CCP-SAS – a new community consortium for the atomistic
advertisement

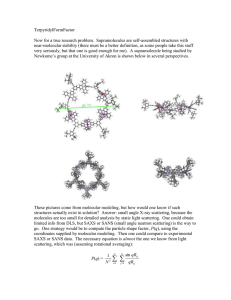
CCP-SAS – a new community consortium for the atomistic modelling of SANS and SAXS data Stephen J. Perkins1, David W. Wright1, Joseph E. Curtis2 and the CCP-SAS consortium3 1 Department of Structural and Molecular Biology, Darwin Building, University College London, Gower Street, London WC1E 6BT, U.K. 2 National Institute of Standards and Technology Center for Neutron Research, National Institute of Standards and Technology, Gaithersburg, Maryland, U.S.A. 3 Funded by a joint EPSRC (EP/K039121/1) and NSF (CHE-1265821) grant; http://www.ccpsas.org/ Email: s.perkins@ucl.ac.uk The major infrastructural investment worldwide in multiuser X-ray synchrotrons and neutron sources during the last two decades has been immensely successful in allowing external (university and commercial) users to perform data collection on ever more challenging and important systems. There is however a need for the hardware investment to be matched by software developments. For example, macromolecular crystallography exploits the fruits of CCP4 software, and the result has been an explosion of new crystal structures. The equivalent revolution is now needed to advance chemical biology and soft condensed matter research based on solution X-ray and neutron scattering. In solution scattering, huge advances have been made in high throughput and accuracy/precision of the experimental measurements. However, the software used to analyse, and in particular to model, this data has lagged behind. The curve-fit approaches from the Svergun group have been most worthwhile for visualizing and rationalizing many problems. To advance the state of the art, and to realize the full benefit of the instrumental investments for novel applications and address different classes of problems, links to atomistic modelling approaches are needed. This requires integrating the SANS and SAXS data with user-friendly, high-throughput, molecular modelling software, in order to reveal how these structures change in time and space under varying experimental conditions. We are funded by the EPSRC and the NSF in a joint UK/USA collaboration called CCP-SAS. The goal is to produce a new generation of open source software that facilitates atomistic modelling based on SANS/SAXS data. We will describe current examples of successful atomistic modelling using our packages SCT/SCTPL (antibodies, complement) and SASSIE (HIV Gag, antibodies), and indicate new directions that will become possible once the CCP-SAS package is developed. At UCL, SCT/SCTPL have already resulted in over 70 structures deposited in the Protein Data Bank; SCT calculates theoretical scattering curves from atomistic models, and SCTPL compares them with experimental SANS and SAXS data (1,2). Analytical ultracentrifugation data for additional constraints are also incorporated within this framework (1,2). SCT/SCTPL will be integrated with the SASSIE packages at NIST (3) that provide conformational sampling. In order that our code has a life after the current project funding, and is usable worldwide, we plan to make the software open source, easy to install and stable to use. We are planning outreach activities to encourage greater use of atomistic modelling. References 1. 2. 3. Perkins, S.J. et al. (2011) Analytical ultracentrifugation combined with X-ray and neutron scattering: Experiment and modelling. Methods. 54(1), 181-99. DOI: 10.1016/j.ymeth.2011.01.004 Perkins, S. J., Okemefuna, A. I., Nan, R., Li, K. & Bonner, A. (2009). Constrained solution scattering modelling of human antibodies and complement proteins reveals novel biological insights. J. Roy. Soc. Interface. 6, S679-S696. http://dx.doi.org/10.1098/rsif.2009.0164.focus J. E. Curtis, S. Raghunandan, H. Nanda, S. Krueger (2012) SASSIE: a program to study intrinsically disordered biological molecules and macromolecular ensembles using experimental scattering restraints. Comp. Phys. Comm. 183, 382-389