Genome-Wide Identification of the AP2/ERF Gene Family Involved in
advertisement

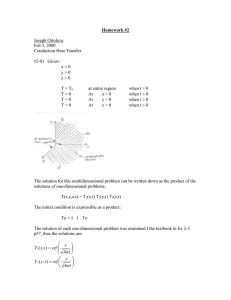
Published March 11, 2016 o r i g i n a l r es e a r c h Genome-Wide Identification of the AP2/ERF Gene Family Involved in Active Constituent Biosynthesis in Salvia miltiorrhiza A. J. Ji, H. M. Luo, Z. C. Xu, X. Zhang, Y. J. Zhu, B. S. Liao, H. Yao, J. Y. Song,* and S. L. Chen* Abstract Tanshinones and phenolic acids are the major bioactive constituents in the traditional medicinal crop Salvia miltiorrhiza; however, transcription factors (TFs) are seldom investigated with regard to their regulation of the biosynthesis of these compounds. Here a complete overview of the APETALA2/ethylene-responsive factor (AP2/ERF) transcription factor family in S. miltiorrhiza is provided, including phylogeny, gene structure, conserved motifs, and gene expression profiles of different organs (root, stem, leaf, flower) and root tissues (periderm, phloem, xylem). In total, 170 AP2/ERF genes were identified and divided into five relatively conserved subfamilies, including AP2 (25 genes), DREB (61 genes), ethylene responsive factor (ERF; 79 genes), RAV (4 genes), and Soloist (1 gene). According to the distribution of bioactive constituents and the expression patterns of AP2/ERF genes in different organs and root tissues, the genes related to the biosynthesis of bioactive constituents were selected. On the basis of quantitative real-time polymerase chain reaction (qRT-PCR) analysis, coexpression analysis, and the prediction of cis-regulatory elements in the promoters, we propose that two genes (Sm128 and Sm152) regulate tanshinone biosynthesis and two genes (Sm008 and Sm166) participate in controlling phenolic acid biosynthesis. The genes related to tanshinone biosynthesis belong to the ERF-B3 subgroup. In contrast, the genes predicted to regulate phenolic acid biosynthesis belong to the ERF-B1 and ERF-B4 subgroups. These results provide a foundation for future functional characterization of AP2/ERF genes to enhance the biosynthesis of the bioactive compounds of S. miltiorrhiza. Published in Plant Genome Volume 9. doi: 10.3835/plantgenome2015.08.0077 © Crop Science Society of America 5585 Guilford Rd., Madison, WI 53711 USA This is an open access article distributed under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/). the pl ant genome ju ly 2016 vol . 9, no . 2 S alvia miltiorrhiza Bunge, a well-known traditional Chinese medicine, is widely used to treat cardiovascular diseases in Asia, the United States, and some European countries; the medicine exhibits strong antidementia, neuroprotective, antioxidant, and anti-inflammatory activities (Zhou, Zuo, et al., 2005; Hügel and Jackson, 2014). According to their chemical properties, the medicinal active constituents of S. miltiorrhiza are divided into two groups: hydrophilic compounds (e.g., salvianolic acid A, salvianolic acid B, Danshensu) and lipophilic compounds (e.g., tanshinone I, IIA, and IIB; cryptotanshinone) (Wang, Morris-Natschke et al., 2007). In the past 20 yr, many genes that encode key enzymes in the biosynthesis pathways of these active compounds have been cloned and functionally analyzed (Huang et al., 2008a; Huang et al., 2008b; Liao et al., 2009; Xiao et al., 2009; Yan et al., 2009; Kai et al., 2010; Kai et al., 2011; Song and Wang, 2011; Cui et al., 2015). Genomic and transcriptomic studies of S. miltiorrhiza have also A.J. Ji, H.M. Luo, Z.C. Xu, X. Zhang, B.S. Liao, H. Yao, and J.Y. Song, Inst. of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100193, China; J. Y. Song, Chongqing Institute of Medicinal Plant Cultivation, Chongqing 408435, China; Y. J. Zhu and S.L. Chen, Inst. of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing 100700, China. Received 27 Apr. 2015. Accepted 18 Oct. 2015. *Corresponding authors (jysong@implad. ac.cn; slchen@icmm.ac.cn). Abbreviations: ADS, amorpha-4, 11-diene synthase; ALDH1, aldehyde dehydrogenase 1; AP2/ERF, APETALA2/ethylene-responsive factor; cDNA, complementary DNA; DREB, dehydration-responsive element binding proteins; ERF, ethylene responsive factor; GT, glandular trichome; NJ, neighbor-joining; qRT-PCR, quantitative real-time polymerase chain reaction; RAV, related to ABI3/VP1; SRA, Sequence Read Archive; TF, transcription factor. 1 of 11 progressed rapidly (Ma et al., 2012; Luo et al., 2014; Wang et al., 2014; Zhang et al., 2015). In particular, the genome of S. miltiorrhiza has been completely assembled (SRA [Sequence Read Archive] accession: SRP051524). Increased knowledge regarding the functional genomics and physiological properties of S. miltiorrhiza has made it an ideal model medicinal plant (Shao and Lu, 2013; Li and Lu, 2014). Compared with the great progress in cloning the enzymes of the synthetic pathway of active compounds in S. miltiorrhiza, little is known about the mechanisms by which TFs regulate tanshinone and phenolic acid biosynthesis (Zhang et al., 2013). The AP2/ERF TF family, one of the largest families of TFs in the plant kingdom, covers five subfamilies: AP2 (APETALA2), ERF (ethylene-responsive factor), DREB (dehydration-responsive element binding proteins), RAV (related to ABI3/VP1), and Soloist ( Sakuma et al., 2002; Yamasaki et al., 2013; Licausi et al., 2013). This TF family has been reported to be involved in secondary metabolism, and the members all belong to the ERF-B3 subgroup. Moreover, several AP2/ERF TFs function as core regulators that positively regulate the biosynthesis of artemisinin (Yu et al., 2012; Lu et al., 2013), which is a terpenoid. ERF-B3 proteins interact with the GCC box element of target genes and regulate their expression. This regulation affects the biosynthesis and accumulation of secondary metabolites (Ohme-Takagi and Shinshi, 1995; Sears et al., 2014). Identification of GCC box elements in promoters of pathway genes may help to confirm the interaction between AP2/ERF genes and the pathway genes. AP2/ERF TFs also regulate alkaloids in Catharanthus roseus (Menke et al., 1999; van der Fits and Memelink, 2001) and Nicotiana tabacum (Shoji et al., 2010). Therefore, we proposed that AP2/ERF TFs may be involved in tanshinone and phenolic acid biosynthesis. The genome-wide identification of AP2/ERF TFs has been documented in several plants such as Arabidopsis thaliana (Sakuma et al., 2002; Nakano et al., 2006), Oryza sativa (Nakano et al., 2006), maize (Zhuang et al., 2010), Glycine max (Zhang et al., 2008), Prunus mume (Du et al., 2013), Brassica rapa ssp. pekinensis and Vitis vinifera ( Zhuang et al., 2009; Licausi et al., 2010; Song et al., 2013). In this study, we performed a genome-wide survey and a systematic characterization of the AP2/ERF family. In addition, the expression patterns of AP2/ERF genes in different organs (root, stem, leaf, flower) and root tissues (periderm, phloem, xylem) were revealed by means of RNA-seq data and qRT-PCR detection. Furthermore, using the phylogenetic tree and coexpression analyses, we proposed four candidate AP2/ERF genes that may be involved in regulating tanshinone and phenolic acid biosynthesis. Overall, systematic analysis of the AP2/ERF family will provide a new gene resource for the metabolic engineering of the secondary metabolites of S. miltiorrhiza. 2 of 11 MATERIALS AND METHODS Plant Materials and Treatment In this study, S. miltiorrhiza Bunge (line 99-3), which was cultivated in the experimental field at the Institute of Medicinal Plant Development (China), was used. Roots, stems, leaves, flowers, periderm, phloem, and xylem were collected to analyze the expression patterns of AP2/ERF genes. All of the sample materials were immediately frozen in liquid nitrogen and stored at -80°C. Database Search and Gene Identification The AP2/ERF sequences of A. thaliana were downloaded from the Arabidopsis Information Resource (http://Arabidopsis.org/). A search of the S. miltiorrhiza genome database (SRA accession: SRP051524) was conducted to obtain all AP2/ERF family members. The AP2-domain hidden Markov model (HMM: PF00847) was used to screen the S. miltiorrhiza genome database for all of the gene sequences that are homologous to AP2/ERF. The predicted genes were corrected manually by comparison with other plant AP2/ERFs by means of the BLASTx algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi). All AP2/ERF protein sequences were then confirmed with PROSITE (http://prosite.expasy.org/), and the conserved AP2 domains of each family member were obtained. The sequence features of the conserved AP2 domains were analyzed by alignment by means of DNAMAN software. Phylogenetic, Gene Structure, and Conserved Motif Analyses The AP2-domain amino acid sequences of S. miltiorrhiza and A. thaliana were aligned with MEGA 5.0 software. Neighbor-joining (NJ) tree construction, gene structures analysis, and conserved motif detection were performed as described in a previous study (Xu et al., 2013). The orthologous proteins were obtained by BLAST analysis of the candidate AP2/ERF TFs against the A. thaliana genome. Synteny analysis was performed by comparing the homology and order of the genes between S. miltiorrhiza scaffolds and A. thaliana chromosomes. The BLASTp cutoff was set as E-value < 1× 10−10. Gene Expression Analysis and Cis-element Prediction The RNA-seq reads from two replicates of four organs (root, stem, leaf, flower) and three replicates of three tissues (periderm, phloem, xylem) were generated by the Illumina HiSeq 2000 and 2500 platforms, respectively (SRA accessions: SRR1640458, SRP051564, and SRP028388). The expression patterns of the AP2/ERF genes in the different organs (root, stem, leaf, flower) and root tissues (periderm, phloem, xylem) were analyzed with TopHat and Cufflinks (Trapnell et al., 2012). The 1500-bp promoter sequences upstream of the transcription start site (ATG) of 72 and 29 genes encoding key enzymes involved in the biosynthesis of tanshinones and phenolic acids, respectively, were obtained from the pl ant genome ju ly 2016 vol . 9, no . 2 whole-genome sequences. The cis-elements targeted by AP2/ERF TFs were investigated with the PLACE database (http://www.dna.affrc.go.jp/PLACE/signalscan.html). RNA Isolation and qRT-PCR Total RNA was isolated from each sample with an RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. The quality and integrity of the RNA were analyzed by electrophoresis and a NanoDrop 2000C spectrophotometer. The first cDNA strand was synthesized using a FastQuant RT Kit (Tiangen Biotech). The specific primers for qRT-PCR were designed with Primer Premier 6.0, and the amplicon length was set between 130 and 180 bp (Table S1). The qRT-PCR reagents, instrument, and method were identical to those in our previous study (Luo et al., 2014). SmActin was used as the reference gene (Yang et al., 2010). Three key genes for enzymes—SmDXS2 (1-deoxy-D-xylulose 5-phosphate synthase 2), SmCPS1 (copalyl diphosphate synthase 1), and SmRAS1 (rosmarinic acid synthase 1)— were tested as positive controls. Each sample was tested in triplicate. To detect expression differences of the candidate genes among various tissues, one-way ANOVA was performed using IBM SPSS 20 software. P < 0.01 was considered highly significant. Gene coexpression analysis of the candidate genes was performed by means of Pearson’s correlation test, and candidate genes with the Pearson coefficients greater than 0.8 were considered to be coexpressed. RESULTS Identification and Phylogenetic Analysis of the AP2/ERF Family in S. miltiorrhiza In total, 170 AP2/ERF genes in S. miltiorrhiza that encode at least one AP2 domain were identified. The lengths of the genomic DNA, coding sequence, and protein; the AP2 domain sequence; and the number of introns are summarized in Table S2. According to the number and sequence features of the AP2 domains, the AP2/ERF family of S. miltiorrhiza was divided into five subfamilies—AP2, DREB, ERF, RAV, and Soloist—and these subfamilies had 25, 61, 79, 4, and, 1 members, respectively (Fig. S1). The DREB and ERF subfamilies were the most dominant in S. miltiorrhiza. Based on the similarities of the AP2-domain amino acid sequences, the 140 members of the largest two subfamilies were further assigned to 12 subgroups: DREB (A1-A6) and ERF (B1-B6). A multiple sequence alignment between S. miltiorrhiza and A. thaliana was performed using the AP2 domain sequences from each subfamily, and it indicated that 31, 34, 70, 86, and 59 conserved amino acid residues were found in the DREB, ERF, AP2, RAV, and Soloist subfamilies, respectively (Fig. S2). An unrooted phylogenetic tree was constructed to confirm these classifications and to analyze their phylogenetic relationships based on the alignments of the AP2 domain sequences of S. miltiorrhiza and A. thaliana (Fig. 1). The analysis showed that the tree was separated into 10 clades, including the AP2, DREB, ERF, RAV, and Soloist subfamilies. Clade 1 and clade 2 represented the AP2 and Soloist subfamilies, respectively. Clades 3 and 4 represented the DREB subfamily, and clade 6 represented the RAV subfamily. Clades 5 and 7–10 indicated the ERF subfamily. To confirm the subgroups of the DREB/ERF subfamilies, we constructed a deep phylogenetic tree using the AP2 domain sequences of the DREB/ERF subfamilies in S. miltiorrhiza and V. vinifera (Fig. S3) because the AP2/ERF members have been well classified in V. vinifera (Zhuang et al., 2009; Licausi, et al., 2010). The classification of the DREB/ERF subgroups of S. miltiorrhiza was consistent with that of V. vinifera. Gene Structure and Conserved Motifs of SmAP2s Gene structure was analyzed using the online Gene Structure Display Server, and the exon-intron structures are shown in Figures 2 and S4. The number of introns in the AP2/ERF genes varied among the subfamilies. The AP2 and Soloist subfamilies contained the most introns (5 to 10), whereas most DREB/ERF and RAV subfamily members possessed no introns. The positions of introns also differed among the subfamilies. Fewer than three introns were found within the AP2-R1 or AP2-R2 domain of the AP2 subfamily. The two DREB members both had one intron before the conserved AP2 domain. Most intron positions in the ERF subfamily were identical to those in the DREB subfamily. Some conserved motifs outside the AP2 domain may be involved in nuclear localization, transcription activity, and protein-protein interactions (Ikeda and Ohme-Takagi, 2009; Causier et al., 2012; Tiwari et al., 2012). We therefore investigated the conserved motifs of all AP2/ERF proteins in S. miltiorrhiza, and the results are shown in Figures 2 and S4. Twenty-six conserved motifs were identified, and most members in the same subgroup possessed one or more motifs together outside the AP2 domain (Table S3). Motifs 1–8, 10, and 20 corresponded to the AP2 domain region, and motifs 21 and 23 corresponded to the B3 domain region in the RAV subfamily. The remaining 14 motifs were selectively distributed among particular subfamilies and subgroups, illustrating the sequence similarity among proteins in the same group. For example, the AP2 subfamily contained three motifs (9, 16, 26), and these motifs were not observed in the other subfamilies. Motifs 14, 15, 17 18, 24, and 25 were specific to the ERF subfamily. Among these motifs, motif 17 was only observed in the B3 subgroup, and motif 15 was shared by both B5 and B6 members. The proteins within a subgroup that shared the same motifs may have similar functions. Expression Pattern of SmAP2s in S. miltiorrhiza Tanshinones and phenolic acids, the major bioactive compounds of S. miltiorrhiza, are distributed primarily in the roots (Ma et al., 2012; Di et al., 2013). Our previous study showed that the highest tanshinone content is in the periderm followed by the phloem and that the lowest content is in the xylem (Xu et al., 2015). In contrast, the highest phenolic acid content is in the phloem and xylem, ji et al .: genome - wi de analysis of ap 2 in salvia mi ltiorrhiz a 3 of 11 Fig. 1. An unrooted phylogenetic tree was constructed with the AP2/ERF (APETALA2/ethylene-responsive factor) domain amino acid sequences of S. miltiorrhiza and A. thaliana. A neighbor-joining tree was constructed using MEGA5 with 1000 bootstrap replicates. The tree was divided into 10 groups that contained the AP2, DREB (dehydration-responsive element binding proteins), ERF, RAV (related to ABI3/VP1), and Soloist subfamilies. The photo in the center of phylogenetic tree presents the plant Salvia miltiorrhiza. and the lowest content is in the periderm. Moreover, previous studies have also revealed that the expression patterns of the TFs that regulate secondary metabolism pathways are similar to the expression patterns of key enzymes and the distribution of secondary metabolites in A. annua (Yu et al., 2012; Lu et al., 2013). We therefore investigated the expression profiles of the AP2/ERF family obtained from seven sources of transcriptome data, including libraries from different organs (root, stem, leaf, flower) and root tissues (periderm, phloem, xylem) of S. miltiorrhiza (Table S4). Heat maps were also constructed (Fig. 3 and Fig. S5), 4 of 11 and they showed the different expression patterns of all AP2/ERF genes across the various organs and tissues. Excluding the genes that could not be detected in the transcriptome analyses, the expression of 120 AP2/ ERF genes in at least one of four organs and three tissues was detected, including 14 genes in the AP2 subfamily, 47 genes in the DREB subfamily, 55 genes in the ERF subfamily, 3 genes in RAV subfamily, and the 1 Soloist gene (Table S4). Among these genes, 38, 8, 1, 12, 12, 21, and 18 AP2/ERF genes were expressed relatively highly in the root, stem, leaf, flower, periderm, phloem, and xylem, the pl ant genome ju ly 2016 vol . 9, no . 2 Fig. 2. The features of the gene structures and the distribution of conserved motifs of the ethylene responsive factor (ERF) subfamily of S. miltiorrhiza. (A) Phylogenetic relationships among ERF subgroups. (B) The distribution of the conserved motifs of each ERF protein. (C) Exon/intron structures of the ERF genes of S. miltiorrhiza. respectively. Thirty-four of the 38 genes in the root belong to the DREB and ERF subfamilies, while 11 of the 12 genes in periderm are ERF members. Interestingly, 26 genes that were markedly expressed in phloem and xylem are also members of the DREB and ERF subfamilies. In total, expression profiling provided a valuable resource for the ji et al .: genome - wi de analysis of ap 2 in salvia mi ltiorrhiz a 5 of 11 Fig. 3. The expression profiles of S. miltiorrhiza AP2/ERF (APETALA2/ethylene-responsive factor) genes in the root, stem, leaf, and flower. (A), (B), (C) and (D) indicate the expression patterns of the AP2, ERF, DREB (dehydration-responsive element binding proteins), and RAV (related to ABI3/VP1)-Soloist subfamilies, respectively. The color bar at the top represents the expression values. The genes in white were not expressed in these four organs, and the values for exon model per million mapped reads were zero. identification of candidate genes involved in the regulation of bioactive compound biosynthesis in S. miltiorrhiza. Identification of Candidate SmAP2s Genes Related to Tanshinone Biosynthesis On the basis of the RNA-seq data of the different organs and tissues, 11 SmAP2s genes (Sm021, Sm026, Sm032, Sm082, Sm086, Sm108, Sm109, Sm128, Sm139, Sm152, 6 of 11 and Sm168) were highly expressed in root and periderm and were identified as potential regulatory factors related to tanshinone biosynthesis. Using further analysis with qRT-PCR (Fig. S6), we concluded that Sm026, Sm082, Sm086, Sm108, Sm128, and Sm139 showed significant expression in the root. Sm026, Sm108, Sm128, Sm139, and Sm152 showed the highest expression in the periderm tissue. The expression of these five genes showed a the pl ant genome ju ly 2016 vol . 9, no . 2 Fig. 4. Expression patterns of selected AP2/ERF (APETALA2/ethylene-responsive factor) genes related to tanshinone (A) and phenolic acid (B) biosynthesis. Relative expression of the selected AP2/ERF genes in R (root), S (stem), L (leaf), F (flower), R1 (periderm), R2 (phloem), and R3 (xylem) was examined by quantitative real-time polymerase chain reaction. The characters on the x axis indicate different organs and tissues. The y axis shows the relative level of gene expression. The S. miltiorrhiza actin gene was used as an internal reference. SmDXS2, SmCPS1, and SmRAS1 were tested as positive controls. We performed a one-way ANOVA with IBM SPSS 20 software. Asterisks represent significant differences. P < 0.01 was considered highly significant. similar pattern with the control terpenoid synthase genes (SmCPS1 and SmDXS2) (Fig. S6). Moreover, phylogenetic tree and gene coexpression analyses were performed to narrow the candidate genes, which were most likely to be related to secondary metabolism. We observed the phylogenetic relationships using the AP2 domain sequences of the candidate TFs and the functional TFs associated with secondary metabolism—ORCA2, ORCA3, AaERF1, AaERF2, NtERF32, NtERF189, NtORC1, and NtJAP1 (Fig. S7) (De Sutter, Vanderhaeghen, et al., 2005, Lu, Zhang, et al., 2013, Menke, Champion, et al., 1999, Sears, Zhang, et al., 2014, Shoji, Kajikawa, et al., 2010, van der Fits and Memelink, 2001, Yu, Li, et al., 2012)—with four methods (NJ, minimum evolution, maximum likelihood, and unweighted pair group method with arithmetic mean [UPGAMA]). The tree was grouped into three clades in the NJ tree (Fig. S7A). Sm108, Sm128, Sm139, and Sm152 proteins were clustered with all of the functional TFs in clade 1, and Sm026 was grouped in clade 2. Sm108, Sm128, Sm139, and Sm152 all belong to the ERF-B3 subgroup, whereas Sm026 is a member of the DREB-A6 subgroup. The phylogenetic relationships of the other trees were similar with the NJ tree. According to this result, we speculate that several members of the ERF-B3 subgroup are the main regulators of tanshinone biosynthesis. Meanwhile, gene coexpression analysis revealed that the expression profiles of Sm026, Sm128, and Sm152 were coexpressed with those of the control terpenoid synthase genes (SmCPS1 and SmDXS2) (Table S5). Overall, these results suggested that Sm128 and Sm152 might be involved in tanshinone biosynthesis (Fig. 4). Finally, to identify the cis-elements of AP2/ERF TFs, the promoter sequences of the genes encoding enzymes in the tanshinone biosynthesis pathway were analyzed (Table S6). Seven cis-elements were observed for AP2/ ERF TFs. RAV1AAT was identified in the promoter regions of all studied genes. The GCC box was the ciselements of ERF-B3 subgroup. Interestingly, many of the pathway genes—including AACT6 (acetyl-CoA C-acetyltransferase 6), HMGS1 (hydroxymethylglutarylCoA synthase 1), HMGR1 (hydroxymethylglutarylCoA reductase), HMGR2 (3-hydroxy-3-methyl glutaryl coenzyme A reductase 2), PMK (5-phosphomevalonate kinase), IPI1 (isopentenyl diphosphate isomerase 1), GGPPS5 (geranylgeranyl diphosphate synthase 5), CPS1, KSL1 (kaurene synthase-like 1), KSL5 (kaurene synthase-like 5), KSL7 (kaurene synthase-like 7) and KSL8 (kaurene synthase-like 8)—all had GCC box sites in their promoter regions. This finding indicated that the expression of these genes might be regulated by AP2/ERF TFs. We also performed synteny analysis by comparing the sequences of candidate genes (Sm008, Sm128, Sm152, and Sm166) with the A. thaliana genome. Only one syntenic block was identified (Fig. S8). Scaffold 766 showed a consistent relation with chromosome 4 of A. thaliana. Compared with A. thaliana, more gene insertions occurred in scaffold 766. No tanshinone or phenolic acid biosynthetic genes were found in scaffold 766. Moreover, although AT4G18450 was the best-hit gene of Sm152 in A. thaliana, AT4G18450 has not yet been functionally analyzed. Therefore, this result provides insight for the characterization of gene functions. Identification of Candidate SmAP2 Genes Related to Phenolic Acid Biosynthesis We used the RNA-Seq data to screen 10 genes (Sm008, Sm038, Sm042, Sm068, Sm098, Sm121, Sm136, Sm163, ji et al .: genome - wi de analysis of ap 2 in salvia mi ltiorrhiz a 7 of 11 Sm165, and Sm166) for their potential involvement in the regulation of phenolic acid biosynthesis. The qRT-PCR results further revealed that the expression of Sm008, Sm038, Sm042, Sm098, Sm121 and Sm166 was highest in the phloem and xylem and lowest in the periderm, and their expression profiles were consistent with those of the positive control SmRAS1 (Fig. S6). Phylogenetic tree and gene co-expression analyses were also performed to narrow the candidate genes. The NJ tree (Fig. S7 A) showed that the candidate TFs were distributed in all three clades. Sm008 and Sm166 were closely related to the functional TFs in clade 1. The other four TFs were slightly farther from the secondary metabolism-related TFs in clades 2 and 3. The genes predicted to be involved in phenolic acid biosynthesis were from different subgroups (Sm008 from ERF-B4; Sm038 and Sm098 from DREB-A1; Sm042 from DREB-A4; Sm121 from DREB-A6; and Sm166 from ERF-B1). Only Sm008 and Sm166 were from the ERF subfamily. Meanwhile, gene co-expression analysis revealed that Sm008 and Sm166 were co-expressed with the control rosmarinic acid synthase gene (SmRAS) (Table S5). These data therefore suggested that Sm008 and Sm166 might be involved in phenolic acid biosynthesis (Fig. 4). Promoter analysis of genes encoding enzymes in the phenolic acid biosynthesis pathway was also performed using online PLACE software (Table S7). In contrast to the tanshinone biosynthesis genes, the candidate genes related to phenolic acid biosynthesis possessed far fewer cis-elements for AP2/ERF TFs. Similar to the genes involved in tanshinone biosynthesis, most of the genes in phenolic acid biosynthesis studied possessed a RAV1AAT site. The GCC-box site was detected in the promoter regions of 4CL3 (4-coumarate:CoA ligase 3), 4CL-like2 (4-coumarate:CoA ligase–like 2), 4CL-like4 (4-coumarate:CoA ligase–like 4), TAT1 (tyrosine aminotransferase 1), HPPR1 (4-hydroxyphenylpyruvate reductase 1), RAS1, HCT1 (hydroxycinnamoyltransferase 1), and CYP98A78, all of which are involved in phenolic acid biosynthesis (Huang, et al., 2008a; Huang, et al., 2008b; Xiao et al., 2011; Di et al., 2013; Wang et al., 2014). Thus, AP2/ERF TFs might regulate these genes to affect phenolic acid accumulation via these genes. DISCUSSION We comprehensively surveyed the phylogeny, gene structure, conserved motifs, and expression profiles of the AP2/ERF TF family in S. miltiorrhiza, a highly economical and medicinally valuable plant, to obtain further information for AP2/ERF TFs. In total, 170 AP2/ERF genes were identified from the S. miltiorrhiza genome database (538 Mb). Compared with the numbers of AP2/ERF genes in other plants—A. thaliana (147 genes/genome size: 125 Mb; Sakuma et al., 2002), rice (164 genes/genome size: 466 Mb; Nakano et al., 2006), soybean (148 genes/genome size: 1115 Mb; Zhang et al., 2008), V. vinifera (149 genes/genome size: 487 Mb; Zhuang et al., 2009; Licausi et al., 2010), Chinese cabbage 8 of 11 (291 genes/genome size: 485 Mb; Song et al., 2013)—the number of AP2/ERF TFs in S. miltiorrhiza is similar (with the exception of Chinese cabbage), indicating that the number of AP2/ERF TFs is conserved. In addition, this result shows that the number of AP2/ERF genes is not related to the genome size. A comparison of the organization of the AP2/ERF gene family of S. miltiorrhiza with that of other species clearly showed that DREB and ERF are the prominent subfamilies. Chinese cabbage has 291 AP2/ERF genes, whereas the AP2, RAV, and Soloist subfamilies have only 49, 14, and 1 members, respectively, a distribution similar to that for A. thaliana. This finding supports the evolution of AP2/ERF TFs because the members of these three subfamilies have more complex gene structures, such as greater numbers of introns and additional domain insertions, and this complexity might have impaired the homing processes, leading to a lower duplication rate (Magnani et al., 2004). In addition, the phylogenetic tree comparing AP2 domain sequences between S. miltiorrhiza and V. vinifera further confirmed the classification of the DREB and ERF subgroups. As expected, the distribution of DREB/ERF members is similar within the two plants. These two species have no A3 subgroup, whereas the B3 subgroup contains the largest number of members, with 37 in V. vinifera and 28 in S. miltiorrhiza. Phytohormones such as jasmonate, salicylate, and ethylene effectively increase the contents of secondary metabolites (Bennett and Wallsgrove, 1994; Qian et al., 2006; Kai et al., 2012; Namdeo, 2007). Some ERF-B3 members have been reported to integrate jasmonate, alicylate, and ethylene hormonal signal-transduction pathways (Brown et al., 2003; Lorenzo et al., 2003; Van der Does et al., 2013). Therefore, considering the 18 members in A. thaliana, we predict that the B3 members in V. vinifera (37 genes) and S. miltiorrhiza (28 genes) may be involved in the diversity of secondary metabolites. Genome-wide identification will provide a more comprehensive understanding of the classification, gene structures, and phylogenetic relationships of the AP2/ERF TF family, thus further elucidating the relationship between structure and gene function. AP2/ERF TFs can be divided into transcriptional activator and repressor proteins. These proteins contain several conserved motifs; however, most of them have not yet been studied. We detected one of the most studied repression motif (ERF-associated amphiphilic repression [EAR] motif) in the ERF-B1 (Sm011, Sm054, Sm105, Sm122, Sm165, and Sm166), DREB-A5 (Sm051 and Sm052) and AP2 (Sm057, Sm081, and Sm106) subfamilies. The EAR motif can effectively suppress the activation of transcription by interacting with other corepressors (Ohta et al., 2001). Another motif named BRD (B3 repression domain), which has an RLFGV sequence, was found in two of RAV members (Sm020 and Sm133); the TFs that contain the BRD motif also exhibited repressive activity (Ikeda and Ohme-Takagi, 2009). Moreover, the strong activation motif EDLL was found in 13 members (Sm001, Sm013, Sm014, Sm021, Sm031, Sm074, Sm083, Sm115, the pl ant genome ju ly 2016 vol . 9, no . 2 Sm139, Sm152, Sm156, Sm159, and Sm164) of the ERF-B3 subgroup. This motif, which has been used to alter flowering time in Arabidopsis, was considered a potential tool for targeted gene activation in a study of various developmental pathways (Tiwari et al., 2012). In addition, some of the DREB-A1 (Sm002, Sm003, Sm098, Sm111, and Sm129) and A4 (Sm086, Sm095, and Sm123) members contain a conserved LWSY motif in the C-terminal region; this motif was reported in OsDREB1A/B/C and in AtCBF3/ DREB1A, which were induced by drought, high-salt, and cold conditions (Dubouzet et al., 2003). In metabolic-engineering research, a core TF can activate several key enzymes of a metabolic pathway, leading to a large increase in the production of the desired compound (van der Fits and Memelink, 2001; Yu et al., 2012). Moreover, genetic manipulation of TF regulation is much easier than more transformations or co-transformation of several key enzymes. Thus, one of the primary objectives of this study was to predict the candidate AP2/ERF TFs that may regulate the biosynthesis of tanshinones and phenolic acids, which are medicinal compounds that have been commonly used to treat cardiovascular diseases for a thousand years. Secondary metabolites usually accumulate in a tissue-specific manner. For example, the biosynthesis and accumulation of artemisinin occur in glandular trichomes (GTs; Olsson et al., 2009). Gossypol accumulates primarily in pigment glands in cotton (Xu et al., 2004). The root of S. miltiorrhiza is used as a medicine and is the major site for tanshinone and phenolic acid accumulation. Additionally, most of the genes encoding the enzymes in this biosynthetic pathway are also highly expressed at the accumulation site. These genes include ADS (amorpha4,11-diene synthase), CYP71AV1, DBR2 (double bond reductase 2), and ALDH1 (aldehyde dehydrogenase 1) in the GTs of A. annua (Olsson et al., 2009) as well as SmDXS2 and SmCPS1 in the root of S. miltiorrhiza (Ma et al., 2012). The spatiotemporal transcriptional regulation of metabolic pathway genes is tightly controlled at different levels involving many regulatory TFs. Previous studies have shown that the expression profiles of TFs are usually consistent with the production of secondary metabolites in different tissues and are similar to the expression profiles of the enzyme genes. In A. annua, both AaERF1 and AaERF2 are highly expressed in inflorescences, similar to ADS and CYP71AV1 (Yu et al., 2012). AaORA from A. annua is a trichome-specific transcription factor, and AaORA exhibits similar expression patterns to those of ADS, CYP71AV1, and DBR2 in different tissues (Lu et al., 2013). Using this result as a screening principle, we used RNA-seq data to select 11 and 10 genes that potentially regulate the biosynthesis of tanshinones and phenolic acids, respectively. The qRT-PCR method was also used to confirm the expression patterns of these genes. The results indicate that five and six genes that potentially regulate the biosynthesis of tanshinones and phenolic acids, respectively, were expressed similarly to pathway genes among different tissues. Finally, combined with phylogenetic tree and gene coexpression analyses, two genes (Sm128 and Sm152) and two genes (Sm008 and Sm166) were predicted to be involved in secondary metabolism in S. miltiorrhiza. All of the following AP2/ERF TFs involved in the regulation of secondary metabolism studied to date belong to the ERF-B3 subgroup, including ORCA2 and ORCA3 from C. roseus (Menke et al., 1999; van der Fits and Memelink, 2001); AaORC, AaERF1, and AaERF2 from A. annua ( Yu et al., 2012; Lu et al., 2013); and NIC-2 locus ERF TFs in N. tabacum (Shoji et al., 2010). Two candidate genes (Sm128 and Sm152) involved in the biosynthesis of tanshinones belong to the ERF-B3 subgroup, revealing the significance of their potential functions. Importantly, other ERF subgroups are likely to regulate the biosynthesis of phenolic acid. No studies thus far have examined the roles of AP2/ERF TFs in regulating phenolic acid biosynthesis. Moreover, our study revealed that the Sm008 and Sm166 candidate genes were coexpressed with the key enzyme gene (SmRAS) with Pearson correlation value of 0.948 and 0.899, respectively. In addition, the promoter sequences of most genes in phenolic acid metabolic pathways possess the cis-elements of AP2/ERF TFs. Therefore, the functions of these genes are worth studying. Supplemental Information Available Supplemental information is included with this article. Figure S1. AP2/ERF transcription factor distribution in S. miltiorrhiza. Figure S2. Comparison of the AP2/ERF domain sequences of the AP2 (A), ERF (B), DREB (C), RAV (D), and Soloist (E) subfamilies proteins between A. thaliana and S. miltiorrhiza. The blue background indicates the conservation of amino acid residues (100%). The pink background indicates the conservation of amino acid residues (75%). Figure S3. Phylogenetic tree constructed using the AP2/ERF domain sequences of the DREB and ERF subfamilies of S. miltiorrhiza and V. vinifera. Figure S4. The features of the gene structures and the distribution of the conserved motifs of the AP2 (A), DREB (B), RAV (C), and Soloist (D) subfamilies of S. miltiorrhiza. (a) Phylogenetic relationships among each subgroups. (b) The distribution of the conserved motifs of each protein. (c) Exon/intron structures of the AP2/ ERF genes of S. miltiorrhiza. Figure S5. The expression profiles of S. miltiorrhiza AP2/ERF genes in the periderm, phloem and xylem. A, B, C, and D indicate the expression patterns of the AP2, ERF, DREB, and RAV-Soloist subfamilies, respectively. The color bar at the top represents the expression values. The genes in white were not expressed in these four organs and the FPKM (fragments per kilobase of exon model per million mapped reads) values were zero. Figure S6. Expression patterns of selected AP2/ERF genes related to tanshinone (A) (five genes) and phenolic acid (B) (six genes) biosynthesis. Relative expression of the selected AP2/ERF genes in R (root), S (stem), L (leaf), F (flower), R1 (periderm), R2 (phloem), and R3 (xylem) was ji et al .: genome - wi de analysis of ap 2 in salvia mi ltiorrhiz a 9 of 11 examined by qRT-PCR. The characters on the x axis indicate different organs and tissues. The y axis shows the fold changes in gene expression. The S. miltiorrhiza actin gene was used as an internal reference. SmDXS2, SmCPS1, and SmRAS1 were tested as positive controls. We performed one-way ANOVA using IBM SPSS 20 software. Asterisks representing significant differences from this comparison. P < 0.01 was considered highly significant. Figure S7. Phylogenetic tree constructed using the AP2 domain sequences of the 11 candidate TFs of S. miltiorrhiza and other medicinal plant AP2s associated with secondary metabolism. The phylogenetic tree was constructed using the neighbor-Joining (A), minimum evolution (B), UPGMA (C), and maximum likelihood (D) method. Figure S8. Synteny relationships between S. miltiorrhiza Sm152 and A. thaliana AT4G18450 loci. The colored boxes represent the different types of genes. Table S1. Primer sequences of the candidate genes related to the biosynthesis of tanshinones and phenolic acids used for qRT-PCR analyses. Table S2. Complete list of the AP2/ERF transcription factors identified in the S. miltiorrhiza genome. Table S3. Conserved motifs identified from the AP2/ ERF genes in S. miltiorrhiza. Table S4. The average FPKM values of 170 AP2/ERF genes in different organs and tissues. Table S5. Coexpression analysis of candidate genes and pathway genes using Pearson’s correlation test. Table S6. Cis-acting regulatory elements targeted by AP2/ERF TFs in the promoters of the key enzyme genes of the tanshinone metabolic pathway. Table S7. Cis-acting regulatory elements targeted by AP2/ERF TFs in the promoters of the key enzyme genes of the phenolic acid metabolic pathway. Acknowledgments This work was supported by the National Natural Science Foundation (grant nos. 81373916 and 81573398) and the Program for Innovative Research Team at the Institute of Medicinal Plant Development (grant no. IT1304). References Bennett, R.N., and R.M. Wallsgrove. 1994. Secondary metabolites in plant defence mechanisms. New Phytol. 127:617–633. doi:10.1111/j.1469-8137.1994.tb02968.x Brown, R.L., K. Kazan, K.C. McGrath, D.J. Maclean, and J.M. Manners. 2003. A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 132:1020–1032. doi:10.1104/ pp.102.017814 Causier, B., M. Ashworth, W. Guo, and B. Davies. 2012. The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 158:423–438. doi:10.1104/pp.111.186999 Cui, G., L. Duan, B. Jin, J. Qian, Z. Xue, G. Shen, et al. 2015. Functional divergence of diterpene syntheses in the medicinal plant Salvia miltiorrhiza. Plant Physiol. 169: 1607–1618. doi:10.1104/pp.15.00695 De Sutter, V., R. Vanderhaeghen, S. Tilleman, F. Lammertyn, I. Vanhoutte, M. Karimi, et al. 2005. Exploration of jasmonate signalling via automated and standardized transient expression assays in tobacco cells. Plant J. 44:1065–1076. doi:10.1111/j.1365-313X.2005.02586.x Di, P., L. Zhang, J. Chen, H. Tan, Y. Xiao, X. Dong, et al. 2013. 13C tracer reveals phenolic acids biosynthesis in hairy root cultures of Salvia miltiorrhiza. ACS Chem. Biol. 8:1537–1548. doi:10.1021/cb3006962 10 of 11 Du, D., R. Hao, T. Cheng, H. Pan, W. Yang, J. Wang, et al. 2013. Genomewide analysis of the AP2/ERF gene family in Prunus mume. Plant Mol. Biol. Rep. 31:741–750. doi:10.1007/s11105-012-0531-6 Dubouzet, J.G., Y. Sakuma, Y. Ito, M. Kasuga, E.G. Dubouzet, S. Miura, et al. 2003. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33:751–763. doi:10.1046/j.1365-313X.2003.01661.x Huang, B., Y. Duan, B. Yi, L. Sun, B. Lu, X. Yu, et al. 2008a. Characterization and expression profiling of cinnamate 4-hydroxylase gene from Salvia miltiorrhiza in rosmarinic acid biosynthesis pathway. Russ. J. Plant Physiol. 55:390–399. doi:10.1134/S1021443708030163 Huang, B., B. Yi, Y. Duan, L. Sun, X. Yu, J. Guo, et al. 2008b. Characterization and expression profiling of tyrosine aminotransferase gene from Salvia miltiorrhiza (Dan-shen) in rosmarinic acid biosynthesis pathway. Mol. Biol. Rep. 35:601–612. doi:10.1007/s11033-007-9130-2 Hügel, H.M., and N. Jackson. 2014. Danshen diversity defeating dementia. Bioorg. Med. Chem. Lett. 24:708–716. doi:10.1016/j.bmcl.2013.12.042 Ikeda, M., and M. Ohme-Takagi. 2009. A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 50:970–975. doi:10.1093/ pcp/pcp048 Kai, G., P. Liao, H. Xu, J. Wang, C. Zhou, W. Zhou, et al. 2012. Molecular mechanism of elicitor-induced tanshinone accumulation in Salvia miltiorrhiza hairy root cultures. Acta Physiol. Plant. 34:1421–1433. doi:10.1007/s11738-012-0940-z Kai, G., P. Liao, T. Zhang, W. Zhou, J. Wang, H. Xu, et al. 2010. Characterization, expression profiling, and functional identification of a gene encoding geranylgeranyl diphosphate synthase from Salvia miltiorrhiza. Biotechnol. Bioprocess Eng. 15:236–245. doi:10.1007/s12257-009-0123-y Kai, G., H. Xu, C. Zhou, P. Liao, J. Xiao, X. Luo, et al. 2011. Metabolic engineering tanshinone biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. Metab. Eng. 13:319–327. doi:10.1016/j.ymben.2011.02.003 Li, C., and S. Lu. 2014. Genome-wide characterization and comparative analysis of R2R3-MYB transcription factors shows the complexity of MYB-associated regulatory networks in Salvia miltiorrhiza. BMC Genomics 15:277. doi:10.1186/1471-2164-15-277 Liao, P., W. Zhou, L. Zhang, J. Wang, X. Yan, Y. Zhang, et al. 2009. Molecular cloning, characterization and expression analysis of a new gene encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase from Salvia miltiorrhiza. Acta Physiol. Plant. 31:565–572. doi:10.1007/s11738-008-0266-z Licausi, F., F.M. Giorgi, S. Zenoni, F. Osti, M. Pezzotti, and P. Perata. 2010. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genomics 11:719. doi:10.1186/1471-2164-11-719 Licausi, F., M. Ohme-Takagi, and P. Perata. 2013. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 199:639–649. doi:10.1111/nph.12291 Lorenzo, O., R. Piqueras, J.J. Sánchez-Serrano, and R. Solano. 2003. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15:165–178. doi:10.1105/ tpc.007468 Lu, X., L. Zhang, F. Zhang, W. Jiang, Q. Shen, L. Zhang, et al. 2013. AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea. New Phytol. 198:1191–1202. doi:10.1111/nph.12207 Luo, H., Y. Zhu, J. Song, L. Xu, C. Sun, X. Zhang, et al. 2014. Transcriptional data mining of Salvia miltiorrhiza in response to methyl jasmonate to examine the mechanism of bioactive compound biosynthesis and regulation. Physiol. Plant. 152:241–255. doi:10.1111/ppl.12193 Ma, Y., L. Yuan, B. Wu, X’e. Li, S. Chen, and S. Lu. 2012. Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J. Exp. Bot. 63:2809–2823. doi:10.1093/jxb/err466 Magnani, E., K. Sjolander, and S. Hake. 2004. From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell 16:2265–2277. doi:10.1105/tpc.104.023135 Menke, F.L., A. Champion, J.W. Kijne, and J. Memelink. 1999. A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and the pl ant genome ju ly 2016 vol . 9, no . 2 elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J. 18:4455–4463. doi:10.1093/emboj/18.16.4455 Nakano, T., K. Suzuki, T. Fujimura, and H. Shinshi. 2006. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140:411–432. doi:10.1104/pp.105.073783 Namdeo, A. 2007. Plant cell elicitation for production of secondary metabolites: a review. Pharmacogn. Rev. 1:69–79. Ohme-Takagi, M., and H. Shinshi. 1995. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7:173–182. doi:10.1105/tpc.7.2.173 Ohta, M., K. Matsui, K. Hiratsu, H. Shinshi, and M. Ohme-Takagi. 2001. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13:1959–1968. doi:10.1105/tpc.13.8.1959 Olsson, M.E., L.M. Olofsson, A.-L. Lindahl, A. Lundgren, M. Brodelius, and P.E. Brodelius. 2009. Localization of enzymes of artemisinin biosynthesis to the apical cells of glandular secretory trichomes of Artemisia annua L. Phytochemistry 70:1123–1128. doi:10.1016/j.phytochem.2009.07.009 Qian, Z.G., Z.J. Zhao, Y. Xu, X. Qian, and J.J. Zhong. 2006. Novel chemically synthesized salicylate derivative as an effective elicitor for inducing the biosynthesis of plant secondary metabolites. Biotechnol. Prog. 22:331–333. doi:10.1021/bp0502330 Sakuma, Y., Q. Liu, J.G. Dubouzet, H. Abe, K. Shinozaki, and K. Yamaguchi-Shinozaki. 2002. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydrationand cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290:998–1009. doi:10.1006/bbrc.2001.6299 Sears, M.T., H. Zhang, P.J. Rushton, M. Wu, S. Han, A.J. Spano, et al. 2014. NtERF32: A non-NIC2 locus AP2/ERF transcription factor required in jasmonate-inducible nicotine biosynthesis in tobacco. Plant Mol. Biol. 84:49–66. doi:10.1007/s11103-013-0116-2 Shao, F., and S. Lu. 2013. Genome-wide identification, molecular cloning, expression profiling and posttranscriptional regulation analysis of the Argonaute gene family in Salvia miltiorrhiza, an emerging model medicinal plant. BMC Genomics 14:512. doi:10.1186/1471-2164-14-512 Shoji, T., M. Kajikawa, and T. Hashimoto. 2010. Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell 22:3390– 3409. doi:10.1105/tpc.110.078543 Song, J., and Z. Wang. 2011. RNAi-mediated suppression of the phenylalanine ammonia-lyase gene in Salvia miltiorrhiza causes abnormal phenotypes and a reduction in rosmarinic acid biosynthesis. J. Plant Res. 124:183–192. doi:10.1007/s10265-010-0350-5 Song, X., Y. Li, and X. Hou. 2013. Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genomics 14:573. doi:10.1186/1471-2164-14-573 Tiwari, S.B., A. Belachew, S.F. Ma, M. Young, J. Ade, Y. Shen, et al. 2012. The EDLL motif: a potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J. 70:855–865. doi:10.1111/j.1365313X.2012.04935.x Trapnell, C., A. Roberts, L. Goff, G. Pertea, D. Kim, D.R. Kelley, et al. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7:562–578. doi:10.1038/ nprot.2012.016 Van der Does, D., A. Leon-Reyes, A. Koornneef, M.C. Van Verk, N. Rodenburg, L. Pauwels, et al. 2013. Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25:744–761. doi:10.1105/ tpc.112.108548 Van der Fits, L., and J. Memelink. 2001. The jasmonate-inducible AP2/ERFdomain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J. 25:43–53. doi:10.1046/j.1365-313x.2001.00932.x Wang, B., W. Sun, Q. Li, Y. Li, H. Luo, J. Song, et al. 2014. Genome-wide identification of phenolic acid biosynthetic genes in Salvia miltiorrhiza. Planta 241:711–725. doi:10.1007/s00425-014-2212-1 Wang, X., S.L. Morris-Natschke, and K.H. Lee. 2007. New developments in the chemistry and biology of the bioactive constituents of Tanshen. Med. Res. Rev. 27:133–148. doi:10.1002/med.20077 Xiao, Y., P. Di, J. Chen, Y. Liu, W. Chen, and L. Zhang. 2009. Characterization and expression profiling of 4-hydroxyphenylpyruvate dioxygenase gene (Smhppd) from Salvia miltiorrhiza hairy root cultures. Mol. Biol. Rep. 36:2019–2029. doi:10.1007/s11033-008-9413-2 Xiao, Y., L. Zhang, S. Gao, S. Saechao, P. Di, J. Chen, et al. 2011. The c4h, tat, hppr and hppd genes prompted engineering of rosmarinic acid biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. PLoS ONE 6:e29713. doi:10.1371/journal.pone.0029713 Xu, W., F. Li, L. Ling, and A. Liu. 2013. Genome-wide survey and expression profiles of the AP2/ERF family in castor bean (Ricinus communis L.). BMC Genomics 14:785. doi:10.1186/1471-2164-14-785 Xu, Y.-H., J.-W. Wang, S. Wang, J.-Y. Wang, and X.-Y. Chen. 2004. Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-d-cadinene synthase-A. Plant Physiol. 135:507–515. doi:10.1104/pp.104.038612 Xu, Z., R.J. Peters, J. Weirather, H. Luo, B. Liao, X. Zhang, et al. 2015. Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J. 82:951–961. doi:10.1111/tpj.12865 Yamasaki, K., T. Kigawa, M. Seki, K. Shinozaki, and S. Yokoyama. 2013. DNA-binding domains of plant-specific transcription factors: Structure, function, and evolution. Trends Plant Sci. 18:267–276. doi:10.1016/j. tplants.2012.09.001 Yan, X., L. Zhang, J. Wang, P. Liao, Y. Zhang, R. Zhang, et al. 2009. Molecular characterization and expression of 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) gene from Salvia miltiorrhiza. Acta Physiol. Plant. 31:1015–1022. doi:10.1007/s11738-009-0320-5 Yang, Y., S. Hou, G. Cui, S. Chen, J. Wei, and L. Huang. 2010. Characterization of reference genes for quantitative real-time PCR analysis in various tissues of Salvia miltiorrhiza. Mol. Biol. Rep. 37:507–513. doi:10.1007/s11033-009-9703-3 Yu, Z.X., J.X. Li, C.Q. Yang, W.L. Hu, L.J. Wang, and X.Y. Chen. 2012. The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant 5:353–365. doi:10.1093/mp/ssr087 Zhang, G., M. Chen, X. Chen, Z. Xu, S. Guan, L.C. Li, et al. 2008. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.). J. Exp. Bot. 59:4095–4107. doi:10.1093/jxb/ern248 Zhang, S., P. Ma, D. Yang, W. Li, Z. Liang, Y. Liu, et al. 2013. Cloning and characterization of a putative R2R3 MYB transcriptional repressor of the rosmarinic acid biosynthetic pathway from Salvia miltiorrhiza. PLoS ONE 8:e73259. doi:10.1371/journal.pone.0073259 Zhang, X., H. Luo, Z. Xu, Y. Zhu, A. Ji, J. Song, et al. 2015. Genome-wide characterisation and analysis of bHLH transcription factors related to tanshinone biosynthesis in Salvia miltiorrhiza. Sci. Rep. 5:11244. doi:10.1038/srep11244 Zhou, L., Z. Zuo, and M.S.S. Chow. 2005. Danshen: An overview of its chemistry, pharmacology, pharmacokinetics, and clinical use. J. Clin. Pharmacol. 45:1345–1359. doi:10.1177/0091270005282630 Zhuang, J., D.-X. Deng, Q.-H. Yao, J. Zhang, F. Xiong, J.-M. Chen, et al. 2010. Discovery, phylogeny and expression patterns of AP2-like genes in maize. Plant Growth Regul. 62:51–58. doi:10.1007/s10725-010-9484-7 Zhuang, J., R.-H. Peng, Z.-M.M. Cheng, J. Zhang, B. Cai, Z. Zhang, et al. 2009. Genome-wide analysis of the putative AP2/ERF family genes in Vitis vinifera. Sci. Hortic. (Amsterdam, Netherlands) 123:73–81. doi:10.1016/j.scienta.2009.08.002 ji et al .: genome - wi de analysis of ap 2 in salvia mi ltiorrhiz a 11 of 11