Non-centered PCA - Broad Institute
advertisement
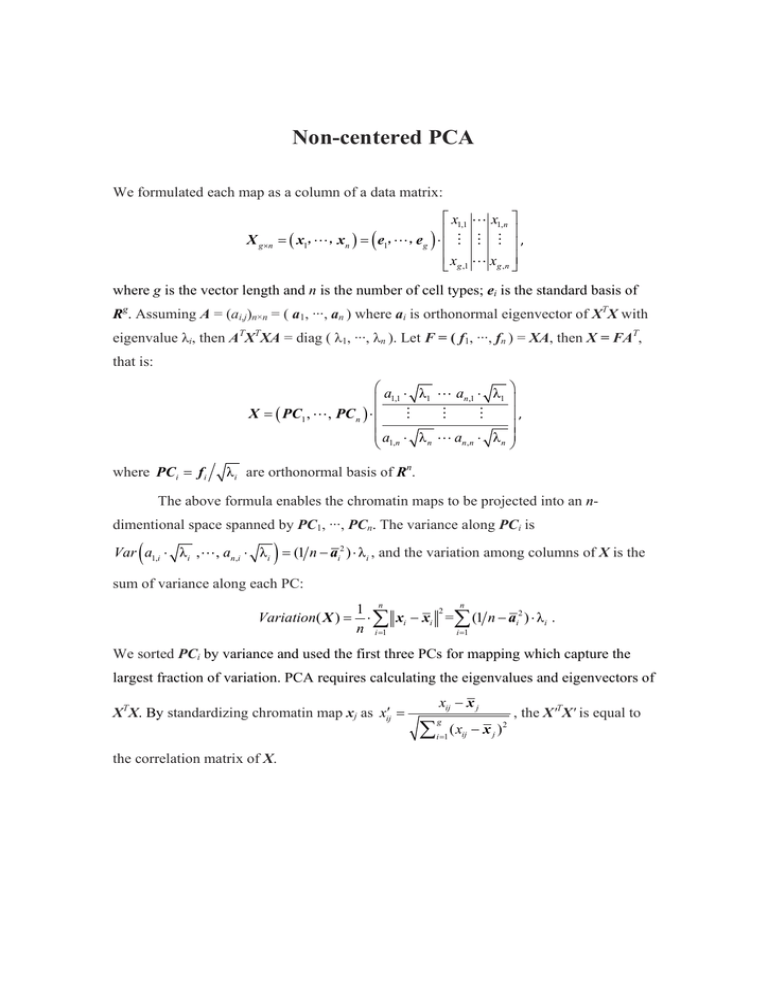
Non-centered PCA We formulated each map as a column of a data matrix: ⎡ x1,1 " x1,n ⎤ ⎢ ⎥ = ( x1, ",xn ) = ( e1, ",e g ) ⋅ ⎢ # # # ⎥ , ⎢ xg ,1 " xg ,n ⎥ ⎣ ⎦ X g ×n where g is the vector length and n is the number of cell types; ei is the standard basis of Rg. Assuming A = (ai,j)n×n = ( a1, ···, an ) where ai is orthonormal eigenvector of XTX with eigenvalue λi, then ATXTXA = diag ( λ1, ···, λn ). Let F = ( f1, ···, fn ) = XA, then X = FAT, that is: ⎛a ⋅ λ "a ⋅ λ ⎞ 1 n ,1 1 ⎜ 1,1 ⎟ X = ( PC1 , " , PC n ) ⋅⎜ # # # ⎟ , ⎜ ⎟ ⎜ a1,n ⋅ λ n " an ,n ⋅ λ n ⎟ ⎝ ⎠ where PCi = f i λ i are orthonormal basis of Rn. The above formula enables the chromatin maps to be projected into an ndimentional space spanned by PC1, ···, PCn. The variance along PCi is ( ) Var a1,i ⋅ λ i ," , an,i ⋅ λ i = (1 n − ai2 ) ⋅ λ i , and the variation among columns of X is the sum of variance along each PC: Variation( X ) = n 1 n 2 ⋅ ∑ xi − xi = ∑ (1 n − ai2 ) ⋅ λ i . n i =1 i =1 We sorted PCi by variance and used the first three PCs for mapping which capture the largest fraction of variation. PCA requires calculating the eigenvalues and eigenvectors of XTX. By standardizing chromatin map xj as xij′ = the correlation matrix of X. xij − x j ∑ g i =1 ( xij − x j ) 2 , the X'TX' is equal to