1 free parameters. Because all similarity parameters are restricted to be between zero and 1, we use logistic-transformed
similarity parameters. Likewise, because response biases must always be
positive, we use an exponential transform of response bias parameters. The
function ex is positive for a real values of x.
6.4. SIMILARITY CHOICE MODEL
153
nll.scm=function(par,dat) {
#par is concatanation of I(I-1)/2 logistic transformed similarity
#parameters and (I-1) exponential transformed response bias parameters
#dat is confusion matrix
par.e=plogis(par[1:(I*(I-1)/2)])
par.b=exp(par[((I*(I-1)/2)+1):((I*(I-1)/2)+I-1)])
beta=par2beta(par.b)
eta=par2eta(par.e)
return(nll.scm.first(eta,beta,dat))
}
The following code shows the analysis of a sample confusion matrix. The
first line reads sample data into the matrix dat.
I=3
dat=matrix(scan(),ncol=3,byrow=T)
49 1 15
6 22 5
17 1 35
par=c(.1,.1,.1,1,1) #eta’s=.1, beta=1
par[1:3]=qlogis(par[1:3]) #probit transformed similarities
par[4:5]=log(par[4:5]) #exponential transform response biases
g=optim(par,nll.scm,dat=dat,control=list(maxit=11000))
The results may be found by transforming parameters:
># similarities
> par2eta(plogis(g$par[1:3]))
[,1]
[,2]
[,3]
[1,] 1.00000000 0.07529005 0.38558239
[2,] 0.07529005 1.00000000 0.07982558
[3,] 0.38558239 0.07982558 1.00000000
># response biases
154
CHAPTER 6. MULTINOMIAL MODELS
> par2beta(exp(g$par[4:5]))
[1] 1.0000000 0.2769761 0.7926932
From this data, it may be seen that a and c are more similar to each other
than either is to b. In addition, there is a tendency to have greater response
bias to a and c and not to b.
Problem 6.4.1 (Your Turn)
Decide if SCM is appropriate for the letter confusion data in the above
R code by comparing it with the general multinomial model with a
nested likelihood ratio test.
155
6.4. SIMILARITY CHOICE MODEL
Problem 6.4.2 (Your Turn)
Rouder (2001, 2004) tested the appropriateness of SCM by manipulating the number of choices in a letter identification task. He reasoned
that if SCM held, the similarity between letters should not be a function of the choice set. Consider the following fictitious data for four
and two choice condition:
Response
Stimulus
a
b
c
d
Four-Choice Condition
A B C
D
49 5 1
2
12 35 7
1
1 5 22
11
1 6 18
19
Two-Choice Condition
A
B
83
10
15
76
Decide if the similarly between A and B is the same across the two
conditions. Consider the following steps:
1. Estimate a general SCM model with separate similarities across
both conditions. This can be done by separately finding estimates
for the four-choice SCM model for the four-choice condition and
the two-choice SCM model for the two-choice condition. What
is the joint log likelihood of these parameters across both conditions?
2. Estimate a model in which there is one ηa,b across both conditions.
What is the log likelihood of this common ηa,b model?
3. With these two log likelihoods, it is straightforward to construct
a likelihood ratio test of the common parameter. Construct a
likelihood ratio test and report the result.
156
6.5
CHAPTER 6. MULTINOMIAL MODELS
SCM and Dimensional Scaling
The SCM models similarity between two stimuli. A related concept is mental
distance. Mental distance is the inverse of similarity; two items are considered
close to each other in a mental space if they are highly similar, and hence,
easily confused. Using distances, it is possible to build a space of mental
representation. To better motivate the concept of a space of representation,
consider the three sets of stimuli in Figure 6.4. The top left panel shows
a sequence of nine lines, labeled A, B,...,I. These lines differ in a single
dimension—length. It is reasonable to suspect the mental representation
of this set varies on a single dimension. The left, middle column shows a
hypothetical unidimensional representation. The mental representation of
all of the stimuli are on a straight line. The smallest and largest stimuli
are disproportionately far from the others indicating that these stimuli are
least likely to be confused. The bottom left shows an alternative mental
representation in two dimensions. The primary mental dimension is still
length, but there is a secondary dimension that indicates how central or
peripheral the stimuli are. The top center panel shows a different set of
stimuli: set of circles with inscribed diameters. These stimuli differ on two
dimensions: size and the angle of the diameter. It is reasonable to suspect
that the mental representation of this set varies on two dimensions as well.
A hypothetical mental space is shown below these stimuli. In the figure,
there is greater distance between stimuli on the size dimension than on the
angle dimension, indicating that size is more salient than angle. The right
panel shows select letters in Asomtavruli, an ancient script of the Georgian
language. It is reasonable to suspect that the mental representation of this set
spans many dimensions. The goal of multidimensional scaling is to identify
the number of dimensions in the mental representation of a set of stimuli and
the position of each stimulus within this set.
Dimensional models of mental space may be formulated as restrictions on
SCM. We follow Shepard (1957) who related distance to similarity as
ηi,j = exp(−di,j ),
(6.15)
where di,j is the distance between stimuli i and j. In SCM, the greatest
similarity is of an item to itself, which is set to 1.0. This level of similarity
corresponds to a distance of zero. In SCM, as items become less confusable,
similarity decreases toward zero. According to Equation (6.15), as items become less confusable, distance increases. In fact, SCM can be parameterized
157
6.5. SCM AND DIMENSIONAL SCALING
C
B
A
F
E
D
I
H
G
ABCDE F GH I
Line Segments
A
B
C DE F G
Asomtavruli Letters
Circles with embedded diameters
H
I
Length
Unidimensional Representation
E
CD F G
B
A
H
Length
I
Two dimensional Representation
A
B
C
D
E
F
G
H
I
Angle
Size
Two dimensional Representation
Figure 6.4: Top: Three types of stimuli of increasing dimensionality. Bottom: Hypothetical mental spaces for line segments and circles with inscribed
diameters.
158
CHAPTER 6. MULTINOMIAL MODELS
General
Multinomial Model
SCM
(I−1 Dimensions)
K−Dimension
SCM
I(I−1) Parameters
For 9 Stimuli: 72 Parameters
I(I−1)/2 + (I−1) Parameters
For 9 Stimuli: 44 Parameters
K−1
( Σ i ) +K(I−K) + (I−1) Parameters
i=1
Two−Dimension
SCM
2(I−1)+(I−2) Parameter
One−Dimension
SCM
2(I−1) Parameters
For 9 stimuli: 23 Parameters
For 9 Stimuli: 16 Parameters
Figure 6.5: Hierarchical relationship between models of mental distance.
with distance instead of similarity. This distance-based SCM model is given
as
exp(−di,j )βj
.
(6.16)
pi,j = PJ
k=1 exp(−di,k )βk
Figure 6.5 describes the nested models approach to dimensional models.
At the top level is the general multinomial model. The most general restriction is an SCM model with distance parameters that obey three restrictions:
1. the distance between any item and itself is di,i = 0; 2. distance is symmetrical e.g., di,j = dj,i, and 3. the shortest distance between any two points is a
straight line, e.g., di,j ≤ di,k + d + j, k. An SCM model that obeys these three
restrictions contain distances that can be represented in at most an I − 1
dimension space. Lower dimensions models are restrictions on the distances
and are represented as submodels. At most restrictive model, with the fewest
parameters, is the one in which all the distances are constrained such that
the mental space is constrained to a single dimension. This model, in fact,
is the easiest to analyze and we consider it next.
6.5. SCM AND DIMENSIONAL SCALING
6.5.1
159
Unidimensional SCM Model
For a unidimension model, we may represent the position of the ith item as
a single number, xi . The distance between any two item is
dij = |xi − xj |,
where |x| is the absolute value of x. The following SCM model relates identification performance to the one-dimensional mental representations of lines.
exp(−|xi − xj |)βj
.
pi,j = P
k exp(−|xi − xk |)βk
(6.17)
Fortunately, only minor changes to the SCM code are needed to analyze
this model. There are I positions xi , but the first one of these may be set to
0 without any loss. Therefore, there are only I − 1 free position parameters.
Likewise, there are I − 1 free response biases. The first step is modifying the
mapping from free parameters to similarities:
par2eta.1D=function(par.e)
{
x=c(0,par.e) #set first item’s position at zero
d=as.matrix(dist(x,diag=T,upper=T)) #type d to see distances
return(exp(-d)) #returns similarities
}
The function dist() is a built-in R function for computing distances from
positions. It returns data in a specific format that is not useful to us; consequently, we use the function as.matrix() to represent the distances as a
matrix.
The last step is to modify the nll to send the appropriate parameters to
par2eta.1D() and par2beta(). Because all real numbers are valid positions,
we do not transform parameters.
nll.scm.1D=function(par,dat) {
par.e=par[1:(I-1)]
par.b=par[I:(2*I-2)]
beta=par2beta(par.b)
eta=par2eta.1D(par.e)
return(nll.scm.first(eta,beta,dat))
}
160
CHAPTER 6. MULTINOMIAL MODELS
Problem 6.5.1 (Your Turn)
Fit the one-dimensional SCM model to the following confusion matrix
for the identification of line segments:
Item
a
b
c
d
e
f
g
h
A B C
84 8 7
14 45 23
8 10 36
2 8 17
3 3 6
2 4 8
0 2 5
0 0 2
Response
D E
1 0
9 5
19 15
31 21
28 34
18 18
6 7
0 3
F G H
0 0 0
4 0 0
7 3 2
9 8 4
14 5 7
32 11 7
9 49 22
5 5 85
Optimization may be improved by first using an optim() call to get
good starting values for an nlm() call. Plot the resulting positions.
Notice that the spacing is not quite uniform. This enhanced distance
between the first and second item and the ultimate and penultimate
item is common in one-dimensional absolute identification and is called
the bow effect (Luce, Nosofsky, Green, Smith, 1982).
6.5.2
Multidimensional SCM Models
To test the appropriateness of the one-dimensional SCM model, we can embed the one-dimensional model within a higher-dimension SCM model. In
the most general case, each item is represented by a point in a higher dimensional space. The location of the ith item can be denoted by (xi,1 , xi,2 , ...xi,M ),
where M is the number of dimensions in the mental representation.
The construction of distance is more complex with multiple dimensions.
One general form of distance is called Minkowski distance.
161
6.5. SCM AND DIMENSIONAL SCALING
Definition 37 (Minkowski Distance) The Minkowski distance between any two points is
di,j =
M
X
m=1
|(xi,m − xj,m )|r
!1/r
.
(6.18)
To understand Minkowski distance, lets take a simple case of two stimuli which exist in two dimensions. The stimuli’s positions are (x1 , y1 ) and
(x2 , y2 ). For these two stimuli and for r = 2, Equation (6.18) reduces to
d1,2 =
q
(x1 − x2 )2 + (y1 − y2 )2 .
This is the familiar formula for the distance of a straight line between two
points. When r = 2, the distance is called the Euclidean distance. Figure 6.6 shows the Euclidean distance between two points as well as two other
distances. When r = 1, the distance is called the city-block distance. For
the example with two points in two dimensions, city-block distances are computed with only vertical and horizontal lines. Much like navigating in a dense
city, diagonal lines are not admissible paths between points. The city-block
distance in the figure is 7. The maximum distance occurs asr → ∞. The
distance is maximum difference between the points on a single dimension.
In the figure, the differences are 4 (x-direction) and 3 (y-direction). The
maximum difference is 4. We use the Euclidean distance here although researchers have proposed the city-block distance for certain classes of stimuli
(e.g., Shepard, 1986)
The following code implements a two-dimensional SCM model for the
line lengths. There are I points, each with two position parameters. The
first item may be placed at (0, 0). Also, the y-coordinate of the second point
may also be placed at 0 without loss. There are, therefore, 2(I − 1) − 1
free position parameters in I − 1 free response biases. The total, therefore,
is 3(I − 1) − 1 free parameters. The function par2eta.2D() converts the
2(I − 1) − 1 position parameters to similarities:
par2eta.2D=function(par.e)
{
x=c(0,par.e[1:(I-1)]) #x-coordinate of each of the I points
162
CHAPTER 6. MULTINOMIAL MODELS
City−Block Distance: 7
5
Euclidean Distance: 5
3
Maximum Distance: 4
4
Figure 6.6: Three distance measures between 2 points.
y=c(0,0,par.e[I:(2*(I-1)-1)]) #y-coordinate of each of the I points
points=cbind(x,y)
d=as.matrix(dist(points,"euclidian",diag=T,upper=T))
return(exp(-d))
}
The cbind() function makes a matrix by binding rows together. In the
above code, it makes a matrix of 2 columns and I rows; the first and second
column is the x-coordinate and y-coordinate of each point, respectively. The
negative log likelihood function is
nll.scm.2D=function(par,dat)
{
par.e=par[1:(2*(I-1)-1)]
par.b=par[(2*(I-1)):(3*(I-1)-1)]
beta=par2beta(par.b)
eta=par2eta.2D(par.e)
return(nll.scm.first(eta,beta,dat))
}
Optimization is performed as follows:
par=c(1:7,rep(0,6),rep(1/8,7))
start=optim(par,nll.scm.2D,dat=dat)
g.2D=nlm(nll.scm.2D,start$par,dat=dat,iterlim=200)
6.5. SCM AND DIMENSIONAL SCALING
163
Problem 6.5.2 (Your Turn)
Decide if the one-dimensional restriction of the two-dimensional SCM
model is appropriate for the line-length data with a likelihood ratio
test.
One of the drawbacks of the SCM models is that the parameters grow
quickly with increasing numbers of items. With sixteen items, for example,
a three-dimensional SCM model has 56 parameters. While this is not a
prohibitive number in some contexts, it is difficult to achieve stable ML with
the methods we have described.
Fortunately, there are standard, high performance multidimensional scaling techniques (Cox and Cox, 2001; Shepard, Romney, and Nerlove, 1972;
Torgeson, 1958). The mathematical bases of these techniques are outside
the scope of this book. Instead, we describe their R implementation. When
distances are known, the function cmdscale() provides a Euclidean representation. An example is built into R and may be found with ?cmdscale.
For psychological applications, however, we rarely know mental distances.
There are a few common alternatives. One alternative is to simply ask people
the similarity of items, two at a time. These data can be transformed to
distances using Equation 6.15. One critique of this approach is that similarity
data is treated as a ratio scale. For example, suppose a participants rates
the similarity between items i and j as a “2” and that between items k and
l as a “4.” The implicit claim is that k is twice as similar to l as i is to j.
This may be too strong; instead, it may be more prudent to just consider the
ordinal relations, e.g., k is more similar to l than i is to j. Fortunately, there
is a form of multidimensional scaling, called non-metric multidimensional
scaling, that is based on these ordinal relations. In R, two methods work
well: either sammon() and isoMDS(). Both of these functions are in the
package “MASS” (Venebles & Ripley, 2002) The package is load with the
library() command; e.g., library(MASS).
164
6.6
CHAPTER 6. MULTINOMIAL MODELS
Generalized Context Model of Categorization
The Generalized Context Model (Nosofsky, 1986) describes how people categorize novel stimuli. According to the the models, categories are represented
by exemplars. For example, shoes are represented by a set of stored mental
exemplars of shoes. They are not represented by a set of verbal rules; such
as “a shoe is a piece of footware that is longer than it is high.” GCM posits
that a novel piece of footwear will be classified as a shoe as opposed to boot
if it is more similar to the stored exemplars of shoes than it is to the stored
exemplars of boots.
GCM has primarily been tested with novel rather than natural categories.
Consider an experiment in which the Asomtavruli letters in Figure 6.4 are
assigned by the experimenter to two different categories. Suppose left-column
letters are assigned to Category A and right-column letters are assigned to
Category B. Further suppose that a participant has been shown these six
stimuli and their category labels numerous times and has well learned the
pairings. GCM describes how participants will classify previously unseen
stimuli, such as middle-column letters.
In GCM, it is assumed that a to-be-classified item and the exemplars are
represented as points in a multidimensional space. The position of to-beclassified item is denoted by y = (y1 , .., yM ); the position of the ith exemplar
is denoted xi = (xi,1 , .., xi,M ). Exemplars belong to specific categories; let sets
A and B denote the exemplars that belong to categories A and B, respectively.
In the Asomtavruli letter example, the letters in the left column belong to
A; those in the right column belong to B. The similarity between the tobe-classified item and the ith exemplar is determined by their Minkowski
distance:
X
ηy,i = exp(−( αm |ym − xi,m |r )1/r ), r = 1, 2.
(6.19)
m
The exponent r is usually assumed before hand. The new parameters in
Equation 6.19 are (α1 , .., αm ), which differentially weights the importance of
the dimensions. The motivation for the inclusion of these parameters is that
participants may stress differences in some dimensions more than others in
categorization. These weights are relative, and the first one may be set to
1.0 without any loss.
When to-be-classified item y is presented, it elicits activity for the cate-
6.6. GENERALIZED CONTEXT MODEL OF CATEGORIZATION 165
gories. This activation depends on the overall similarity of the item to the
exemplars for specific categories. The activation for category A is
X
a=
ηy,i ,
i∈A
where the sum is over all exemplars of Category A. Activation for category
B is given analogously:
b=
X
ηy,i .
i∈B
The probability that stimulus y is placed in category A may be given as:
p(y, A) =
aβA
,
aβA + bβA
where βA and βB are response biases.
In fitting SCM, researchers do not let the position parameters of the exemplars of the to-be-classified stimuli be free. Instead, these are estimated
prior to the categorization experiment through either a similarity ratings task
or an absolute identification experiment. The data from these supplementary tasks are submitted to a multidimensional scaling routine (see previous
section) to yield distances. The remaining free parameters are the weights
α and response bias β. GCM is attractive because one can predict categorization from the identification data with a minimal number of additional
parameters.
166
CHAPTER 6. MULTINOMIAL MODELS
Problem 6.6.1 (Your Turn)
Let’s use Asomtavruli letters to test GCM. The stimuli in our experiment are sixteen Georgian letters which exist in a four dimensional
space as follows:
Letter
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
x1
-0.167
-0.445
-0.308
-0.648
-0.254
0.001
0.013
0.441
0.014
0.251
-0.357
-0.218
0.466
0.549
-0.082
-0.218
Coordinates
x2
x3
0.136 0.042
0.265 -0.008
0.325 -0.046
0.120 0.138
-0.048 0.354
-0.746 -0.080
-0.357 -0.887
-0.064 0.261
-0.206 -0.065
0.492 0.157
0.387 -0.267
0.402 -0.218
0.113 -0.054
0.289 -0.047
-0.181 0.782
0.402 -0.218
x4 Category
-0.088
A
-0.218
A
0.241
A
0.350
A
0.428
A
0.033
B
0.203
B
0.309
B
0.671
B
-0.102
B
-0.084
?
-0.044
?
-0.124
?
-0.052
?
-0.075
?
-0.044
?
In our experiment, a participant learns to classify the Letters 1 to 5 to
Category A and Letters 6 to 10 to Category B. They then are repeatedly
tested on Letters 11 through 16 without any feedback. The table below
contains the number of times each letter was placed in each category:
Letter
11
12
13
14
15
16
Response Category
A
B
28
22
26
24
16
34
21
29
23
27
30
20
There are 6 pieces of data and 4 free parameters (α2 , α3 , α4 , βB ). Estimate these parameters with a Euclidean distance GCM model and
decide if the model fits well by comparing it to a general binomial
model (the general model is binomial as there are only two choices in
the categorization task).
6.7. GOING FORWARD
6.7
167
Going Forward
In this and the preceding chapters, we have shown how substantive models
may be analyzed as restrictions on general binomial and multinomial models.
The data for these models are frequency counts of responses. Frequency count
data of this type are called categorical data; models of categorical data are
called categorical models. The focus of the next two chapters is on continuous
variables such as response time. Once again, we embed substantive models
as restrictions on nested models whenever it is feasible. In the next chapter,
we discuss the normal model which is the basis for ANOVA and regression.
Following that, we discuss substantive and statistical models of response
time. The last chapter is devoted to hierarchical models.
168
CHAPTER 6. MULTINOMIAL MODELS
Chapter 7
The Normal Model
The first six chapters focused on substantive models for binomial and multinomial data. The binomial and multinomial are ideal for data that is discrete,
such as the frequency of events. Psychologists often deal with data which is
more appropriately modeled as continuous. A common example is the time
to complete a task in an experiment, or response time (RT). Because RT
may take any positive value, it is appropriate to model it with a continuous
random variable. In some contexts, even discrete data may be more conveniently modeled with continuous RVs. One example is intelligence quotients
(IQ). IQ scores are certainly discrete, as there are a fixed number of questions
on a test. Even though it is discrete, it is typically modeled as a continuous
RV. The most common model of data in psychology is the normal. In this
chapter we cover models based on the normal distribution.
7.1
The Normal-Distribution Model
IQ scores are typically modeled as normals. Consider the case in which we
obtain IQ test scores from a set of participants. The ith participant’s score,
i = 1, .., I, is denoted by random variable Yi which is assumed to be a normal:
Yi ∼ Normal(µ, σ 2 ), i = 1, .., I.
(7.1)
Parameters µ and σ 2 are the mean and variance of the distribution. The goal
is to estimate these parameters. One way to do this is through maximum
likelihood.
169
170
CHAPTER 7. THE NORMAL MODEL
The first step is expressing the likelihood. Accordingly, we start with
the probability density function (pdf). The pdf for the normally distributed
random variable Yi is
(y−µ)2
1
fYi (yi ) = √
e− 2σ2 .
2πσ
We must then express the joint pdf for all I random variables. Because the
observations are independent, the joint pdf is obtained by multiplying
fY1 ,..yI (y1, .., yI ) =
PI (y −µ)2
1
− i=1 i 2
2σ
e
.
(2π)I/2 σ I
The likelihood is obtained by rewriting the pdf as a function of the parameters.
PI (yi −µ)2
L(µ, σ 2 ; y1 , .., yI ) = σ −I e− i=1 2σ2 ,
where the term in 2π has been omitted. Log-likelihood is
l(µ, σ 2 ; y1 , .., yI ) = −I log σ + −
I
X
(yi − µ)2
.
2σ 2
i=1
The next step in estimating mu and σ 2 is maximizing log-likelihood.
This may be done either numerically or with calculus methods. The calculus
methods yield the following estimators:
µ̂ =
σˆ2 =
PI
i=1
PI
yi
(7.2)
I
i=1 (yi
I
− µ)2
(7.3)
The estimator for µ, µ̂, is the sample mean. Under the assumption that Yi
are normal random variables, the sampling distribution of µ̂ is also normal:
µ̂ ∼ Normal(µ, σ 2 /I).
Figure 7.1 shows the relationship between normally-distributed data Yi
and the normally-distributed estimate µ̂. In the Figure, the higher variance
probability density function is the distribution of data and has a mean of 100
and a standard deviation of 15. The lower variance density is the distribution
of the
√ sample mean of 9 observations. The standard error of µ̂ this case is
15/ 9 = 5.
171
0.04
0.00
Density
0.08
7.1. THE NORMAL-DISTRIBUTION MODEL
60
80
100
120
140
Intellegence Quotient (IQ)
Figure 7.1: The distribution of the IQ population and the distribution of the
sample mean, with N = 9.
172
CHAPTER 7. THE NORMAL MODEL
The maximum likelihood estimator for σˆ2 , however, is not the sample
variance. Sample variance, s2 , is defined as:
2
s =
PI
− µ)2
.
I −1
i=1 (yi
The difference between s2 and σˆ2 is in the denominator; the former involves a
factor of I −1 while the latter involves a factor I. The practical consequences
of this difference are explored in the following exercise.
Problem 7.1.1 (Your Turn)
Consider the model for IQ, Yi ∼ Normal(µ = 100, σ 2 = 100), where
i = 1, .., 10.
1. Use the simulation method to explore the sampling distributions
of the ML estimator (σˆ2 ) and the classic estimator s2 . How different are they? Compare them to the distribution of a χ2 random
variable with 9 degrees of freedom. What do you notice?
2. Compute the RMSE and bias for each estimate. Which is more
efficient?
7.2
Comparing Two Means
The main appeal of the normal model is that methods of inference are well
known. Consider, for example, a hypothetical investigation of ginko root
extract on intelligence. Participants may be split into two groups: those
receiving ginko root extract (the treatment group) and those receiving a
placebo pill (the control group). After receiving their respective ginko extract
or the placebo, participants take an intelligence test. The resulting scores
may be denoted Yi,j , where i indicates participant and j indicates the group
(either treatment or control).
The normal model is
Yi,j ∼ Normal(µj , σ 2 )
(7.4)
Density
173
0.000 0.010 0.020
7.2. COMPARING TWO MEANS
60
80
100
120
140
Intellegence Quotient (IQ)
Figure 7.2: The independent-groups t test model.
The model is shown graphically in Figure 7.2. The main question is
whether the treatment affected intelligence. The questions is assessed by
asking whether µtreatment equals µcontrol .
This question may certainly be answered with a likelihood ratio test. It
is more standard, however, to use a t-test1 . The rationale for a t-test may be
found in several texts of statistics for behavioral scientists (e.g., Hays, 1994).
We focus on the R implementation.
treatment=c(103,111,112,89,120,123,92,105,87,126)
control=c(81,114,105,75,104,98,114,106,92,122)
t.test(treatment,control,var.equal=T)
The output indicates the t value, the degrees of freedom for the test, and
the p value. The p value denotes the probability of observing a t value as
1
The two tests are in fact equivalent in this case.
174
85
95
105
115
CHAPTER 7. THE NORMAL MODEL
Control
Treatment
Figure 7.3: Hypothetical data with confidence intervals.
extreme or more extreme than the one observed under the null hypothesis
that the true means are equal. In this case, the p value is about 0.39. We
cannot reject the null hypothesis that the two groups have the same mean.
It is common when working with the normal model to plot µ̂, as shown in
Figure 7.3. The bars denote confidence intervals (CIs) rather than standard
errors. Confidence intervals have an associated percentile range and the 95%
confidence interval is typically used. The interpretation of CI, like much
of standard statistics, is rooted in the concept of repeated experiments. In
the limit that an experiment is repeated infinitely often, the true value of
the parameter will lie in the 95% CI for 95% of the replicates. Confidence
intervals are constructed with the t distribution as follows.
#for treatment condition
mean(treatment)+c(-1,1)*qt(.025,9)*sd(treatment)/sqrt(10)
#for control condition
7.3. FACTORIAL EXPERIMENTS
175
mean(control)+c(-1,1)*qt(.025,9)*sd(treatment)/sqrt(10)
Alternatively, R will compute the confidence intervals for you if you use the
t.test() function. Reporting CI’s or standard errors are equally acceptable
in most contexts. We recommend CI’s over standard errors in general, though
we tend to make exceptions when plotting CI’s clutters a plot more than
plotting standard errors.
Problem 7.2.1 (Your Turn)
Compare t test with ML estimation. Using the model in Equation 7.4,
construct a nested ML test to test the hypothesis that two group means
are equal. Use 10 subjects, and examine the rejection rates when the
difference between the two group means are 0, .5, 1, and 1.5 group
standard deviations apart. Do the tests perform differently?
7.3
Factorial Experiments
The t test works well when there are two groups to compare. In the example
above, there was one factor manipulated and that factor had two groups. If
we wanted to manipulate another factor, or add another treatment, the t test
would be inappropriate.
Suppose that in addition to the effect of ginko root, we were also interested in how the exposure to certain types of music effects IQ. There has
been some suggestion that the music of Mozart increases IQ for a short time
(Rauscher, Shaw, & Ky, 1993). This controversial effect has been dubbed
the ”Mozart effect”. We could design an experiment in which we manipulated both treatment with ginko root and exposure to music. This would
produce 2 × 2 = 4 groups. The four groups are shown in Figure 7.4. This
design is called a factorial design because every combination of the factors is
represented.
We can think of running this experiment in two ways. In a betweensubjects design, each participant is a member of only one group. In a withinsubjects design, each participant will typically be involved in every combina-
176
CHAPTER 7. THE NORMAL MODEL
Figure 7.4: A factorial design with 2 factors.
7.3. FACTORIAL EXPERIMENTS
177
tion. Both designs have advantages; for the purposes of this section we will
consider the between-subjects design.
For an experiment design like the one described above, the most widely
used model for analysis is ANOVA. The details and theory behind ANOVA
analyses is covered in elementary statistics texts (Hays, 19xx). The basic
ANOVA model for the 2-factor, between-subjects design above is
IQijk ∼ Normal(µjk , σ 2 )
µjk = µ0 + αj + βk + (αβ)jk
(7.5)
(7.6)
The IQ score of the ith participant in the jth ginko root treatment and
the kth music treatment is distributed as a normal. This normal has a mean
which depends on the condition. The terms µ0 , αj , and βk are the grand mean
and the effects of the ginko and music treatments respectively. Typically we
P
P
apply the constraint that αk = βk = 0. The last term, (αβ)jk is called
the interaction term, and describes any deviation from the additivity of the
treatment effects. For instance, ginko may only be effective in the presence
of music. For each effect, the null hypothesis is all treatment means are the
same.
R has functions for ANOVA analyses built in. Consider the experimental
design above. Data for a hypothetical experiment using this design is in the
file factorial.dat. Download this file into your working directory, then use the
following code to load and analyze it.
dat=read.table(’factorial.dat’,header=T)
summary(aov(IQ~Ginko*Music,data=dat))
The result should be a table like the one in Table 7.1. The p values in
the last column tell us the probability under the null hypothesis (that all
treatment means are the same) of getting an F statistic as large as the one
obtained. If p is less than our prespecified alpha, we reject the null and
conclude that all treatment means are not equal. In this case, there is a
significant effect of ginko root, but not of exposure to music.
To see this graphically, consider Figure 7.5. The boxplot shows that the
median scores of the groups receiving ginko are higher than the groups not
receiving it, which accords with the ANOVA analysis. It also allows us to
quickly check the equal-variance assumption of the ANOVA test; in this case
there does not appear to be any violation of this assumption.
178
CHAPTER 7. THE NORMAL MODEL
Df Sum Sq Mean Sq F value Pr(¿F)
1
1690.0
1690.0 5.9455 0.01982 *
1
28.9
28.9 0.1017 0.75168
1
547.6
547.6 1.9265 0.17368
36 10233.0
284.2
Ginko
Music
Ginko:Music
Residuals
—
Signif. codes: 0 ’***’ 0.001 ’**’ 0.01 ’*’ 0.05 ’.’ 0.1 ’ ’ 1
Table 7.1: ANOVA table for the hypothetical IQ data.
IQ
140
120
100
80
NG.NM
G.NM
NG.M
G.M
Group
Figure 7.5: A boxplot of the IQ data.
7.4. CONFIDENCE INTERVALS IN FACTORIAL DESIGNS
7.4
179
Confidence Intervals in Factorial Designs
Confidence intervals in factorial designs are more difficult to use. Because
of the variety of possible deigns (between- and within-subjects, fixed- and
random-effects models) it is impossible to prescribe one method that will
work for every design. Also, the quantity on which the confidence interval
is placed can vary. One can place confidence intervals on group means,
differences between groups, or standardized effect sizes. The choice depends
on how you wish to use the intervals; if they are for inference, you may
wish to put the confidence intervals on effect sizes. If the intervals are for
graphical purposes, you may want to put them on group means instead.
For the purposes of simplicity, we will deal with only the latter approach.
Confidence intervals on effect sizes is discussed in depth in (cites).
One simple way to put confidence intervals on the groups in the betweensubjects designs like the design above is to extend the approach with the t
test to each group. We can compute a sample mean and standard deviation
for each group and use them to build a individual confidence intervals. A
better approach, however, is to use the estimate of σ 2 , the error variance,
provided by the ANOVA analysis. This is the mean squared error in the
ANOVA table. The 95% confidence interval for the ijth group is then
CIij =
"
s
MSE
X̄ ± t.025 (errordf )
Nij
#
(7.7)
where Nij is the number of subjects in the ijth group.
Applying this to the data and plotting,
gmeans=tapply(dat$IQ,list(dat$Ginko,dat$Music),mean)
size=qt(.025,36)*sqrt(284.2/10)
g=barplot(gmeans,beside=T,ylim=c(70,130),xpd=F)
errbar(g,gmeans,size,.3,1)
The resulting plot is shown in Figure 7.6. Two things are apparent from this
plot. First, with only 10 participants in each group, our ability to localize
means with 95% confidence is limited; the confidence intervals are fairly wide.
Second, the mean of the two dark bars (no ginko) is less than the mean of the
light bars (ginko treatment). The pattern of means is similar to the pattern
of medians from the boxplot in Figure 7.5.
180
70
90
110
130
CHAPTER 7. THE NORMAL MODEL
NM
M
Figure 7.6: Barplot of the IQ data with 95% confidence intervals.
181
7.5. REGRESSION
7.5
7.5.1
Regression
Ordinary Least-Squares Regression
Regression is perhaps the most popular method of assessing the relationships
between variables. We provide a brief example of how to do regression in R.
The example comes from an experiment by Gomez, Perea & Ratcliff (in
press) who asked participants to perform a lexical decision task. In this task,
participants decide whether a string of letters is a valid English word. For
example, the string cafe is a valid English word while the string mafe in not.
In this task, it has been well established that responses to common valid
words are faster than the rare common words. The goal is to explore some
of the possible functional relationships between word frequency and response
time (RT). Word frequency is a measure of the number of times of word
occurs for every million words of text in magazinge (Kucera & Francis, 1968).
Gomez et al. collected over 9,000 valid observations across 55 participants
and 400 words. The basic data for this example are the mean RTs for each
of the 400 words computed by averaging across participants. The following
code loads the data and draws a scatter plot of RT as a function of word
frequency. Before running it, download the file regression.dat and set it in
your working directory 2 .
dat=read.table(’regression.dat’,header=T)
colnames(dat) #returns the column names of dat
plot(dat$freq,dat$rt,cex=.3,pch=20,col=’red’)
There are four hundred points, with each coming from a different word.
From the scatter plot in the left panel of Figure 7.7, it seems evident that as
frequency increases, RT decreases.
The linear regression model is
RTi ∼ Normal(β0 + β1 fj , σ 2 ).
(7.8)
RTj and fj denote the mean RT to the jth item and the frequency of the
jth item, respectively. Parameters beta0 and β1 are the intercept and slope
of the best fitting line, respectively. The model is written equivalently as
RTi = β0 + β1 fi + ǫi ,
2
How to in Windows
182
CHAPTER 7. THE NORMAL MODEL
14
0.9
1.0
1.1
6
0.5
0.6
0.7
0.8
0.9
0.8
0.7
0.6
0.5
Response Time
1.0
1.1
2
5
10
15
Word Frequency
20
0.0
1.0
2.0
3.0
log(Word Frequency)
183
7.5. REGRESSION
where epsilon is taken to be independent and identically distributed normals
centered at zero; i.e.,
ǫ ∼ Normal(0, σ 2 ).
The standard approach to estimating this model is based on least-squares
regression (Pedhauzer). The code for analysis is
g=lm(dat$rt~dat$freq)
summary(g)
abline(g)
The first line fits the regression model. The commandlm() stands for “linear
model,” and is extremely flexible. The argument is the specification of the
model. The output is stored in the object g. The command summary(g)
returns a summary of the fit. Of immediate interest are the estimates βˆ0 and
βˆ1 , which are .753 seconds and -.008 seconds per frequency units, respectively.
Also reported are standard errors and a test of whether these statistics are
significantly different than zero. The model accounts for 21.4% of the total
variance in the data (multiple R2 = .214). The last line adds the regression
line in the plot. This line has intercept βˆ0 and slope βˆ1 .
184
CHAPTER 7. THE NORMAL MODEL
Problem 7.5.1 (Parameter Estimates in Regression)
1. Are slopes and intercept estimates biased? Consider the case
in which we are evaluating the effects of therapy in behavioral
therapy on IQ for autistic three-year olds. Suppose for each participant, the change in IQ, denoted Yi , is related to the number
of days in therapy, denoted X as follows:
Yi ∼ Normal(1 + .05Xi, σ 2 = 2).
Let’s assume that an experimenter observes 50 children. Let’s
assume that the time in therapy, Xi , is distributed as a normal:
Xi ∼ Normal(100, 10).
To answer the question, simulations should proceed as follows.
For each replicate experiment, first draw Xi values for all 50 children from a normal. Then, for each value Xi , draw a value of Yi
from the above regression equation. Then, analyze the 50 pairs
of (Xi , Yi ) with lm(). Save the intercept and slope estimates.
Repeat the process until you have a reasonable estimate of the
sampling distribution of slope and intercept.
2. The results should indicate that the slope and intercept estimators are unbiased. Does the bias depend on the distribution of
Xi ? Assess the bias when Xi is distributed as a binomial with
N = 120 and p = .837.
7.5.2
Nonparametric Smoothing
Although the regression line appears reasonable for the Gomez et al., there
may be more parsimonious models. One sensible method of checking the
validity of a regression line is to fit a nonparametric regression model. A
nonparametric model does not assume a particular form for the data, but
instead fits a smooth curve. One nonparametric regression model is lowess
185
7.5. REGRESSION
(Cleveland, 1981). In a small interval, lowess fits the points with a polynomial regression line. Points further away from the center of the interval
are weighted less than those near the center. These polynomial fits are done
across the range of the points, and a kind of ”average” line is constructed.
In this way, a fit to the points is generated without recourse to parametric
assumptions.
In R, lowess is by the commands lowess() or loess(). The syntax of the
former is somewhat more convenient and we use it here. The nonparametric
smoothing line may be added to the plot by
lines(lowess(dat$freq,dat$rt),lty=2)
The nonparametric smooth is the dotted line (and is produced with the
option lty=2). The regression line overestimates the nonparametric smooth
in the middle of the range while underestimating it in the extremes. The
nonparametric smooth is a more faithful representation of the data. The
discordance between the nonparametric line and the regression line indicates
a misfit of the regression model.
RT is frequently modeled as the logarithm of word frequency, e.g.,
RTi ∼ Normal(β0 + β1 log fj , σ 2 ).
Figure 7.7, right panel, shows this relationship along with regression line and
lowess nonparametric smooth. The figure is created with the following code:
logfreq=log(dat$freq)
plot(logfreq,dat$rt,cex=.3,pch=20,col=’red’)
w=seq(2,20,4) # tick marks for axis drawn in next statemet
axis(3,at=log(w),labels=w) #create axis on top
g=lm(dat$rt~logfreq)
summary(g)
abline(g)
lines(lowess(logfreq,dat$rt),lty=2)
The new element in the code is the use of an axis on top of the figure, and
this is done through the axis() command. The intercept is .775 seconds
and the slope is -.051 second per log unit frequency. The slope indicates
how many seconds faster RT is when word frequency is multiplied by e, the
natural number (e ≈ 2.72).
186
CHAPTER 7. THE NORMAL MODEL
This interpretation of slope is awkward. If slope is multiplied by log 2, the
interpretation is the amount of time saved for each doubling of RT. In this
case, the amount is .035 sec. The model accounts for 25.0% of the variance,
which is somewhat more than the 21.4% accounted for by the linear model.
Moreover, the regression line is closer to the nonparametric smooth for this
model than it is for the linear model indicating an improved fit.
7.5.3
Multiple Regression
In the lexical decision application, Gomez et al. manipulated two other
factors than may affect RT: word length and neighborhood size. Words were
either 4 or 5 letters in length. The neighborhood of a word refers to all words
that look very similar. It is conventionally operationlized as the collection
of words that differ from the target by only one letter, e.g.; hot is in the
neighborhood of hat. The size of the neighborhood for a word refers to the
number of neighbors. For Gomez et al.’s words, these varied from 0 to 26.
Researchers are often interested in how the three variables account for RT.
This problem is termed a multiple regression problem as there are multiple
predictors of the dependent measure. The model is
RTi ∼ Normal(β0 + β1 fi + β2 ni + β3 δ(li − 5)
(7.9)
This can be accomplished in R with the following code:
dat$length=as.factor(dat$length)
summary(lm(rt~freq+length+neigh,data=dat))
Interpretation of the parameters is analogous to that in the simpler onepredictor regression case. For details on multiple regression, consult any
introductory statistics text.
7.5.4
Maximum Likelihood
The maximum likelihood approach provides for very similar estimates to the
least squares approach for ordinary and multiple regression. The likelihood
for the multiple regression is
187
7.5. REGRESSION
Problem 7.5.2 (Your turn)
• The linear and logarithm models of the effect of word frequency
on RT are not nested. Use the AIC statistic to compare them.
• Consider the power model:
RTj ∼ Normal(α + βfjγ , σ 2 ).
Estimate the parameters with maximum likelihood. Compare
this model with the log and linear models through AIC. Note,
this model is a generalization of the linear one. The linear model
holds if γ = 1. Check if the linear model restriction holds with a
likelihood ratio test.
7.5.5
A z-ROC Signal-Detection Application
One advantage of the ML approach over the least-squares is that it is more
flexible. Many advanced normal models, such as hierarchical linear models
and structural equation models, are analyzed with likelihood-based techniques. Here we show the superiority of ML over a more common leastsquares approach for the analysis of the free-variance signal detection model.
In Section x.x, we discussed z-ROCs as a common graphical representation for analyzing the signal detection model. The z-ROC plot is a plot
of Φ−1 (pˆh ) as a function of Φ−1 (pˆf ). According to the free-variance model
ph = Φ((d′ − c)/σ) and pf = Φ(−c).Substituting yields
Φ−1 (ph ) =
d′ Φ−1 pf
+
.
σ
σ
This last equation describes a line with slope of 1/sigma and intercept of
d′ /σ. Based on this fact, researchers have used linear regression of z-ROC
plots to estimate σ and d′ (e.g., Ratcliff, Shue & Grondlund, 1993). Specifi-
188
CHAPTER 7. THE NORMAL MODEL
cally:
d̂′ =
bˆ0
bˆ1
σ̂ = 1/bˆ1
To see how the regression technique is implemented, consider an experiment with a payoff manipulation. Payoff is manipulated through three level,
and for each level, there are 100 signal and noise trias. The number of hits
across the three conditions are 50, 70, 84, respectively; the number of false
alarms are 16, 30, and 50, respectively. The following code plots a zROC,
fits a line, and provides estimates of d′ and σ.
hit.rate=c(69,89,98)/100
fa.rate=c(12,23,38)/100
z.hit=qnorm(hit.rate)
z.fa=qnorm(fa.rate)
plot(z.fa,z.hit)
g=lm(z.hit~z.fa)
lines(g)
summary(g)
coef=g$coefficients #first element is intercept, second is slope
est.sigma=1/coef[2]
est.dprime=coef[1]/coef[2]
Estimates of the intercept and slope are 2.58 and 1.79, respectively; corresponding estimates of d′ and σ are 1.44 and .558, respectively.
We use the simulation method to assess the accuracy of this approach vs.
the ML approach. Consider a five-condition payoff experiment with true d′ =
1.3, true criteria c = (.13, .39, .65, .91, 1.17) and true σ = 1.2. Assume there
are 100 noise and 100 signal trials for each of the five conditions. For each
replicate experiment, data were generated and then d′ and σ were estimated
two ways: 1. from least squares regression, and 2. from the maximum
likelihood method discussed in Section x.x. Figure 7.8 shows the results.
The top panel shows a histogram of estimates of d′ for the two methods. The
likelihood estimators are more accurate; the regression method is subject to
an overestimation bias. The middle panel shows the same for σ; once again,
the likelihood method is more accurate. These differences in bias are not
7.5. REGRESSION
189
severe and it is reasonable to wonder if they are systematic. The bottom row
shows scatter plots. The x-axis value is the likelihood estimate for a replicate
experiment; the y-axis value is the regression method estimate. For every
replicate experiment, the regression method yielded greater estimates of d′
and σ showing that there is a systematic difference between the methods. In
sum, the ML method is more accurate because it does not suffer the same
degree of systematic over estimation.
Problem 7.5.3 (Your Turn)
Do the simulation described in the previous paragraph, then make a
figure similar to Figure 7.8 to present the results.
Why are the likelihood estimates better than the regression ones? The
regression method assumes that the regressor variable, the variable on the xaxis, is known with certainty. Consider the example in Your Turn in which we
regressed IQ gain (denoted Yi ) onto the number of days of treatment (denoted
Xi ) for autistic children. The x-axis variable, time in treatment, is, for each
participant, a constant that is known to exact precision. This statement
holds true regardless of the overall distribution of these times. This precision
is assumed in the regression model. Let’s explore a violation.. Suppose, due
to some sloppy bookkeeping, we had error-prone information about how long
each individual was in therapy. Let Xi′ be the length recorded, and let’s
assume Xi′ ∼ Normal(Xi , 5). We wish to recover the slope and intercepts
when we regress X onto Y . Since we don’t have true values Xi , we use our
error-prone values X ′ instead. You will show, in the following problem, that
the estimates of slope and intercept are biased. The bias in slope is always
toward zero; that is, estimated slopes are always flattened with respect to
the true slope.
Let’s return to the signal detection problem. Let’s say we wanted to study
subliminal priming. Subliminal priming is often operationalized as priming
in the absence of prime detectability (cite). For instance, imagine a paradigm
where trials are constructed as in Figure 7.9. The task is determine where a
given number is less-than or greater-than 5. With the sequence in Figure 7.9,
there are two possible types of trial: participants can be instructed to respond
190
3.0
2.0
Density
0.0
1.0
LS
1.0
2.0
ML
0.0
Density
3.0
CHAPTER 7. THE NORMAL MODEL
1.0
2.0
1.0
2.0
d’
Density
ML
0.5
1.5
2.5
sigma
3.5
0.0 0.5 1.0 1.5 2.0
0.0 0.5 1.0 1.5 2.0
Density
d’
LS
0.5
1.5
2.5
3.5
sigma
Figure 7.8: A comparison of maximum likelihood and least squares methods
of estimating signal detection parameters. The top row shows histograms of
1000 estimates of d′ with ML and LS, respectively; the bottom row shows
1000 estimates of σ with ML and LS. Blue (solid) vertical lines represent
mean estimates, and red (dashed) vertical lines represent true values.
191
7.5. REGRESSION
Foreperiod
578ms
Forward Mask
#####
Time
66ms
Prime
2
22ms
Backward Mask
#####
66ms
Target
6
200ms
Until
Response
Figure 7.9: The distribution of the IQ population and the distribution of the
sample mean, with N = 9.
to the prime or the target. When participants respond to the target, the
prime can actually affect decision to the prime. If both numbers are on the
same side of 5, responses are faster than if they are not.
In a subliminal priming paradigm, the primes are displayed fast enough
that it is difficult to see them. If there is a priming effect in the target
task when detectability in the prime task is 0, we call this subliminal priming. Although this is conceptually straightforward, actually establishing that
detectability is 0 is difficult.
One method proposed by Greenwald et al (cite) uses linear regression to
establish subliminal priming. In this method, priming effects are regressed
onto detectability (typically measured by d′ ), and a non-zero y intercept is
taken as evidence for subliminal priming. An example of this is shown in
Figure 7.10.
192
20
15
10
5
0
Priming Effect (RT)
25
30
CHAPTER 7. THE NORMAL MODEL
0.0
0.4
0.8
Detection (d’)
193
7.5. REGRESSION
Problem 7.5.4 (Your Turn)
Let’s examine the effects of a random predictor on the regression
method in subliminal priming through simulation. In order to do this,
we will assume that there is no subliminal priming; instead assume that
the relation ship between detectability and priming effect is
γi = d′i
d′i ≥ 0
(7.10)
(7.11)
In this case, the priming effect for the ith subject is linearly related to
detectability. Follow these steps:
1. Sample 15 true d′ from a normal with mean .5 and standard
deviation .3. Set any negative d′ values to 0. Assume that c =
d′ /2.
2. Sample each participant’s priming effect from a normal with mean
γi and standard deviation .1. Call these γ̂i .
3. Sample 50 signal and 50 noise trials for each person, and estimate
d′ . Call these d̂′i
4. Use linear regression to estimate the slope and intercept predicting γ̂i from d̂′i . Save the slope and intercept estimates and the p
values. Repeat these steps 1000 times.
5. Are the slope and intercept estimates biased? What is the true
type-I error rate for the intercept if you use p < .05 to reject the
null? What does this mean for inferences of subliminal priming?
Several of the examples in this chapter have involved specific instances
where traditional models such as least squares qregression may provide biased
estimates or incorrect inferences. This focus is not intended to dissuade
researchers from using traditional techniques. Rather, we believe that they
are important tools in research. It is important, however, to use the proper
tool in the proper situation. Whenever you decide to use a model, be sure
that you understand the assumptions of the model and ask whether they are
194
CHAPTER 7. THE NORMAL MODEL
appropriate for your application. Sometimes equal-variance normal errors
are not appropriate, and a more sophisticated technique is warranted. Other
times, ANOVA or regression will serve well. It is up to the researcher to
make an informed decision regarding the best analysis for the situation.
Chapter 8
Response Times
The main emphasis of this chapter and next is the modeling of response time.
Response time may be described as the time needed to complete a specified
task. Response time typically serves as a measure of performance. Stimuli
that yield lower response times are thought to be processed more mental
facility than those that yield higher response times. In this sense, response
time is often used analogously to accuracy, with the difference being that a
lower RT corresponds to better performance. Although this direct use of RT
is the dominant mode, there are a growing number of researchers who have
used other characteristics of RT to draw more detailed conclusions about
processing. This chapter and the next provide some of the material needed
to study RT models. The goal of this chapter is to outlay statistical models
of response time; the goal of the next is to briefly introduce a few process
models.
8.1
Graphing Distributions
Response time varies from response choice in that it is modeled as a continuous random variable. It is helpful to consider methods of looking at
or examining distributions of continuous data. Consider the simple case in
which a single participant observes stimuli in two experimental conditions.
We model the data in each condition as a sequence of independent and identicaly distributed random variables. For the first conditions, the observations
may be modeled as realization of a sequence independent and identically
195
196
CHAPTER 8. RESPONSE TIMES
distributed random variables X1 , .., XN ; e.g.,
iid
X1 , .., XN ∼ X
where N is the number of observations. Likewise, for the second condition,
the data may be modeled as
iid
Y1 , .., YM ∼ Y
The question at hand is to compare the distributions with graphical methods. Data in the first condition are given as
x1=c(0.794, 0.629, 0.597, 0.57, 0.524, 0.891, 0.707, 0.405, 0.808, 0.733,
0.616, 0.922, 0.649, 0.522, 0.988, 0.489, 0.398, 0.412, 0.423, 0.73,
0.603, 0.481, 0.952, 0.563, 0.986, 0.861, 0.633, 1.002, 0.973, 0.894,
0.958, 0.478, 0.669, 1.305, 0.494, 0.484, 0.878, 0.794, 0.591, 0.532,
0.685, 0.694, 0.672, 0.511, 0.776, 0.93, 0.508, 0.459, 0.816, 0.595)
The data in second conditon are given as
x2=c(0.503, 0.5, 0.868, 0.54, 0.818, 0.608, 0.389, 0.48, 1.153, 0.838,
0.526, 0.81, 0.584, 0.422, 0.427, 0.39, 0.53, 0.411, 0.567, 0.806,
0.739, 0.655, 0.54, 0.418, 0.445, 0.46, 0.537, 0.53, 0.499, 0.512,
0.444, 0.611, 0.713, 0.653, 0.727, 0.649, 0.547, 0.463, 0.35, 0.689,
0.444, 0.431, 0.505, 0.676, 0.495, 0.652, 0.566, 0.629, 0.493, 0.428)
Figures 8.1 through ?? shows several different methods of plotting the
data. Each of these methods shows a different facet of the data. We highlight
the relative advantage and disadvantage of each. In exploring data, we often
find it ueful to make multiple plots of the same data so that we may more
fully understand them.
8.1.1
Box Plots
The top left panel shows box plots of data in both conditions. Box plots
are a very good way of gainng an initial idea about distributions. The thick
middle line of the plot corresponds to the median; the box corresponds to
the 25th and 75th percentiles. The height of this box, the distance between the 25th and 75th percentile, is called the interquartile range. The
197
1.2
1.0
0.8
0.6
0.4
Response Time (sec)
8.1. GRAPHING DISTRIBUTIONS
Condition 1
Condition 2
Figure 8.1: Boxplots of two distributions.
whiskers extend to the most extreme point 1.5 times the inter-quartiel range
past the box. Observations outside the whiskers are denotes with a small
circles; these should be considered extreme observations. One advantage
of histograms is that several distributions can be compared at once. For
the displayed plots, it may be seen that the RT data in the second condition is quicker and less variable than that in the first condition. Boxplots are drawn in R with the boxplot() command; Figure 8.1 was drawn
with the command boxplot(x1,x2,names=c("Condition 1","Condition
2"),ylab="Response Time (sec)").
8.1.2
Histograms
The top panel of the Figure 8.2 shows histograms of the distributions for
each condition separately. We have plotted relative area histograms as these
converge to probability density functions (see Chapter 4). The advantage of
histograms are that they provide a detailed and intuitive approximation of
the desnity function. There are two main disadvantages to histograms: First,
it is difficult to draw more than one distribution’s histogram per plot. This
fact often makes it less convenient to compare distributions with histograms
than other graphical methods. For example, the comparison between the
two distributions is easier with two boxplots (Figure 8.1) than with two
histograms. The second disadvantage is that the shape of the histogram
depends on the choice of boundaries for the bins. The bottom row shows
what happens if the bins are chosen to finely (panel C) or too coarsely (panel
D). An alternative appoach to histograms is advocated by Ratcliff (1979)
who chooses bins with equal area instead of equal width. An example for
198
2.0
CHAPTER 8. RESPONSE TIMES
B
2.0
0.0
1.0
Density
1.0
0.0
0.5
Density
1.5
3.0
A
0.4
0.6
0.8
1.0
1.2
1.4
0.4
0.6
1.2
1.4
1.5
0.5
1.0
D
0.0
2
4
Density
6
C
0
Density
1.0
Response Time
8
Response Time
0.8
0.4
0.6
0.8
1.0
1.2
1.4
Response Time
0.4
0.6
0.8
1.0
1.2
Response Time
Figure 8.2: A & B: Histograms for the first and second conditions, respectively. C & D: Histogram for Condition 1 with bins chosen too finely (5 ms)
and too coarsely (400 ms), respectively.
Condition 1 is shown in Figure 8.3. Here, each bin consists of area .2 but
the width changes with height. Equal-area histograms may be drawn in R
by setting bins widths with the quantile command. The figure was drawn
with hist(x1,prob=T,main="",xlab="Response Time",xlim=c(.3,1.5),
breaks=quantile(x,seq(0,1,.2))).
8.1.3
Smoothed Histograms
Figure 8.4 (left) provides an example of smoothed or kernel-density histograms. One heuritic for smoothing is to consider a line drawn smoothly
between the midpoints of each bin. The key is the smoothness, the line is
1.4
199
1.0
0.0
0.5
Density
1.5
2.0
8.1. GRAPHING DISTRIBUTIONS
0.4
0.6
0.8
1.0
1.2
1.4
Response Time
0.8
0.4
Condition 1
Condition 2
0.0
Probability
1.0
2.0
Condition 1
Condition 2
0.0
Density
3.0
Figure 8.3: Equal-area alternative histogram for Condition 1.
0.2
0.4
0.6
0.8
1.0
1.2
Response Time (sec)
1.4
0.4
0.6
0.8
1.0
1.2
1.4
Response Time (sec)
Figure 8.4: Left: Smoothed histograms. Right: Empirical cumulative distribution functions.
constrained to change gradually. Smoothed histograms are drawn by a more
complex algorithm, but the heuristic is sufficeint for our purposes. The advantage of smoothed histograms is that they approximate density functions
and that several may be presented on a single grpah, facilitating comparsion.
The disadvantage is that smoothed histograms always have some degree of
distortion when compared to the true density function. They tend to be a bit
more variable than the true density and they cannot capture sharp changes.
The density() function smoothes histograms. The plot in the left panel is
drawn with plot(density(x1),xlim=c(.2,1.5),lty=2,lwd=2,main="",xlab="Response
Time (sec)") and the line is added with lines(density(x1),lwd=2).
200
8.1.4
CHAPTER 8. RESPONSE TIMES
Empirical Cumulative Distribution Plots
Figure 8.4 (right) provides an example of an empirical cumulative distribution
plot. These plots have several advantages. First, several distributions may be
displayed in a single plot fascilitating comparisons. Second, the analyst does
not need to make any choices as he or she does in drawing histograms (size
and placement of bins) or smoothed histograms (the amount of smoothing).
Third, empirical CDFs provide a more detailed display of the distribution
than box plots. The disadvantage is that these plots are not easy to interpret
at first. It takes experience to effectively read these graphs. We find that
after viewing several of these, they become nearly as intuitive as histograms.
Often, these graphs are very useful for exploring data and observing how
models fit and misfit data. Given these advantages, an empirical CDF plot
is a strong candidate for data representation in many contexts.
8.2
Descriptive Statistics About Distributions
In the previous section we described how to graphically display distributions.
There are also desciptive statistics used to descirbe distribution. In this
section we describe the two most common sets of these statistics: moments
and quantiles.
8.2.1
Moments
Moments are a standard set of statistics used to describe a distribution. The
terminology is borrowed from mechanics in which the moments of an object
described where its center of mass was, its momenton when spun, and how
much wobble it had when spun. Moments are numbered, that is, we speak
of the first moment, the second moment, etc. of a distribution. The first
moment of a distribution is its expected value; i.e., the first moment of RV
X is E(X) (see Eq. xx and yy). The first moment, therefore, is an index
of the center of middle of a distribution and it is estimated with the sample
mean. Higher moments come in two flavors, central and raw, and are defined
as:
Definition here (Richard?) [ Raw moments are given as E(X n ) where n
is an integer. The second and third raw moments, for example, are given by
E(X 2 ) and E(X 3 ). Central moments are g ]
8.3. COMPARING DISTRIBUTIONS
201
Central moments are far more common than raw moments. In fact, central moments are so common that they are sometimes refered to simply as the
moments, and we do so here. The second moment is given by E(X −E(X)2 ),
which is also the variance. Variance indexes the spread or dispersion of a
distributoin and is estimated with the sample variance (Eq. ). The third
moment and fourth moments are integral for the computation of skewness
and kurtosis, defined below.
Defn here: (Richard).
Skewness indexes assymetries in the shape of a distribution. Figure xx
provides an example. The normal distribution (solid line) is symmetric
around its mean. The corresponding skewness value is 0. The distribution
with the long right tails all have positive skewness values. If these distributions had skewed the other direction and had long left tails, the skewness
would be negative.
Kurtosis is especially useful for characterizing symmetric distributions. A
normal has.
In R, sample mean and variance is given by mean() and var(), respectively. Functions for skewness and kurtosis are not built in, but may defined
as:
RCODE of skewness and kurtosis.
8.2.2
Quantiles
In Chapter 3, we explored and defined quantile functions. One method of
describing a distribution is to simply list its quantiles. For example, it is not
characterize a distribution by its .1, .3, .5, .7, and .9 quantiles. This listing is
analogous to box plots and portrays, in list form, the same basic information.
8.3
Comparing Distributions
The main goal in analysis of RT is not to simply describe distributions but
to also compare them. We find it useful to consider three properties when
comparing distributions: location, scale, and shape:
202
8.3.1
CHAPTER 8. RESPONSE TIMES
Location
Figure 8.5 shows distributions that differ only in location. The left, center,
and right panels show the effects of locaton changes on probability density
functions, cumulative distribution functions, and quantile functions, respectively. The effect is most easily described with reference to probability and
cumulative distribution functions: a location effect is a shift or translation
of the entire distribution. Location changes are easiest to see in the quantile functions; the lines are parallel over all probabilities. (In the figure, the
lines may not appear parallel, but this appearance is an illusion. For every
probability, the top line is .2 seconds larger than the bottom one.) It is
straightforward to express location changes formally. Let X and Y be two
random variables with density functions denoted by f and g, respectively,
and cumulative distribution functions denoted by F and G. If X and Y
differ only in location, then the following relations hold:
Y
g(t)
G(t)
−1
G (p)
=
=
=
=
X + a,
f (t − a),
F (t − a),
a + F −1 (p),
for location difference a. Location is not the same as mean. It is true that if
two distributions differ in only location, than they differ in mean by the same
amount. The opposite, however, does not hold. There are several changes
that result in changes in mean besides location.
8.3.2
Scale
Scale refers to the dispersion of a distribution, but it is different than variance. Figure 8.6 shows three distributions that differ in scale. The top row
shows two zero-centered normals that differ in scale. The panels, from leftto-right, show density, cuulative distribution, and quantile functions. For
the normal, scale describes the dispersion from the mean (depicted with a
vertical dotted line in the density plot). The middle row show the case for
a Weibull distribution. The Weibull is a reasonable model for RT as it is
unimodal and skewed to the right; it is discussed subsequently. The depicted
Weibull distributions have the same location and shape; they differ only in
scale. For this distribution, scale describes the dispersion from the lowest
203
0.0
0.5
1.0
1.5
1.0
0.2
0.6
Time (ms)
0.8
0.4
0.0
1.0
0.0
Density
2.0
Cumulative Probability
8.3. COMPARING DISTRIBUTIONS
0.0
Time (sec)
0.5
1.0
1.5
Time (sec)
0.0
0.4
0.8
Cumulative Probability
Figure 8.5: Distributions that differ in location are shifted. The plots show,
from left to right, locaton changes in density, cumulative distribution, and
quanitle functions, respectively. The arrowed line at the right shows a difference of .2 sec; the distance separating the quantile functions for all probability
values.
value (depicted with a vertical line) rather than from the mean. The bottom
panel shows scale changes in an ex-Gaussian distribution, another popular
RT distribution that is described subsequently. For this distribution the scale
describes the amount of dispersion from a value that is near the mode (depicted with a vertical line). The quantile plot of the ex-Gaussian is ommited
as we do not know of a convenient expression for this function.
Y
= a + bX,
t−a
1
,
f
g(t) =
b b t−a
G(t) = F
,
b
G−1 (p) = a + bF −1 (p).
8.3.3
Shape
Shape is a catch-all category that refers to any change in distributions that
cannot be described as location and scale changes. Figure 8.7 shows some
examples of shape changes. The left panel shows two distributions; the one
with the dotted line is more symmetric than the one with the solid one.
204
0
1
2
3
Score
−3
−3
−2
−1
0.0
2.0
0.6
0.8
1.0
Time (ms)
0.2
0.0
0.5
1.0
1.5
0.0
0.2
0.4
0.6
0.8
0.8
Cumulative Probability
0.4
Cumulative Probability
1.5
0.4
0.0
4
3
2
1.0
Time (sec)
0.2
Cumulative Probability
Time (sec)
1
Density
3
0.8
1.5
0
0.5
2
0.4
Cumulative Probability
1.0
Time (sec)
0.0
1
0.0
4
3
2
Density
1
0.5
0
Score
0
0.0
−1
1
2
0.8
3
Score
1.0
−1
0.6
−2
0.4
Cumulative Probability
−3
0.0
0.4
0.0
Density
0.8
CHAPTER 8. RESPONSE TIMES
0.0
0.5
1.0
1.5
2.0
Time (sec)
Figure 8.6: Distributions that differ in scale. The plots show, from left to
right, scale changes in density, cumulative distribution, and quanitle functions, respectively. The top, middle, and bottom rows show scale changes for
the normal, Weibull, and ex-Gaussian distributions, respectively.
1.0
205
0.3
0.1
0.2
Density
1.0
Density
2
0.0
0.5
1.0
Time (sec)
1.5
0.0
0.0
1
0
Density
2.0
3
0.4
4
3.0
8.3. COMPARING DISTRIBUTIONS
0.0
0.5
1.0
1.5
2.0
−4
Time (sec)
−2
0
2
Score
Figure 8.7: Distributions that differ in shape.
The center and right panel shows more subtle shape differences. The center
panel shows the case in which the right tail of the dotted-line distribution
is stretched relative to the right tail fo the solid-line distribution. There
is no comparable stretching on the left side of the distributions. Because
the stretching is inconsistent, the effect is a shape change and not a scale
change. The right panel shows two symmetric distribution. Nonetheless,
they are different with the solid one having more mass in the extreme tails
and in the center. There is no stretching that can take one distribution into
the other; hence they have different shape.
8.3.4
The Practical Consequences of Shape
In some sense, shape is the most important of the three relationships in the
location, scale, shape trichotomy. From a statistical point of view, it often
makes little sense to compare the location and scale of distributions that
vary in shape. For example, it would make little sense to compare the effect
of scale across two conditions if the distribution in the first conditions was
normal and in the second condition was exponential. The ordering of scales
has no meaning when the shape changes. Conversely, comparisons of scale
and licensed whenever there is shape invariance. From a psychological point
of view, this type of constraint makes sense. Shape serves as an indicator of
mental processing: distributions with different shapes may indicate a change
in strategy or cognitive architecture. For example, the distribution of RT
from serial processing and parallel processing often vary in shape. It would
4
206
CHAPTER 8. RESPONSE TIMES
be fairly meaningless to talk about changes in overall scale across conditions
which differ in architecture. If two distributions vary in shape, than this
variability warrants further exploration. Why and how they differ are important clues to how the conditions affect mental processing. If shape does
not change, then the analyst may study how scale changes across conditions.
8.4
Quantile-Quantile Plots
Quantile-quantile plots (also called QQ plots) are epecially useful for assessing whether distributions differ in location, scale, or shape. QQ plots are
formed by plotting the quantiles of one distribution against that of another.
An example is constructed by considering the case presented in the beginning
of the chapter (page xx). Here a single participant provided 50 RTs in each
of two conditions. The data in the first and second condition were denoted
in R by vectors x1 and x2, respectively. The QQ plot is shown in Figure 8.8.
The first step in consrtucting it is sorting RTs from smallest to largest. For
each of the 50 observations there is a point. The first point (marked with
the lower of the three arrows) comes from the smallest RT in each condition;
the y-axis value is the smallest RT in the first condition, the x-axis value is
the smallest RT in the second condition. The next point is from the pair of
next smallest observations and so on. The graph is drawn by
range=c(.2,1.5)
par(pty=s)
plot(sort(x2),sort(x1),xlim=range,ylim=range)
The assignment of distributions to axes in the QQ plot is a matter of personal
choice. We prefer to draw the larger distribution on the y-axis and will
maintain this convention thoughout.
The reason this plot is called a quantile-quantile plot may not be immediately evident. Consider the median of each distribution. Whereas there are
50 observation in each condition, the median is estimated by the 25th observation. The upper arrow denotes the median from both conditions. This
point, therefore, is a median-median point. The middle arrow denotes same
for .25 quanile; the y-axis value is the estimate of the .25 quantile for the first
condition; the x-axis value is the estimate of the .25 quantile for the second
condition. Each point may be thought of likewise, they are plots of quantiles
of one distribution against the corresponding quantiles of another.
207
1.0
0.6
0.2
RT in Condition 1
1.4
8.4. QUANTILE-QUANTILE PLOTS
0.2
0.6
1.0
1.4
RT in Condition 2
Figure 8.8: Qauntile-quantile (QQ) plots of two distributions. The lower,
middle, and upper arrows indicate the smallest RT; the .25 quantile, and the
median, respectively.
One aspect that simplifies the drawing of qqplots in the above example
is that there are the same number of observations in each condition. QQ
plots can still be drawn when there are different numbers and R provides
a convenient function qqplot(). For example, suppose there were only 11
observations in the first condition: z=x1[1:11]. The QQ plot is drawn with
qqplot(x2,z). In this plot, there are 11 points. The function computes the
approriate 11 values from x2 corresponding to the same quantiles as the 11
points in z.
QQ plots graphical depcit the location-scale-shape loci of the effect of
variables. Figure 8.9 shows the relationships. The left column shows the
pdf (top)of two distributions that differ in location. The resulting QQ plot
(bottom) is a straight line with a slope of 1.0. The y-intercept indexes the
degree of location change. The middle colun shows the same for scale. The
QQ plot s a straight line and the slope denotes the scale change. In the figure,
the scale of the slow distribution is twice that of the fast one; the slope of
the QQ plot is 2. The bottom row shows the case for shape changes. If
shape changes, then the QQ plots are no longer straight lines and show some
208
CHAPTER 8. RESPONSE TIMES
degree of curvature. The QQ plot of the sample data x1 and x2 (Figure ??)
indicate that the primary difference between the conditions is a scale effect.
One drawback to QQ plots is that it is often visually difficult to inspect
small effects. Typical effects in subtle tasks, such as in priming, are on the
order of 30 ms or less. QQ plots are not ideally suited to express such small
effects because each axis must encompass the full range of the distribution,
which often encompasses a second or more. The goal then is to produce
a graph that, like the QQ plot, is diagnostic for location, scale, and shape
changes, but for which small effects are readily apperent. The solution is
the delta plot, which is shown in Figure 8.10. Like QQ plots, these plots
are derived from quantiles. The y-axis in these plots are difference between
quantiles; the x-axis is the average between quantiles. The R code to draw
delta plots is
p=seq(.1,.9,.1)
df=quantile(x1,p)-quantile(x2,p)
av=(quantile(x1,p)+quantile(x2,p))/2
plot(av,df,ylim=c(-.05,.25),
ylab="RT Difference (sec)",xlab="RT Average (sec)")
abline(h=0)
axis(3,at=av,labels=p,cex.axis=.6,mgp=c(1,.5,0))
We prefer to also indicate the cumulative probability associated with an
average on delta plots. This is indicated by the top axis and drawn by the
last line in the above R code.
Delta plots provide the same basic information as QQ plots. The topright panel shows the case when two distributions vary in location; the delta
plot is a straight horizontal line. The botom-left panel shows changes in
scale; the straigth line has positive slope. The bottom-right panels shows
changes in shape; the line has curvature.
The choice of average quantile on the x-axis of delta plot seems arbitrary.
A difference choices might be the cumulative probability of the quantiles
and this choice has been used in the literature (xx). It turns out there is a
justification for the use of average quantiles values, and this jusification is
demonstrated in Figure 8.11. First, note that the unfilled points are just the
QQ plot of the sample and their position is identical to Figure ??. These
solid points were obtained by simply rotating
√ the figure through 45 degrees.
The new x-axis value of each point is (1/ 2)(x1 (p) + x2 (p)); the new y-axis
209
0.5
1.0
1.5
3
1
0
1
0
0.0
2
Density
3
2
Density
1.5
1.0
0.0
0.5
Density
2.0
2.5
4
8.4. QUANTILE-QUANTILE PLOTS
0.0
1.0
1.5
0.0
Time (sec)
0.5
1.0
1.5
Time (sec)
1.2
1.0
0.8
0.6
Condition 2
1.0
0.8
0.6
Condition 2
0.8
0.6
0.4
0.4
0.4
0.2
Condition 2
1.0
1.2
1.2
Time (sec)
0.5
0.2
0.4
0.6
0.8
Condition 1
1.0
1.2
0.4
0.6
0.8
1.0
Condition 1
1.2
0.4
0.6
0.8
1.0
Condition 1
Figure 8.9: QQ plots are useful for comparing distributions. Left: Changes
in location affect the intercept of the QQ plot. Middle: Changes in scale
affect the slope of the QQ plot. Right: Changes in shape add curvature to
the QQ plot.
1.2
210
0.3
0.4
0.5
0.6
0.7
0.8
0.9
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
RT Difference (sec)
0.0
0.1
0.2
−0.1
−0.05
RT Difference (sec)
0.05
0.15
0.3
0.25
0.1
CHAPTER 8. RESPONSE TIMES
0.5
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.8
0.9
0.4
0.9
0.1
0.5
0.2
0.3
0.4
0.6
RT Average (sec)
0.5
0.6
0.7
0.7
0.8
0.8
0.9
−0.1
−0.05
RT Difference (sec)
0.05 0.10
RT Difference (sec)
0.1 0.2 0.3
0.4
0.15
0.1
0.6
0.7
RT Average (sec)
0.4
0.5
0.6
RT Average (sec)
0.7
0.8
0.40
0.45
0.50 0.55 0.60
RT Average
Figure 8.10: Delta plots are also useful for comparing distributions. TopLeft: Delta plot of two distributions. Top-Right: Location changes result in
a straight horizontal delta plot. Bottom-Left: Changes in scale are reflected
in an increasing slope in the delta plot. Bottom-Richt: Changes in shape
result in a curved delta plot.
0.65
0.70
211
−2
−1
0
1
2
8.5. STOCHASTIC DOMINANCE AND RESPONSE TIMES
−2
−1
0
1
2
Figure 8.11: A demonstration that delta plots are rotated QQ plots.
√
value is (1/ 2)(x1 (p) − x2 (p)), where x1 (p) and x2 (p) are the observed pth
quantile for
the new y-axis
√ observations x1 and x2 , respectively. Multiplying
√
value by 2 and dividing the new x-axis value by 2 yields the delta plot.
Hence, delta plots retain all of the good features of QQ plots, but are placed
on a more convenient scale.
8.5
Stochastic Dominance and Response Times
One of the most useful concepts in analysis of distributions is stochastic
dominance. If one distribution stochastically dominates another, then the
first isunambiguously bigger than the second. For example, the distributions
in Figure 8.5 show stochastic dominance; the one denoted by dotted lines
dominates the one denoted by solid lines. On contrast, the left and right
panels of Figure 8.7 show violations of stochastic dominance. In both panels,
neither of the compared distributions is unambiguously bigger than the other.
definition here.
Stochastic dominance is implicitly assumed in most statistical tests. For
example, t-tests, ANOVA, and regression stipulate that observations are normally distributed with equal variance. Under the equal variance assumption,
the specified relationship is a location change which is always stochastically
dominant. Stochastic dominance even belies nonparametric tests in statistics.
Without stochastic dominance, it is impossible to say that one distribution
is unambiguously greater than another.
Little attention has been paid to stochastic dominance in experimental
212
CHAPTER 8. RESPONSE TIMES
psychology. Most manipulations appear to obey stochastic dominance; if a
manipulation slows RT, it appears to do so across the distribution. For example Brown and Wagenmachers note that RT mean and standard deviation
tend to covary in several tasks. Even so, it is conceivable that stochastic dominance may be violated in a few priming paradigms. Heathcote, Mewhort,
and Popiel report an intriuging example from a Stroop task. In this task
participants must read the color of the ink that form words. In the concordant condition, the word describe the color; e.g., the word green is written in
green ink, the word red is written in red ink, and so on. In the inconcordant
condition, the word and its color mismatch, e.g.; the word green is written in
red ink. Response times are slower in the inconcordant than concordant conditions. Surprisingly, according to Heathcote, Mewhort, and Popiel (1991),
this effect may violate stochastic dominance. These researchers report that
concordancy speeds the fastest responses while slowing the slowest. This
type of an effect, however, is most likely rare; at least it is rarely reported.
8.6
Parametric Models
The previous RT properties and methods are nonparametric as no specific
form of random variables was assumed. The vast majority of analyses are
parametric; that is, the researcher assumes a particular form. In this book, we
have covered in depth the binomial, multinomial, and normal distributions.
These distributions are ill-suited for modeling RT. We introduce a few more
common choices: the ex-Gaussian, Weibull, log normal, gamma, and inverse
Gaussian. We focus more on how to fit these distributions rather than a
discussion of their successes and failures in the literature. In the following
chapter, we focus on select substantive models of response time.
8.6.1
The Wiebull
The three-parameter Weibull is a standard distribution in statistics with pdf
β
f (t; ψ, θ, β) =
θ
t−ψ
θ
!(
t−ψ
β − 1) exp −
θ
!β
,
θ, β > 0.
The parameters ψ, θ, and β are location, scale, and shape parameters, respectively. Figure 8.12 shows the effect of changing each parameter while
213
8.6. PARAMETRIC MODELS
leaving the remaining ones fixed. This figure provides one rationale for the
Weibull distribution; it is a convenient means of measuring location, scale,
and shape. The location parameter, ψ is the minimum value and the scale
parameter θ describes the dispersion of mass above this point. The Weibull
is flexible with regard to shape. When the shape is β = 1.0, the Weibull
reduces to an exponential. As the shape parameter increases past 1.0, the
Weibull becomes less skewed. At β = 3.43, the Weibull is approximately
symmetric. In general, shapes that characterize RT distributions lie between
β = 1.2 and β = 2.5. The Weibull is stochastically dominant for effects
in location and scale but not for in shape. Therefore, the Weibull is especially useful for modeling manipulations that affect location and scale, but
not those that affect shape. The Weibull has a useful process interpretation
which we cover in the following chapter.
Maximum likelihood estimation of the Weibull is straightforward as R already has built-in Weibull functions (dweibull(), pweibull(), qweibull(),
rweibull()). The log likelihood of a single observation t at parameters
(ψ, θ, β) may be computed by dweibull(t-psi, shape=beta, scale=theta,log=T).
The likelihood of a set of independent and identically distributed observations, such as x (page cc), is given by sum(dweibull(t-psi, shape=beta,
scale=theta,log=T)). Estimation of Weibull parameters is straightforward:
#par = psi,theta,beta
nll.wei=function(par,dat)
return(-sum(dweibull(data-par[1],shape=par[3],scale=par[2],log=T)))
par=c(min(x),.3,2)
optim(par,nll.wei,dat=x)
8.6.2
The lognormal
The lognormal is used similarly to the Weibull. The pdf is
1
(log(t − ψ) − µ)2
√ exp −
f (t; ψ, µ, σ ) =
2σ 2
(t − ψ)σ 2π
2
!
where parameters are (ψ, µ, σ 2 ). Parameter ψ and σ 2 are location and shape
parameters, respectively. Parameter µ is not a scale parameter, but exp µ
is. There reason for the use of µ and σ 2 in this context is as follows: Let
214
CHAPTER 8. RESPONSE TIMES
4
3.0
2.0
2
Weibull
0.0
0
0.0
1
1.0
1.0
Density
Shape
3
Scale
2.0
Location
0.6
1.0
1.4
0.2
0.6
1.0
1.4
0.6
1.0
1.4
3
2
2.0
1.5
Lognormal
0
0.0
1
1.0
0.0
0.6
1.0
1.4
0.2
0.6
1.0
1.4
Inverse
Gaussian
0.2
0.6
1.0
Time (sec)
1.4
0
0
0.0
1
2
2
4
1.5
3
8
0.2
6
1.4
5
1.0
3.0
0.6
4
Density
0.2
Density
0.2
3.0
3.0
0.2
0.2
0.6
1.0
Time (sec)
1.4
0.2
0.6
1.0
1.4
Time (sec)
Figure 8.12: Probability density functions for Weibull, lognormal, and inverse
Gaussian distributions. Left: Changes in shift parameters; Middle: Changes
in scale parameters; Right: Changes in shape parameters.
215
8.6. PARAMETRIC MODELS
Z be a normal random variable with mean µ and variability sigma2 . Let
T, response time, be T = ψ + log T . Then T is distributed as a log normal
with parameters (ψ, µ, σ 2). Figure 8.12 show how changes in parameters
correspond to changes in location, scale, and shape. Like the Weibull, the
log normal is stochastically dominant in location and scale parameters, but
not in the shape parameter. Hence, like the Weibull, it is a good model
for shift and scale change, but not for shape change. The main pragmatic
difference between the log normal and the Weibull is in the left tail of the
distribution. The log normal left tail is more curved and gradual than the
Weibull’s. Because scale is given by exp µ, reasonable values of µ are often
negative. The values of Figure 8.12 range from µ = −1.4 to µ = 1. Values
of shape σ typically range from .4 to 1.0. Negative loglikelihood is given as
nll.lnorm=function(par,dat)
-sum(dlnorm(dat-par[1],mu=par[2],sigma=par[3],log=T))
8.6.3
Inverse Gaussian
The inverse Gaussian, which is also known as the Wald, plays a proinant
role in substantive models of RT. Here, we focus on a location-scale-shape
parameterization given by
f (x|λ, φ) =
s
−((x − ψ)φ − λ)2
λ
.
(x − ψ)−3/2 exp
2π
2λ(x − ψ)
!
Here, parameters (ψ, λ, φ) are location, scale, and shape. The following R
code provides the pdf and negative log-likelihood of the inverse Gaussian.
#2-parameter inverse gaussian
dig=function(x,lambda,phi) #density of IG
sqrt(lambda/(2*pi))*x^{-1.5}*exp(-(x*phi-lambda)^2/(2*lambda*x))
nll.ig=function(par,dat)
-sum(log(dig(dat-par[1],lambda=par[2],phi=par[3])))
Reasonable values for shape parameter φ range from 1 to 20 or so.
One feature of the inverse Gaussian is that it is stochastically dominant in
all three parameters. This feature is in contrast to the Weibull and lognormal
216
CHAPTER 8. RESPONSE TIMES
which are only stochastically dominant in location and scale. Even though
this stochastic dominance seems to be an advantage, it is not necessarily so.
The inverse Gaussian does not change much in shape across wide ranges of
parameter settings. In fact, shape changes describe more subtle behavior of
the tail without much affecting the overall asymmetry of the distribution.
The consequence is that it is often difficult to estimate inverse Gaussian parameters. Figure ?? provides an illustration; here vastly different parameters
give rise to similar distributions. It is evident that it would take large sample sizes to distinguish the two distributions. The inverse Gaussian may be
called weakly identifiable because it takes large sample sizes to identify the
parameters.
8.6.4
ex-Gaussian
The ex-Gaussian is the most popular descriptive model of response time. It
is motivated by assuming that RT is the sum of two processes. For example,
Hohle (1965), who introduced the model, speculated that RT was the sum
of the time to make a decision and a time to execute a response. The first of
these two was assumed to be distributed as an exponential (see Figure 8.13),
the second was assumed to be a normal. The sum of an exponential and
a normal distribution is an ex-Gaussian distribution. The exponential component has a single parameter, τ , which describes its scale. The normal
component has two parameters: µ and σ 2 . The ex-Gaussian, therefore has
three components: µ, σ, and τ . Examples of the ex-Gaussian are provided
in Figure 8.13.
The ex-Gaussian pdf is given by
and programmed in R as
dexg<-function(t,mu,sigma,tau)
{
temp1=(mu/tau)+((sigma*sigma)/(2*tau*tau))-(t/tau)
temp2=((t-mu)-(sigma*sigma/tau))/sigma
(exp(temp1)*pnorm(temp2))/tau
}
With this pdf, ML estimates are easily obtained:
217
2.0
Density
1.0
3
0.0
0
0
1
1
2
Density
4
3
2
Density
5
6
4
8.6. PARAMETRIC MODELS
1.5
0.0
0.5
1.5
0.0
1.0
1.5
3.0
0.5
1.0
1.5
0.0
1.5
3.0
Density
3.0
2.0
0.0
Density
0.0
1.5
1.0
Time (sec)
1.0
2.0
1.0
1.0
Time (sec)
0.5
Time (sec)
0.0
0.5
1.5
0.0
0.0
Time (sec)
0.0
1.0
2.0
Density
3.0
2.0
Density
0.0
0.5
0.5
Time (sec)
1.0
2.0
Density
1.0
0.0
0.0
Density
1.0
Time (sec)
1.0
1.0
Time (sec)
2.0
0.5
1.0
0.0
0.0
0.5
1.0
1.5
Time (sec)
Figure 8.13: Some stuff here.
0.0
0.5
1.0
Time (sec)
1.5
218
CHAPTER 8. RESPONSE TIMES
nll.exg=function(par,dat)
-sum(log(dexg(dat,mu=par[1],sigma=par[2],tau=par[3])))
Hohle originally postulates a tight correspondance between mental processes and ex-Gausian components. Hohle and Taylor tested this correspondance by performing selective influence experiments; unfortunately, no selective influence was identified. In modern examples, the ex-Gaussian is used
descriptively: parameter µ is a measure of central tendency, parameter σ indexes the extent of the right tail, and parameter τ indexes the extent of the
left tail. Most researchers find that manipulations that lengthed RT do so,
primarily, by extending τ . Changes in τ and µ obey stochastic dominance;
changes in σ violate it.
In the standard parameterization, parameters σ and τ are both shape
parameters for the ex-Gaussian and there is no scale parameter. Scale, however, is preserved if changes in σ affect τ to the same degree. For example, if
both τ and σ double, then shape is preserved. Hence, it is possible to define
a new parameter η = στ as the shape of the ex-Gaussian. As η is increased,
the shape of the distribution becomes more skewed. The three parameters of
the ex-Gaussian may be expressed as (µ, σ, η). In this parameterization, the
parameters correspond to the location, scale, and shape of the distribution.
Figure 8.13 bottom row shows the effects of changing any one of these parameters. The distribution is stochastically dominant in location and shape but
stochastically indominant in scale. We do not recommend the ex-Gaussian
because stochastically-dominant scale changes tend to describe the effects of
many manipulations and participant variables. The following code provides
location, scale, and shape estimates for the ex-Gaussian:
nll.exg.lss=function(par,dat)
-sum(log(dexg(dat,mu=par[1],sigma=par[2],tau=par[3]*par[2])))
8.7
8.7.1
Analysis Across Several Participants
Group Level Analysis
Most studies are concerned with analysis across several participants. Response times cannot simply be aggregated across people. To see the harm
219
4
3
2
Density
2
0
0
1
1
Density
3
5
8.7. ANALYSIS ACROSS SEVERAL PARTICIPANTS
0.2
0.4
0.6
0.8
1.0
1.2
0.2
0.4
0.6
1.0
1.2
1.0
1.2
x2
3
1
2
Density
1.0
0
0.0
Density
2.0
4
x1
0.8
0.2
0.4
0.6
0.8
1.0
1.2
0.2
Binned
0.4
0.6
0.8
z−Transformed
Figure 8.14: Some stuff here.
of this practice, consider the example in Figure ??. The top row shows histograms of observations from two individuals. The bottom right shows the
case in which the observations are aggregated, that is, they are grouped together. The resulting distribution is more variable than those for the two
individuals. Moreover, it is bimodal where the distributions for the two
indeividuals are unimodal. In sum, aggregation greatly distorts RT distributions.
We address the question of how to draw a group-level distribution. Defining a group-level distribution is straightforward if shape is assumed not to
vary across participants and far more difficult otherwise. If shape is invariant,
220
CHAPTER 8. RESPONSE TIMES
then the jth participant’s response time distribution, Tj can be expressed as
Tj = ψj + θj Z,
where Z is a base random variable with a location of 0 and a scale of 1.0.
For example, if all participants had a Weibull distribution with shape β =
1.7, then Z would be distributed as a Weibull with density function f (t) =
1.7t.7 exp(−(t1 .7)). In the case that all participants have the same shape, the
group RT, denoted by S, is defined as
S = ψ̄ + θ̄Z.
There are two methods for plotting group-level distribution S without
committing to a particular form of Z. The first is quantile averaging.1 To
quantile-average a set of oberservations, the researcher first estimates quantiles in each distribution. For example, if x1 and x2 are data from two
participants, then we might estimate the deciles of each; e.g.,
decile.p=seq(.1,.9,.1)
x1.d=quantile(x1,decile.p)
x2.d=qunatile(x2,decile.p)
The next step is to average these deciles: group.d=(x1.d+x2,d)/2. Here
group.d are the deciles of the group distribution S that is defined above.
If we wish to know this distribution in greater detail, all we need to do
is increase the number of probabilities for which we compute quantiles.
While qauntile averagin is popular, there is an easier but equivalent method
to drawing group RT distributions when assuming shape invariance. For
each participant, simply take the z-transform of all scores. For example,
z1=(x1-mean(x1))/sd(x1) and z2=(x2-mean(x2))/sd(x2). The z-transformed
data may be aggregated, multiplied by the average standard deviation and
shifted by the average mean. For example,
mean.mean=mean(c(mean(x1),mean(x2))
mean.sd=mean(c(sd(x1),sd(x2)))
1
A related method to quantile averaging is Vincentizing. (cites) Instead of taking a
standard estimate of quantile, the researcher averages all observations in certain range. For
example, the .1 Vincentile might be the average of all observations above the .05 quantile
and below the .15 quantile. We do not recommend Vincentizing on princible. Quantile
estimators are well understood in statistics; vincentiles have never been explored.
8.7. ANALYSIS ACROSS SEVERAL PARTICIPANTS
221
s=mean.sd*(c(z1,z2))+mean.mean
hist(s)
If shape is invariant, then the group distribution are samples from S as
defined above. Quantile averaging and z-transforming yield identical results
in the limit of increasing number of observations per partipant and nearly
identical results with smaller sample sizes. For all practical purposes, they
may be considered equivalent.
The question of how to define a group-level distribution when shape
varies is difficult. A researcher can always perform quantile averaging or ztransforms; but the results are not particularly interpretable without shape
invariance. The problem is that if shape varies, the group-level distribution
will not belong to the same distribution as the individuals. If researchers
suspect that shape changes across people, then specific parametric models
are more appropriate than quantile averaging or z-transforms. The bottomleft plot of Figure 8.14 shows the group-level distribution derived from ztransformed for samples x1 and x2.
A related question is whether group-level distributions are helpful in estimating grouplevel parameters in specific parametric forms. Rouder and
Speckman explored this question in detail. If shape does not vary across
participants, then there is no advantage to drawing group-level distributions
before estimation. The question is more complex when shape does change.
As the number of observations per participant increases, estimating each participant’s parameters is advantageous. However, for small sample sizes, there
is stability in the group-level distribution that may lead to better parameter estimates. Rouder and Speckman show, for example, a large benefit of
quantile-averaging data when fitting a Weibull if there are less than 60-80
observations per participant.
8.7.2
Inference
In the previous section, we described how to draw goup level distributions.
These distributions are useful in exploring the possible loci of effects. In this
section, we discuss a different problem: how to draw the conclusion about
the loci of effects. If the researcher is willing to adopt a parametric model,
such as a Weibull or ex-Gaussian, the problem is well formed. For example,
suppose previous research has indicates that an effect is in parameter τ . We
descrbe how to use a likelihood ratio test for this example. We let Xijk
222
CHAPTER 8. RESPONSE TIMES
denote the response time for the ith participant (i = 1, . . . , I) observing a
stimulus in the jth condition (j = 1, 2) for the kth replicate (k = 1, . . . , K).
The general model is given by
Xijk ∼ ex-Gaussian(µij , σij , τij ).
An effect in τ may be tested against the restriction that τij does not vary
across conditions; i.e., τi = τi1 = τi2 .
Xijk ∼ ex-Gaussian(µij , σij , τi ).
The general model has 6I parameters; the restricted model has 5I parameters. Hence, the likelihood ratio test is I degrees of freedom. We provide the
code and an example of this type of test in the next section.
There is a strong caveat to this test. We model each particpant’s parameters as free to take on any value without constraint. For example, µij may
be any real value; σij and τij may be any positive real value. This approach
seems appropriate, it does not allow for the generalization to new participants. In statistical parlance, each participant’s parameters are treated as
fixed effects. The results of the likelihood ratio test generalize to repeated experiments with these specific participants. It is important to note that they
do not generalize to other participants. It is possible to generalize the model
such that inference can be made to new participants. The resulting model
is a hierarchical or multilevel one (c.f., Rouder et al., 2003). The analysis of
hierarchical models is outside the scope of this book.
A second approach which does provide for generalization to new participants is to model resulting parameter estimates. For example, it is conventional to see a t-test on ex-Gaussian parameters (cites). A paired t-test on
τ would imply the following null model:
τi2 − τi1 ∼ Normal(0, σ 2 ).
Researchers using the t-test should be sensitive to the normality and equalvariance assumptions in the model. If these are not met, an appropriate
nonparametric test, such as the rank-sign test (cite), may be used.
8.8
An example analysis
The existence of serveral options for modeling RT necessitates that the analyst make choices. Perhaps the most productive approach is to let the data
8.8. AN EXAMPLE ANALYSIS
223
themselves guide the choices. To illustrate, we consider the case in which 40
participants each read a set of nouns and verbs. The dependent measure is
the time to read a word, and each participant reads 100 of each type. The
main goal of analysis is to draw conclusions about the effect of nouns vs.
verbs.
Sample data are provided in file chapt6.dat. They may be read in with the
following command: dat=read.table(’chapt6.dat’,header=T). A listing
of dat will reveal 4 columns (subject, part-of-speech, replicate, response time)
and 4000 rows. In this example, we simulated the data from an inverse
Gaussian and built the effect of part-of-speech into scale. Therefore, we
hope to recover a scale effect and not shift or shape effects.
Had this set been obtained from participants, cleaning procedures would
have been needed to remove errors, premature responses, and inordinate
lapses in attention. We find it helpful to define the following vectors to aid
analysis:
sub=dat$sub
pos=dat$pos
rt=dat$rt
8.8.1
Analysis of Means
A quick analysis of means is always recommended. Figure ??, top row,
shows the relevant plots. The left plot is a line plot of condition means for
each participants. The following code computes the mean with a tapply()
command and plots the resultant matrix with a matplot() command:
ind.m=tapply(rt,list(sub,pos),mean) #Matrix of participant-by-pos means
I=nrow(ind.m) #I: Number of participants
matplot(t(ind.m),typ=’l’,col=’grey’,lty=1,axes=F,xlim=c(.7,2.3),
ylab=’Response Time (sec)’)
One new element in this code is the axes=F option in the matplot() statement. This option supresses the drawing of axes. When the axis are automatically plotted, values of 1 and 2 are drawn on the x-axis, which do not
have relavence to the reader. We manually add a more approriate x-axis
with the command axis(1,label=c(’Nouns’,’Verbs’),at=c(1,2)). The
“1” in the first argument indicates the x-axis. The y-axis and box are added
224
CHAPTER 8. RESPONSE TIMES
with axis(2) and box(), respectively. The final element to add are group
means:
grp.m=apply(ind.m,2,mean)
lines(1:2,m1,lwd=2)
The termination points are added with matpoints(t(ind.m),pch=21, bg=’red’).
t.test(ind.m[,1],ind.m[,2],paired=T). The right plot is a graphical display of the effect of part-of-speech. It shows a boxplot of each individual’s
effect along with 95% CI error bars around the mean effect. This graph was
drawn with
effect=ind.m[,2]-ind.m[,1]
boxplot(effect,col=’lightblue’,ylab="Verb-Noun Effect (sec)")
errbar(1.3,mean(effect),qt(.975,I-1)*sd(effect)/sqrt(length(effect)),.2)
points(1.3,mean(effect),pch=21,bg=’red’)
abline(0,0)
As can be seen, there is a 27 ms advantage for nouns; moreover, over 3/4 of
the 40 participants have noun advantage. The significance of the effect may
be confirmed with a t-test (for these data, t(39) = 6.03, p < .001).
8.8.2
Delta Plots
To explore the effect of part-of-speech on distributional relationships, we
drew a delta plot for each individual (Figure ??, bottom-left). We used a
loop to calculate deciles for each individual reading nouns and verbs and
stored the results in matrix noun and verb. The rows index individuals; the
columns index deciles:
decile.p=seq(.1,.9,.1)
noun=matrix(ncol=length(decile.p),nrow=I)
verb=matrix(ncol=length(decile.p),nrow=I)
for (i in 1:I)
{
noun[i,]=quantile(rt[sub==i & pos==1],p=decile.p)
verb[i,]=quantile(rt[sub==i & pos==2],p=decile.p)
}
0.00
−0.15
0.10
0.4
0.6
0.8
1.0
1.2
1.4
1.6
0.04
0.20
Nouns
0.02
Difference of Deciles
0.00
Difference in Deciles (sec)
0.6
0.8
1.0
1.2
−0.02
0.02
0.06
0.10
Verb−Noun Effect (sec)
0.4
Response Time (sec)
8.8. AN EXAMPLE ANALYSIS
225
Verbs
0.45
Average of Deciles (sec)
Figure 8.15: Some stuff here.
0.55
0.65
Average of Deciles
0.75
226
CHAPTER 8. RESPONSE TIMES
ave=(verb+noun)/2
dif=verb-noun
The last two lines are matrices for the delta plot: the difference between
deciles and the average of deciles. These matrices of deciles may be drawn
with the matplot(ave,dif,typ=’l’). The result is a colorful mess. The
problem is that matplot() treats each column as a line to be plotted. We
desire a line for each participant, that is, for each row. The trick is to
transpose the matrices. The approriate command, with a few options to
improve the plot, is
matplot(t(ave),t(dif),typ=’l’,
lty=1,col=’blue’,ylab="Difference in Deciles (sec)",
xlab="Average of Deciles (sec)")
abline(0,0)
These individual delta plots are too noisy to be informative. This fact
necessitates a group-level plot. We therefore assume that participants do not
vary in shape and quantile average deciles across people. Deciles of group
level distributions are given by
grp.noun=apply(noun,2,mean)
grp.verb=apply(verb,2,mean)
A delta plot from these group-level deciles is provided in the bottom-right
panel and drawn with
dif=grp.verb-grp.noun
ave=(grp.verb+grp.noun)/2
plot(ave,dif,typ=’l’,ylim=c(0,.05),
ylab="Difference of Deciles",xlab="Average of Deciles")
points(ave,dif,pch=21,bg=’red’)
abline(0,0)
The indication form this plot is that the effect of part-of-speech is primarily
in scale.
8.8. AN EXAMPLE ANALYSIS
8.8.3
227
Ex-Gaussian Analysis
We fit the ex-Guassian distributional model to each participant’s data.
Xij ∼ ex-Gauss(µij , σij , τij ),
where i = 1, . . . , I indexes participants and j = 1, 2 indexes part of speech.
The code to do so is:
r=0
est=matrix(nrow=I*J,ncol=7)
for (i in 1:I)
for (j in 1:J)
{
r=r+1
par=c(.4,.1,.1)
g=optim(par,nll.exg,dat=rt[sub==i & pos==j])
est[r,]=c(i,j,g$par,g$convergence,g$value)
}
%$
The matrix est has seven columns: a participant label, a condition label,
ML estimates of µ, σ, and τ , an indication of convergence, and a minimized
negative loglikelihood. All optimizations should converge and this facet may
be checked by confirming sum(est[,6]) is zero. Figure 8.16 (left) provides
boxplots on the effect of part-of-speech on each parameter. The overall effects
on µ, σ, and τ are 9.4 ms, 1.5 ms, and 16.6 ms, respectively. The overall
NLL for this model may be obtained by summing the last column; it is
Researchers typically test the effects of the manipulation on a parameter
by the t-test. For example, effect of part-of-speech 0n τ is marginally significant by a t-test (values here). This t-test approach differs from a liklihood
ratio test in two respects. On one hand, it is less princibled as the researcher
is forced to assume that the parameter estimates are normally distributed
and of equal variance. The likelihood ratio test provides for a more princibled alternative as it accounts for the proper sampling distributions of the
parameters. The above code provides estimates of the general models in
which all parameters are free to vary. The overall negative loglikelihood for
the general model may be obtained by summing the last column; it is xx.xx.
The restricted model is given by:
228
Mu
Sigma
0.6
0.2
−0.2
−0.6
Verb−Noun Effect (sec)
0.2
0.1
0.0
−0.2
Verb−Noun Effect (sec)
0.3
CHAPTER 8. RESPONSE TIMES
Tau
Shift
Scale
Figure 8.16: Some stuff here.
One interpretation of these analyses is that the effect of part-of-speech
is primarily in τ (a shape parameter). This interpretation would be incorrect. We generated the data with only scale effects. The achilles heal of
the ex-Gausssian is that scale effects violate stochastic dominance. The inverse Gaussian posits stochastic dominant scale effects. Figure ?? shows the
consequence. When scale changes, the effect on the ex-Gaussian is a large
change in τ and smaller changes in µ and σ. Indeed, this occurs with the
estimates from the data. Therefore, the ex-Gaussian is of limited use because
it frequently leads to the misinterpretation of scale effects as τ effects. We
believe this distribution should be used wth great caution. Moreover, we are
suspicious of many of the reported effects in τ in the literature may be in
fact stochastically-dominant scale effects.
We also performed a Weibull distribution analysis on each individual’s
data. The results are shown in Figure ?? (right). The shift and shape effect
are insignificant; the scale effect is more significant than appears. Scale
effects are multiplicative, hence the ratio is the best way to compare scales.
We computed the ratio of the scale of the verb RT distribution to the scale
of the noun RT distribution for each participant. These scales are shown in
Figure ??. Notice that the spacing is not equidistant; that is ... Significance is
most appropriately assesed on the log scale—in this case, we use the following
model:
!
θiV
log
∼ Normal(µ, σ 2 ),
θiN
where θiN and θiV are the ith participant’s Weibu
Shape
8.8. AN EXAMPLE ANALYSIS
the effect of part-of-speech on scale is significant ().
229
230
CHAPTER 8. RESPONSE TIMES
Bibliography
[1] N. D. Anderson, F. I. M. Craik, and M. Naveh-Benjamin. The attentional demands of encoding and retrieval in younger and older adults: I.
evidence from divided attention costs. Psychology & Aging, 13:405–423,
1998.
[2] W. H. Batchelder and D. M. Riefer. Theoretical and empirical review of
multinomial process tree modeling. Psychonomic Bulletin and Review,
6:57–86, 1999.
[3] R. Brent. Algorithms for Minimization without Derivatives. PrenticeHall, Englewood Cliffs, N.J., 1973.
[4] A. Buchner, E. Erdfelder, and B. Vaterrodt-Plunneck. Toward unbiased
measurement of conscious and unconscious memory processes within the
process dissociation framework. Journal of Experimental Psychology:
General, 124:137–160, 1995.
[5] F. R. Clarke. Constant ratio rule for confusion matrices in speech communications. Journal of the Accoustical Society of America, 29:715–720,
1957.
[6] T. F. Cox and M. A. A. Cox. Multidimensional Scaling, 2nd Edition.
Chapman and Hall / CRC press, Boca Raton, FL, 1994.
[7] A. P. Dempster, N. M. Laird, and D. B. Rubin. Maximum liklihood from
incomplete data via the em algorithm. Journal of the Royal Statistical
Society, Series B, 39:1–38, 1977.
[8] S. Glover and P. Dixon. Likelihood ratios: a simple and flexible statistic
for empirical psychologists. Psychonomic Bulletin & Review, 11:791–
806, 2004.
231
232
BIBLIOGRAPHY
[9] P. Graf and D. L. Schacter. Implicit and explicit memory for new associations in normal and amnesic subjects. Journal of Experimental
Psychology: Learning, Memory, and Cognition, 11:501–518, 1985.
[10] M. J. Hautus and A. L. Lee. The dispersions of estimates of sensitivity
obtained from four psychophysical procedures: Implications for experimental design. Perception & Psychophysics, 60:638–649, 1998.
[11] E. Herman, J. L.; Schatzow. Recovery and verification of memories of
childhood sexual trauma. Psychoanalytic Psychology, 4:1–14, 1987.
[12] R. V. Hogg and A. T. Craig. Introduction to mathematical statistics.
MacMillan, New York, 1978.
[13] X. Hu and W. H. Batchelder. The statistical analysis of general processing tree models with the em algorithm. Psychometrika, 59:21–47,
1994.
[14] L. L. Jacoby. A process dissociation framework: Separating automatic
from intentional uses of memory. Journal of Memory and Language,
30:513–541, 1991.
[15] Gideon Keren and Stan Baggen. Recognition models of alphanumeric
characters. Perception & Psychophysics, 29:234–246, 1981.
[16] A. N. Kolmogorov. Foundations of the theory of probability. Chelsea,
New York, 1950.
[17] E. L. Lehmann. Theory of point estimation. Wadsworth, 1991.
[18] E. F. Loftus. The reality of repressed memories. American Psychologist,
48:518–537, 1993.
[19] R. D. Luce. Individual Choice Behavior. Wiley, New York, 1959.
[20] R. D. Luce. Detection and recognition. In R. D. Luce, R. R. Bush,
and E. Galanter, editors, Handbook of mathematical psychology (Vol.
1). Wiley, New York, 1963.
[21] R. D. Luce. A threshold theory for simple detection experiments. Psychological Review, 70:61–79, 1963.
BIBLIOGRAPHY
233
[22] R. D. Luce, R. M. Nosofsky, D. M. Green, and A. F. Smith. The bow and
sequential effects in absolute identification. Perception & Psychophysics,
32:397–408, 1982.
[23] R. A. J. Matthews. Tumbling toast, Murphy’s law, and fundamental
constants. European Journal of Physics, 16:172–175, 1995.
[24] J. A. Nelder and R. Mead. A simplex method for function minimization.
Computer Journal, 7:308–313, 1965.
[25] R. M. Nosofsky.
Attention, similarity, and the identificationcategorization relationship. Journal of Experimental Psychology: General, 115:39–57, 1986.
[26] W. H. Press, S. A. Teukolsky, W. T. Vetterling, and F. P. Flannery.
Numerical Recipes in C: The art of Scientific Computing. 2nd Ed. Cambridge University Press, Cambridge, England, 1992.
[27] J. Rice. Mathematical statistics and data analysis. Brooks/Cole, Monterey, CA, 1998.
[28] D. M. Riefer and W. H. Batchelder. Multinomial modeling and the
measure of cognitive processes. Psychological Review, 95:318–339, 1988.
[29] J. N. Rouder. Absolute identification with simple and complex stimuli.
Psychological Science, 12:318–322, 2001.
[30] J. N. Rouder. Modeling the effects of choice-set size on the processing
of letters and words. Psychological Review, 111:80–93, 2004.
[31] J. N. Rouder and W. H. Batchelder. Multinomial models for measuring
storage and retrieval processes in paired associate learning. In C. Dowling, F. Roberts, and P. Theuns, editors, Progress in mathematical psychology. Erlbaum, Hillsdale, NJ, 1998.
[32] J. N. Rouder and R. D. Morey. Relational and arelational confidence
intervals: A comment on Fidler et al. (2004). Psychological Science,
16:77–79, 2005.
[33] J. N. Rouder, D. Sun, P. L. Speckman, J. Lu, and D. Zhou. A hierarchical Bayesian statistical framework for response time distributions.
Psychometrika, 68:587–604, 2003.
234
BIBLIOGRAPHY
[34] D. L. Schacter. Perceptual representation systems and implicit memory:
Toward a resolution of the multiple memory systems debate. Erlbaum,
Hillsdale, NJ, 1990.
[35] R. N. Shepard, A. K. Romney, and S. B. Nerlove. Multidimensional
scaling: Theory and applications in the behavioral sciences: I. Theory.
Seminar Press, Oxford, 1972.
[36] Roger N. Shepard. Stimulus and response generation: A stochastic
model relating generalization to distance in a psychological space. Psychometrika, 22:325–345, 1957.
[37] J. G. Snodgrass and J. Corwin. Pragmatics of measuring recognition
memory: Applications to dementia and amnesia. Journal of Experimental Psychology: General, 117:34–50, 1988.
[38] L.R. Squire. Declarative and nondeclarative memory: Multiple brain
systems supporting learning and memory. In D.L. Schacter and E. Tulving, editors, Memory systems 1994. MIT Press, Cambridge, MA, 1994.
[39] W. S. Torgerson. Theory and methods of scaling. Wiley, New York,
1958.
[40] A Tversky. Elimination by aspects: A theory of choice. Psychological
Review, 79:281–299, 1972.

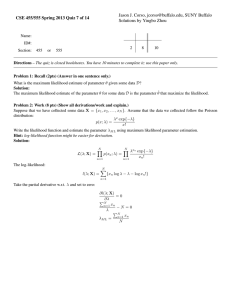
![School Risk Register [DOCX 19.62KB]](http://s2.studylib.net/store/data/014980461_1-ba10a32430c2d15ad9059905353624b0-300x300.png)


