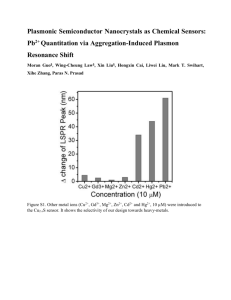
Miniaturized Lead Sensor Based on Lead
advertisement

University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln US Army Research US Department of Defense 1-1-2005 Miniaturized Lead Sensor Based on Lead-Specific DNAzyme in a Nanocapillary Interconnected Microfluidic Device In-Hyoung Chang Joseph J. Tulock Juewen Liu Won-Suk Kim Donald M. Cannon, Jr. See next page for additional authors Follow this and additional works at: http://digitalcommons.unl.edu/usarmyresearch Part of the Operations Research, Systems Engineering and Industrial Engineering Commons Chang, In-Hyoung; Tulock, Joseph J.; Liu, Juewen; Kim, Won-Suk; Cannon, Jr., Donald M.; Lu, Yi; Bohn, Paul W.; Sweedler, Jonathan V.; and Cropek, Donald M., "Miniaturized Lead Sensor Based on Lead-Specific DNAzyme in a Nanocapillary Interconnected Microfluidic Device" (2005). US Army Research. Paper 8. http://digitalcommons.unl.edu/usarmyresearch/8 This Article is brought to you for free and open access by the US Department of Defense at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in US Army Research by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Authors In-Hyoung Chang; Joseph J. Tulock; Juewen Liu; Won-Suk Kim; Donald M. Cannon, Jr.; Yi Lu; Paul W. Bohn; Jonathan V. Sweedler; and Donald M. Cropek This article is available at DigitalCommons@University of Nebraska - Lincoln: http://digitalcommons.unl.edu/usarmyresearch/8 Environ. Sci. Technol. 2005, 39, 3756-3761 Miniaturized Lead Sensor Based on Lead-Specific DNAzyme in a Nanocapillary Interconnected Microfluidic Device IN-HYOUNG CHANG,† JOSEPH J. TULOCK,† JUEWEN LIU,† WON-SUK KIM,† DONALD M. CANNON, JR.,† YI LU,† PAUL W. BOHN,† JONATHAN V. SWEEDLER,† AND D O N A L D M . C R O P E K * ,‡ Department of Chemistry and the Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, 600 South Mathews Avenue, Urbana, Illinois 61801, and Construction Engineering Research Laboratory, U.S. Army Engineer Research and Development Center, Champaign, Illinois 61822 A miniaturized lead sensor has been developed by combining a lead-specific DNAzyme with a microfabricated device containing a network of microfluidic channels that are fluidically coupled via a nanocapillary array interconnect. A DNAzyme construct, selective for cleavage in the presence of Pb2+ and derivatized with fluorophore (quencher) at the 5′ (3′) end of the substrate and enzyme strands, respectively, forms a molecular beacon that is used as the recognition element. The nanocapillary array membrane interconnect is used to manipulate fluid flows and deliver the small-volume sample to the beacon in a spatially confined detection window where the DNAzyme is interrogated using laser-induced fluorescence detection. A transformed log plot of the fluorescent signal exhibits a linear response (r2 ) 0.982) over a Pb2+ concentration range of 0.1-100 µM, and a detection limit of 11 nM. The sensor has been applied to the determination of Pb2+ in an electroplating sludge reference material, the result agreeing with the certified value within 4.9%. Quantitative measurement of Pb2+ in this complex sample demonstrates the selectivity of this sensor scheme and points favorably to the application of such technologies to analysis of environmental samples. The unique combination of a DNAzyme with a microfluidic-nanofluidic hybrid device makes it possible to change the DNAzyme to select for other compounds of interest, and to incorporate multiple sensing systems within a single device for greater flexibility. This work represents the initial steps toward creation of a robust field sensor for lead in groundwater or drinking water. Introduction Bioavailable lead is a toxic element, linked to a variety of adverse health effects (1-3). Since lead is not biodegradable, * Corresponding author phone: (217) 373-6737; fax: (217) 3737222; e-mail: Donald.M.Cropek@erdc.usace.army.mil. † University of Illinois at Urbana-Champaign. ‡ U.S. Army Engineer Research and Development Center. 3756 9 ENVIRONMENTAL SCIENCE & TECHNOLOGY / VOL. 39, NO. 10, 2005 it accumulates in the environment and produces toxic effects in plants and animals, even at low concentrations (4-6). With growing understanding of the health effects of lead, the U.S. government has increasingly become involved in addressing the lead threat, and new regulations have been created for lead. In the Clean Air Act Amendment of 1990, the EPA designated Pb2+ as an “air toxic”, meaning that it may cause serious health and environmental hazards when present as an airborne pollutant (3-7). Also, new guidance from the Department of Defense Directive 4715 (8) requires a high degree of management and monitoring of impact areas on test firing ranges where lead accumulates due to use of lead-containing munitions. In addition, lead has been listed as a pollutant of concern in EPA’s Great Water Program (9) due to its persistence in the environment, potential for bioaccumulation, and toxicity to humans and the environment. These examples demonstrate that the focus of lead monitoring has been extended from high-dose effects for workers in an industrial environment to potential health and ecological hazards in the global habitat and ecosystem. It is therefore essential that sensitive, reliable, and cost-effective analytical methods are developed for the remote monitoring of lead. Miniaturization is currently an important trend in environmental monitoring due to its potential to reduce cost, provide portability, and increase analysis speed. In recent years, considerable interest has been focused on the development of miniaturized microfluidic (lab-on-a-chip) systems (10-13) offering improved analytical performance metrics such as inducing fast and efficient chemical reactions within small volumes and low manufacturing costs. A further benefit of miniaturization is the reduction in reagent and sample consumption and a subsequent reduction in the quantity of waste produced. Several approaches have been published for the separation and simultaneous determination of metal ions based on microfluidic devices (14-21). For example, Jacobson et al. have demonstrated a successful separation of Zn, Cd, and Al with detection limits in the 10-100 ppb range using a capillary electrophoresis (CE) microchip (16). Deng and Collins utilized colorimetric detection, and six heavy metals including Pb2+ were effectively separated and simultaneously determined on a CE microchip with sub parts per billion detection limits after preconcentration by solidphase extraction (21). The use of biosensors for environmental pollution monitoring has been another growing area, as these devices provide rapid, simple, and reliable determination of pollutants at trace levels. Novel lead-specific biosensors have been developed (22-27). Lu et al. (22, 26-28) developed a new biosensor for lead by combining the high selectivity of a DNAzyme with a molecular beacon strategy to achieve sensitive and quantitative fluorescent detection of Pb2+ over a wide concentration range from 10 nM to 10 µM. This study demonstrates the combination of a Pb2+specific DNAzyme biosensor with a multilevel nanofluidicmicrofluidic hybrid device, in which a nanocapillary array membrane is used to control motion of picoliter-volume fluid voxels from the analyte-containing sample stream to the biosensor compartment. These devices employ a membrane containing an array of nanocapillaries located between multilayered microfluidic channels, allowing for the convenient and efficient control of fluids in the device (29-31). In this paper, methods for adapting this lead-selective DNAzyme to the nanofluidic device are explored, an optimal protocol is identified, and the analytical figures of merit including dynamic range, limit of detection, accuracy, and 10.1021/es040505f CCC: $30.25 This article is a U.S. government work, and is not subject to copyright in the United States. 2005 American Chemical Society Published on Web 04/15/2005 FIGURE 1. The sensor is composed of a dual-labeled cleavable substrate DNA whose 5′ and 3′ ends are labeled with a fluorophore (FAM) and a quencher (Dabcyl), respectively, and an enzyme strand whose 3′ end is labeled with a Dabcyl. Initially, the fluorescence of FAM is quenched because of the close proximity of the Dabcyl. In the presence of Pb2+, the substrate DNA is cleaved, resulting in the release of fragments and a concomitant increase in fluorescence. precision are determined. Finally, the microfluidic/DNAzyme molecular beacon is successfully applied to analysis of Pb2+ in an electroplating sludge certified reference material. Experimental Section Materials. Lactic acid and ammonium hydroxide were obtained from Fisher Scientific (Fair Lawn, NJ). HEPES (N(2-hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid)), sodium chloride, and sodium hydroxide were purchased from Aldrich (Milwaukee, WI). Prepolymer and curing agent (Sylgard 184, Dow Corning Corp., Midland, MI) and polycarbonate nuclear track-etched (PCTE) membrane with a hydrophilic wetting layer of poly(vinylpyrrolidine) (Osmonics, Minnetonka, MN) were used in the PDMS (poly(dimethylsiloxane)) chip. These PCTE membranes are 10 µm thick with 200 nm diameter pores at a pore density of 3 × 108 pores/cm2. The lead stock solutions (1000 mg/L) were purchased from Fisher Scientific as an atomic absorption standard solution in 2% HNO3. Working solutions of lower concentration were prepared by serial dilution of the stock solution with a background electrolyte (BGE). The BGE (25 mM lactate, 25 mM HEPES, and 50 mM NaCl) was prepared by dissolving lactic acid, HEPES, and NaCl in deionized water (18.2 MΩ, Milli-Q UV-plus system, Millipore, Bedford, MA). The pH of the electrolyte was adjusted to 7 with ammonium hydroxide. Calibration of the chip sensor was performed using seven different lead concentrations. All reagents were analytical grade or higher. Preparation of DNAzyme. The fluorescently labeled oligonucleotides were purchased from Integrated DNA Technology Inc. (Coralville, IA). The design of lead DNAzymes is described in detail by Lu et al. (28). Briefly, a 3′ end Dabcyl (4-(4′-dimethylaminophenylazo)benzoic acid) labeled enzyme strand, termed 17E-Dy, and a 5′ end FAM (6-carboxyfluorescein) and 3′ end Dabcyl labeled cleavable DNA substrate, termed 17DS-FD, were chosen (Figure 1). The DNAzyme enzyme-substrate complex was prepared with 500 nM concentrations of both 17E-Dy and 17DS-FD for laser-induced fluorescence (LIF) measurements and 2.5 µM concentrations of both the enzyme and substrate for fluorescence microscopy studies. A sample of the enzyme and substrate was heated at 90 °C for 2 min and slowly cooled to 5 °C for 2 h to anneal the strands together and create the complex. Sample Preparation. The electroplating sludge sample was purchased from Resource Technology Corp. (Laramie, WY) and prepared for analysis by USEPA Method 3050B (32). The sludge sample was thoroughly mixed, dried, and ground immediately before use. For the digestion, 2.5 mL of concentrated HNO3 and 10 mL of concentrated HCl were added to 0.1 g of sample, and the mixture was refluxed for 15 min on a hot plate. The digestate was filtered through filter paper (Whatman No. 41), and the filtrate was collected in a volumetric flask. The filter paper and the residue were both washed with 5 mL of hot HCl. These washings were FIGURE 2. Schematic of a three-dimensional nanocapillary array interconnect (NAI) gateable microfluidic device. Both crossed microfluidic channels are identical with dimensions of 50 µm width, 30 µm depth, and 14 mm length. collected in the same flask. The filter paper and residue were removed and placed back in the reflux beaker. A 5 mL portion of concentrated HCl was added, and the mixture was heated at 95 ( 5 °C until the filter paper dissolved. The residue was filtered, and the filtrate was collected in the same flask. The cover and sides of the reflux beaker were washed with HCl, and this solution was also added to the flask. A control sample was prepared by following the entire sample preparation procedure without sludge. The sample was diluted 1:1 using concentrated ammonium hydroxide followed by a 1000-fold dilution with background electrolyte before injection on the microchip. The lead concentration was determined on the basis of the actual weight of the dried sludge sample and the final dilution volumes. Microfluidic Device and Measurement System. Details of the channel layout and fabrication of multilevel microfluidic-nanofluidic hybrid architectures have been provided previously (29). A three-dimensional transport device is depicted schematically in Figure 2. Two identical channels (50 µm wide, 30 µm deep, and 14 mm long) were orthogonally oriented on a PDMS microchip and separated at the intersection by a nanocapillary array interconnect (NAI), the PCTE membrane with 200 nm diameter cylindrical pores. Platinum wires (250 µm diameter, Goodfellow Corp., Berwyn, PA), mounted into reservoirs at the distal ends of the microchannels, were used to apply bias voltages. An eightrelay system, designed to switch electrical contacts between Pt electrodes and high-voltage power supplies (Bertan High Voltage, Hicksville, NY) for different configurations and magnitudes of microfluidic manipulation, was computer controlled via a multifunction data acquisition card (DAQ; National Instruments Corp., Austin, TX) and Labview software (National Instruments Corp.). Fluorescence microscopy was used for signal acquisition using an inverted Olympus epi-illumination microscope (Melville, NY). The CCD camera (Javelin Ultrichip Hi Res, Torrance, CA) output was recorded with a videocassette recorder and a computer-controlled video capture device (ATI Technologies, Markham, Ontario, Canada). Fluorescence was excited with 488 nm radiation from an Ar+ ion laser (Innova 300, Coherent Inc., Santa Clara, CA), which is very close to the FAM 492 nm excitation maximum. The laser light was passed through a set of irises and a neutral density filter (Newport, Irvine, CA) before reaching a dichroic mirror (505DCLP, Chroma Technology Corp., Brattleboro, VT). The excitation light was focused by a 10× objective for a 50 µm diameter area of interrogation. Fluorescence signals were collected by the same lens and dichroic mirror assembly and optically filtered through a 100 µm pinhole and band-pass filter (HQ525/50m, Chroma Technology Corp.) that permits passage of the 518 nm FAM emission maximum before being VOL. 39, NO. 10, 2005 / ENVIRONMENTAL SCIENCE & TECHNOLOGY 9 3757 detected by a photomultiplier tube (PMT) (HC124, Hamamatsu Corp., Bridgewater, NJ). Control of the PMT data collection was achieved through a computer with a Labview program and DAQ card (National Instruments Corp.). All fluorescence signals were collected at the intersection of the crossed channels, i.e., just below the nanocapillary array interconnect membrane. Results and Discussion The goal of this study is to adapt the Pb2+-specific DNAzyme concept to a nanofluidic reagent delivery and detection scheme to realize a robust, sensitive, regenerable platform for sensing of Pb2+. The specific recognition of Pb2+ by the DNAzyme is a necessary, but not sufficient, condition to realize a biosensor. For sensing, the necessary elements of reagent delivery and signal generation and recording must be added. Achieving these objectives requires several advances over the macroscale homogeneous DNAzyme assays, including (a) using solutions compatible with electrokinetic control of fluid motion, (b) accomplishing the molecular recognition reaction efficiently within a small (<100 pL) volume, and (c) regenerating the analytical reagent, i.e., active form of the DNAzyme. After optimization of the composition and device geometry, the overall system is characterized in terms of common analytical figures of merit and validated using a standard reference material. Pb2+-Specific DNAzyme and Electrolyte Optimization. The synthesis and characterization of the Pb2+-specific DNAzyme were described earlier in detail (28). The enzyme DNA strand is a 33-base oligomer, and the substrate DNA strand is a 20-base DNA/RNA chimer with a single RNA base whose specific sequences have been previously determined (22, 28). Briefly, the molecular beacon is constructed by labeling the 5′ end of the cleavable substrate with the fluorophore FAM and the 3′ end with a fluorescence quencher, Dabcyl, and labeling the 3′ end of the enzyme strand with Dabcyl as shown in Figure 1. When the substrate (17DS-FD) is hybridized to the enzyme strand (17E-Dy), the fluorescence of FAM is quenched by inter- and intramolecular Dabcyl. The melting temperature of the uncleaved substrate is designed to be above room temperature (∼34 °C) so that the substrate will not melt from the enzyme strand in this hybridized state. On addition of Pb2+ (the right-hand side of Figure 1), the substrate is cleaved at the RNA base site. Upon cleavage, the melting temperatures of the shorter parts of the substrate are designed to be below room temperature so that it will melt and dissociate from the enzyme strand (28). Thus, FAM is no longer in close proximity to the Dabcyl quenchers, and the fluorescence increases with the concentration of Pb2+. In a previous paper, the substrate cleavage reaction was monitored using fluorescence spectroscopy that illustrated the excellent sensitivity, dynamic range (quantifiable detection in the range of 10 nM < [Pb2+] < 10 µM), and selectivity (at least an 80-fold selectivity enhancement over other divalent metals) for Pb2+ (22). Optimizing the composition of the background electrolyte (BGE) is critical, because the composition affects biosensor performance and detection efficiency. The electrolyte must allow controllable fluidic transfer of lead-containing analyte solution and delivery of the hybridized DNA strands (DNAzyme) and sample to the detection window while also enabling-high efficiency cleavage of the DNA substrate in the presence of Pb2+. Future generations of the device shown in Figure 2 may include four fluidic channels: an injection channel that has a continually refreshed sample stream, a separation channel that can act as a capillary electrophoresis column to isolate the analyte from other matrix components, the nanofluidic gate PCTE membrane, and a detection channel containing the DNAzyme. Solution flow along any channel can be controlled via potential bias application. 3758 9 ENVIRONMENTAL SCIENCE & TECHNOLOGY / VOL. 39, NO. 10, 2005 Injection of a sample band onto the separation (source) channel merely requires the application of the correct bias along the ends of the injection and separation channels. The migration rate of the analyte must be characterized to know its movement along the separation channel and the time interval when the analyte is present at the NAI. Another bias change will divert solution flow through the PCTE membrane and thus introduce the analyte into the detection channel. The analyte is simultaneously removed from potential matrix interferents and established in the appropriate detection electrolyte. Clearly, this proposed device may require one electrolyte for optimal electrophoretic separation and a second electrolyte for effective DNAzyme cleavage. Current research is investigating the ability of the NAI to segregate these cross-channel solutions. While electrophoretic separation is not occurring on the device described in this paper, the future goal is to optimize DNAzyme performance while retaining the possibility of preseparating a metal ion mixture by capillary ion analysis in the microfluidic device. Therefore, the lactate system, developed by Fritz et al. (33) for the separation of metal ions and lanthanides by capillary electrophoresis, became the starting electrolyte. Using the 12 mM lactate system at pH 4, 5, and 7, Pb2+ was separated from other divalent metal ions such as Mn2+, Cd2+, Co2+, Ni2+, and Cu2+ in a laboratory CE (P/ACE, Beckman Coulter Inc., Fullerton, CA) system equipped with indirect UV detection. For the CE study, 8 mM 4-methylbenzylamine was used as a UV reagent, the separation voltage was 30 kV, and a 75 µm i.d. and 60 cm long capillary was used. Unfortunately, the DNAzyme was not active in the presence of Pb2+ in these lactate systems. In an effort to improve the DNA cleavage reaction performance, 50 mM HEPES with 50 mM NaCl (22) was selected as a potential electrolyte. Addition of 50 mM NaCl was found to play a critical role in stabilizing the substrate and enzyme strand hybridization reaction, resulting in improved sensitivity. But CE separation was not optimal in the HEPES buffer. Finally, the BGE was optimized by a combination of lactate and HEPES at 25 mM with 50 mM NaCl. Under this condition, Pb2+ was separated from other metal ions in the laboratory CE system and the DNA cleavage reaction was efficient (700% fluorescence enhancement in the presence of 5 µM Pb2+). The use of higher NaCl concentrations was not pursued to prevent generation of excess current within the microchannels where Joule heating degrades performance and can eventually cause bubble formation. Transport Control and Measurement. The function of the microfluidic device is to introduce and reactively mix the lead-containing sample with the Pb2+-selective DNAzyme. NAI provides a controllable mechanism for fluidic metering and rapid mixing (29-31, 34). In essence, the PCTE membrane acts as an electronically gateable valve with voltagecontrolled fluid transport rates, preventing fluid flow between vertically separated microchannels in the off state, and electrokinetically driving fluid flow across the membrane when the proper voltage scheme is applied to the fourterminal device. In the off state, any diffusion of analyte across the NAI is below our detection limit as shown by the use of fluorescent probes under reverse bias conditions (29, 30). The electrical bias pathways for the sequence of on and off states on the microfluidic device are defined in Figure 3a,b. The source channel (horizontal) was filled with 1 µM Pb2+ solution in BGE, and the receiving channel (vertical) was filled with 2.5 µM hybridized DNA in BGE. The sequential images in Figure 3c-h capture the fluorescence from repeated injections of Pb2+ at the intersection of the cross-channels. Figure 3c shows the background fluorescence of the twochannel system. The dotted white lines indicate the position of the horizontal channel, and fluorescence from the vertical channel is barely observable. In the on state (Figure 3d,e), FIGURE 4. Fluorescence enhancement as a function of lead ion concentration from 0.1, 1, 5, 10, 50, 100, and 200 µM. Error bars represent (1σ (n ) 4). The inset shows the plot of ln(Imax - I) vs [Pb2+], demonstrating excellent linearity (r2 ) 0.982) over the 0.1-100 µM range. FIGURE 3. Electrical bias configurations for fluidic control of lead and DNAzyme within an NAI/microfluidic device: (a) on state and (b) off state. Temporal sequence of fluorescence images at the intersection of the crossed microchannels (c-h). The source channel (horizontal) was filled with 1 µM Pb2+ in BGE (25 mM lactic acid, 25 mM HEPES, 50 mM NaCl), and the receiving channel (vertical) was filled with hybridized DNAzyme in BGE. Dashed lines indicate the position of the horizontal source channel. (c) All reservoirs were floated. (d) Voltage is applied in the on state, causing injection of Pb2+ solution. Lead solution is transferred from the source channel, across the NAI, toward the grounded reservoir. The reaction with DNAzyme produced fluorescence from cleaved DNA in the receiving channel. (e) During on state bias, the DNA cleavage reaction reached equilibrium and a constant fluorescent signal was maintained (the captured image is ∼40 s after on state bias is applied). The Pb2+ plug continued to move to the ground electrode, and cleaved DNA moved toward the positive bias. (f) ∼1 s after off state bias is applied. (g) ∼40 s after off state bias is applied. Cleaved DNA moved toward the positive bias, and the receiving channel was flushed with bulk hybridized DNAzyme solution. (h) Repetitive Pb2+ plug injection after switching back to the on state bias. Pb2+-containing solution is electrostatically driven from the horizontal channel across the NAI to the vertical channel containing the DNAzyme. To accomplish this transfer, positive high voltages were applied to the reservoirs at the two ends of the source channel and the upper reservoir of the receiving channel while grounding the bottom reservoir of the receiving channel. The transfer efficiency of Pb2+ is proportional to the magnitude of applied bias on either end of the source channel (Figure 3a) up to 400 V, showing larger transfer ratios at higher voltages. However, bubble generation due to electrolysis/heating was observed after several repetitive injections at voltages higher than 400 V, thereby establishing an upper limit on the voltage applied to any of the channel arms. With a 200 nm pore diameter NAI, the bias condition illustrated in Figure 3a causes Pb2+ flow from both arms of the source channel through the nanocapillary array toward the bottom reservoir of the receiving channel. As soon as the on state bias is applied (Figure 3d), the Pb2+ plug moves through the NAI toward the ground end of the receiving channel and starts to cleave the substrate DNA, resulting in a significant fluorescence increase. The cleaved, fluorescently tagged DNA has a slight negative charge and FIGURE 5. Typical fluorescence signals during repetitive Pb2+ injection (100 nM in BGE). Detection was performed at the intersection of crossed channels. Electrical bias configurations are described for Figure 3a,b. The on state was maintained for 20 s and switched to the off state for 10 s, repetitively. is thus transported toward the positively biased upper reservoir, giving rise to fluorescence on both sides of the channel intersection. The off state switches the positive bias on the source channel to ground, stopping the injection of Pb2+ into the receiving channel and flushing the cleaved DNA strand to the upper reservoir (Figure 3f) until all cleaved DNA is removed from the viewing image (Figure 3g). Repetitive injections of Pb2+ solution (Figure 3h) show that this microfluidic chip can be used repeatedly. Calibration, Precision, and Detection Limit. Calibration of the DNA biosensor coupled microfluidic system was accomplished by measuring fluorescence intensity (DNAzyme substrate strand cleavage efficiency) as a function of Pb2+ concentration in the range of 100 nM < [Pb2+] < 200 µM. A plot of fluorescence enhancement vs lead concentration is shown in Figure 4. At each measurement, electrical bias was cycled four times between the on and off states. Each datum in the plot represents the average fluorescence enhancement of these four trials as a function of Pb2+ concentration, and the error bars represent (σ. The expression of the best fit for the plot is described as ln(Imax - I) ) -0.0436[Pb2+] + 4.2072, where Imax and I are the maximum fluorescence enhancement (%) and fluorescence enhancement (%) at the lead concentration, respectively. The microfluidic system response has a linear correlation using the above expression over the 100 nM to 100 µM concentration range with a correlation coefficient (r2) of 0.98. This range likely encompasses Pb2+ concentration levels for most environmental samples. Repetitive detection of 100 nM Pb2+ is illustrated in Figure 5. During repetitive injection sequences, signals were reproducible with a coefficient of variation of 3.5% (n ) 5) and the VOL. 39, NO. 10, 2005 / ENVIRONMENTAL SCIENCE & TECHNOLOGY 9 3759 TABLE 1. Certified Metal Content in Electroplating Sludge Reference Material element concn,a mg/kg element concn, mg/kg element concn, mg/kg Al Cr Pb Hg Na 692.5 ( 82.5 79.5 ( 14.1 119344.0 ( 27453.0 (1.4) (1576.2) Ba Cu Mg Ni Zn 173.3 ( 23.5 63169.3 ( 2410.0 (80.0) 193.6 ( 15.0 182.6 ( 41.0 Ca Fe Mn Ag 562.7 ( 33.0 2,698.7 ( 814.5 17.5 ( 2.1 56.4 ( 6.3 a Certified and noncertified values (36). Values in parentheses are not certified. Certified values are determined on a dry weight basis. Uncertainties are 1 standard deviation of the measurement. The uncertainty is obtained from 95% confidence intervals. FIGURE 6. Fluorescence enhancement as a function of lead ion concentration spiked in the electroplating sludge sample for standard addition calibration. Error bars represent (1σ (n ) 4). baseline consistently returned to a constant level. The detection limit was evaluated by repetitive injection of a 50 nM Pb2+ standard solution. From the baseline noise during the off state and the fluorescence intensity of 50 nM Pb2+ during the on state, the detection limit (signal-to-noise ratio of 3:1) was determined to be 11 nM (2.2 ppb), which is lower than the 72 nM (15 ppb) action level in drinking water recommended by the U.S. Environmental Protection Agency (35). These results demonstrate that the combination of electrokinetically actuated measurement cycles on a microfluidic device and a Pb2+-selective DNAzyme produces a device sensitive enough to monitor lead in drinking water or groundwater. Determination of Lead in an Electroplating Sludge Standard Reference Material. To challenge the microfluidic DNAzyme sensor against a complex matrix, it was used for Pb2+ determination in an electroplating sludge standard reference material. The certified metal contents in this material are shown in Table 1. For this assay, the standard addition method was used to account for matrix effects. In this electroplating sludge sample, it was also observed that copper (at a 2-fold higher molar concentration) partially quenched the fluorescence of cleaved DNA, resulting in a systematic error in the quantitative detection of Pb2+. Since the solubility constant of Pb(OH)2 (Ksp ) 2.5 × 10-16) is 3 orders of magnitude larger than that of Cu(OH)2 (Ksp ) 1.6 × 10-19) (37), it was possible to effect quantitative removal of copper in the sludge digestate as a copper hydroxide precipitate at the electrolyte pH of g8. We confirmed that copper ion was removed to an undetectable level using the laboratory CE instrument. The lead ion, on the other hand, gave a quantitative recovery at pH 8, and the enzymatic DNA reaction was even more efficient than at pH 7, showing faster reaction times in the microfluidic device. Other metal ions did not interfere in the quantitative determination of lead. Figure 6 shows the standard addition curve for the determination of lead in the sludge sample. The sludge digestate was prepared as described earlier, but four aliquots were spiked with a 50 mM Pb2+ standard solution to make final added concentrations of 7, 12, 52, and 102 µM Pb2+, 3760 9 ENVIRONMENTAL SCIENCE & TECHNOLOGY / VOL. 39, NO. 10, 2005 producing a five-point calibration including the nonspiked sludge sample. The experiments were conducted in the same manner as the calibration using the lead standard in buffer solution. The calibration plot had a correlation coefficient of 0.9993. The concentration of lead in the standard electroplating sludge reference material was determined to be [Pb2+] ) 125200 ( 3756 mg/kg, a value within 4.9% of the certified value of 119344 mg/kg, indicating the potential for excellent accuracy of this microfluidic/DNAzyme system for Pb2+ determination. Although this DNAzyme is selective for Pb2+ compared to its response for other divalent cations, higher selectivities are required in some applications. This microfluidic device will be further developed so that the sample analytes are separated using on-chip capillary electrophoresis, allowing user-selectable fractions of the sample flow to be introduced to the DNAzyme (30, 38). For such a system, the selectivity for particular ions would be enhanced, since it will be determined by the product of the ability to separate the desired metal cation from interfering metal ions and the selectivity of the DNAzyme molecular recognition agent itself. The sensitivity and robust nature of the DNAzyme can also be improved by immobilizing the DNA within the NAI pores, instead of using it in solution (39). This platform offers the possibility of incorporating multiple sensing locations in one device; thus, by incorporating different metal-ion-selective DNAzymes into a single microfluidic device, multiple species can be determined simultaneously. Acknowledgments This work was supported by the Strategic Environmental Research and Development Program, the Department of Energy under Grant FG02 88ER13949, and the Engineering Research and Development Center Long Term Monitoring Focus Area. Literature Cited (1) Needleman, H. Lead poisoning. Annu. Rev. Med. 2004, 55, 209222. (2) Fassett, J. D.; MacDonald, B. S. The development and certification of standard reference materials (SRMs) to assess and ensure accurate measurement of Pb in the environment. Fresenius’ J. Anal. Chem. 2001, 370, 838-842. (3) Control of Emissions of Hazardous Air Pollutants from Motor Vehicles and Motor Vehicle Fuels; EPA-420/R-00-023; Environmental Protection Agency: Washington, DC, 2000. (4) Rodriguez, B. B.; Bolbot, J. A.; Tothill, I. E. Development of urease and glutamic dehydrogenase amperometric assay for heavy metals screening in polluted samples. Biosens. Bioelectron. 2004, 19, 1157-1167. (5) Shetty, R. S.; Deo, S. K.; Shah, P.; Sun, Y.; Rosen, B. P.; Daunert, S. Luminescence-based whole-cell-sensing systems for cadmium and lead using genetically engineered bacteria. Anal. Bioanal. Chem. 2003, 376, 11-17. (6) Blake, D. A.; Jones, R. M.; Blake, R. C., II; Pavlov, A. R.; Darwish, I. A.; Yu, H. Antibody-based sensors for heavy metal ions. Biosens. Bioelectron. 2001, 16, 799-809. (7) Taking Toxics Out of the Air; EPA/451/K-98-001; Environmental Protection Agency: Washington, DC, 1998; available at http:// www.epa.gov/ttnatw01/ brochure/brochure.html. (8) DoD Directive 4715.1-Environmental Security; Department of Defense: Washington, DC, 1996; available at http:// www.dtic.mil/whs/directives/corres/pdf/d47151_022496/ d47151p.pdf. (9) Deposition of Air Pollutants to the Great Waters; EPA-453/ R-00-005; Environmental Protection Agency: Washington, DC, 2000. (10) Eijkel, J. C. T.; Mello, A. J. D.; Manz, A. A miniaturized total chemical analysis system: µ-TAS. In Organic Mesoscopic Chemistry; Masuhara, H., Shuryver, F. C. D., Eds.; Blackwell Science: New York, 1999; pp 185-219. (11) Jacobson, S. C.; Ramsey, J. M. Microfabricated Chemical Separation Devices: Theory, Techniques and Applications. In High Performance Capillary Electrophoresis; Khaledi, M. G., Ed.; John Wiley & Sons: New York, 1998; pp 613-633. (12) Kopp, M. U.; Crabtree, H. J.; Manz, A. Developments in technology and applications of microsystems. Curr. Opin. Chem. Biol. 1997, 1, 410-419. (13) Effenhauser, C. S.; Bruin, G. J. M.; Paulus, A. Integrated chipbased capillary electrophoresis. Electrophoresis 1997, 18, 22032213. (14) Reay, R. J.; Flannery, A. F.; Storment, C. W.; Kounaves, S. P.; Kovacs, G. T. A. Microfabricated electrochemical analysis system for heavy metal detection. Sens. Actuators, B 1996, 34, 450-455. (15) Tsukagoshi, K.; Hashimoto, M.; Nakajima, R.; Arai, A. Application of microchip capillary electrophoresis with chemiluminescence detection to an analysis for transition-metal ions. Anal. Sci. 2000, 16, 1111-1112. (16) Jacobson, S. C.; Moore, A. W.; Ramsey, J. M. Fused quartz substrates for microchip electrophoresis. Anal. Chem. 1995, 67, 2059-2063. (17) Kutter, J. P.; Ramsey, R. S.; Jacobson, S. C.; Ramsey, J. M. Determination of metal cations in microchip electrophoresis using on-chip complexation and sample stacking. J. Microcolumn Sep. 1998, 10, 313-319. (18) Collins, G. E.; Lu, Q. Radionuclide and metal ion detection on a capillary electrophoresis microchip using LED absorbance detection. Sens. Actuators, B 2001, 76, 244-249. (19) Collins, G. E.; Lu, Q. Microfabricated capillary electrophoresis sensor for uranium (VI). Anal. Chim. Acta 2001, 436, 181-189. (20) Lu, Q.; Collins, G. E. Microchip separations of transition metal ions via LED absorbance detection of their PAR complexes. Analyst 2001, 126, 429-432. (21) Deng, G.; Collins, G. E. Nonaqueous based microchip separation of toxic metal ions using 2-(5-bromo-2-pyridylazo)-5-(N-propylN-sulfopropylamino)phenol. J. Chromatogr., A 2003, 989, 311316. (22) Li, J.; Lu, Y. A highly sensitive and selective catalytic DNA biosensor for lead ions. J. Am. Chem. Soc. 2000, 122, 1046610467. (23) Tauriainen, S.; Karp, M.; Chang, W.; Virta, M. Luminescent bacterial sensor for cadmium and lead. Biosens. Bioelectron. 1998, 13, 931-938. (24) Deo, S.; Godwin, H. A. A selective, ratiometric fluorescent sensor for Pb2+. J. Am. Chem. Soc. 2000, 122, 174-175. (25) Babkina, S. S.; Ulakhovich, N. A. Amperometric biosensor based on denatured DNA for the study of heavy metals complexing with DNA and their determination in biological, water and food samples. Bioelectrochemistry 2004, 63, 261-265. (26) Liu, J.; Lu, Y. A colorimetric lead biosensor using DNAzymedirected assembly of gold nanoparticles. J. Am. Chem. Soc. 2003, 125, 6642-6643. (27) Lu, Y.; Liu, J.; Li, J.; Bruesehoff, P. J.; Pavot, C. M.-B.; Brown, A. K. New highly sensitive and selective catalytic DNA biosensors for metal ions. Biosens. Bioelectron. 2003, 18, 529-540. (28) Liu, J.; Lu, Y. Improving fluorescent DNAzyme biosensors by combining inter- and intramolecular quenchers. Anal. Chem. 2003, 75, 6666-6672. (29) Kuo, T.-C.; Cannon, D. M., Jr; Shannon, M. A.; Bohn, P. W.; Sweedler, J. V. Hybrid three-dimensional nanofluidic/microfluidic devices using molecular gates. Sens. Actuators, A 2003, 102, 223-233. (30) Kuo, T.-C.; Cannon, D. M., Jr.; Chen, Y.; Tulock, J. J.; Shannon, M. A.; Sweedler, J. V.; Bohn, P. W. Gateable nanofluidic interconnects for multilayered microfluidic separation systems. Anal. Chem. 2003, 75, 1861-1867. (31) Cannon, D. M., Jr.; Kuo, T.-C.; Bohn, P. W.; Sweedler, J. V. Nanocapillary array interconnects for gated analyte injections and electrophoretic separations in multilayer microfluidic architectures. Anal. Chem. 2003, 75, 2224-2230. (32) Acid Digestion of Sediments, Sludges and Soils; Method 3050B; Environmental Protection Agency: Washington, DC, 1996; available at http://www.epa.gov/sw-846/pdfs/3050b.pdf. (33) Shi, Y.; Fritz, J. S. Separation of metal ions by capillary electrophoresis with a complexing electrolyte. J. Chromatogr. 1993, 640, 473-479. (34) Kuo, T.-C.; Kim, H.-K.; Cannon, D. M., Jr.; Shannon, M. A.; Sweedler, J. V.; Bohn, P. W. Nanocapillary arrays effect mixing and reaction in multilayer fluidic structures. Angew. Chem. 2004, 116, 1898-1901. (35) Lead and Copper Rule Minor Revision; EPA 815-F-899-010; Environmental Protection Agency: Washington, DC, 1999. (36) Certificate of Analysis; CRM 010-100 Electroplating Sludge No. 2; Resource Technology Corp.: Laramie, WY, 2003. (37) Skoog, D. A.; West, D. M. Analytical Chemistry, 3rd ed.; Holt, Rinehart and Winston Press: New York, 1979. (38) Tulock, J. J.; Shannon, M. A.; Bohn, P. W.; Sweedler, J. V. Microfluidic separation and gateable fraction collection for mass-limited samples. Anal. Chem. 2004, 76, 6419-6425. (39) Swearingen, C. B.; Wernette, D. P.; Cropek, D. M.; Lu, Y.; Sweedler, J. V.; Bohn, P. W. Immobilization of a catalytic DNA molecular beacon on Au for Pb(II) detection. Anal. Chem., in press. Received for review August 26, 2004. Revised manuscript received February 3, 2005. Accepted March 1, 2005. ES040505F VOL. 39, NO. 10, 2005 / ENVIRONMENTAL SCIENCE & TECHNOLOGY 9 3761