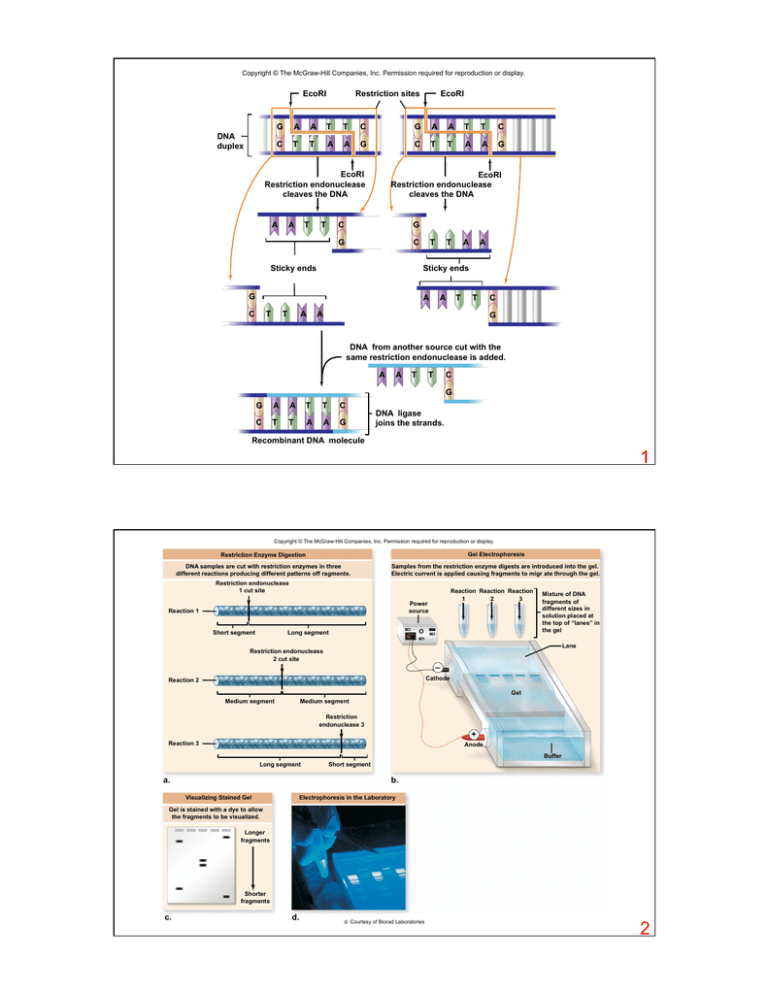
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
EcoRI
DNA
duplex
Restriction sites
EcoRI
G
A
A
T
T
C
G
A
A
T
T
C
C
T
T
A
A
G
C
T
T
A
A
G
EcoRI
Restriction endonuclease
cleaves the DNA
A
A
T
T
EcoRI
Restriction endonuclease
cleaves the DNA
C
G
G
C
Sticky ends
T
A
T
T
A
A
Sticky ends
G
C
A
T
A
T
T
A
C
G
DNA from another source cut with the
same restriction endonuclease is added.
A
A
T
T
C
G
G
A
A
T
T
C
C
T
T
A
A
G
DNA ligase
joins the strands.
Recombinant DNA molecule
1
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
Restriction Enzyme Digestion
Gel Electrophoresis
DNA samples are cut with restriction enzymes in three
different reactions producing different patterns off ragments.
Samples from the restriction enzyme digests are introduced into the gel.
Electric current is applied causing fragments to migr ate through the gel.
Restriction endonuclease
1 cut site
Reaction Reaction Reaction
1
2
3
Power
source
Reaction 1
Short segment
Long segment
Mixture of DNA
fragments of
different sizes in
solution placed at
the top of “lanes” in
the gel
Lane
Restriction endonuclease
2 cut site
–
Cathode
Reaction 2
Gel
Medium segment
Medium segment
Restriction
endonuclease 3
+
Reaction 3
Anode
Buffer
Long segment
a.
Short segment
b.
Visualizing Stained Gel
Electrophoresis in the Laboratory
Gel is stained with a dye to allow
the fragments to be visualized.
Longer
fragments
Shorter
fragments
c.
d.
d. Courtesy of Biorad Laboratories
2
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
A Plasmid Vector
Restriction
endonuclease
Foreign
DNA
lacZ gene
Transform
No DNA
inserted
Medium contains
ampicillin and X-gal
Ampicillin
resistance
gene
Restriction enzymes
cuts within
the lacZ gene
Foreign DNA
and DNA ligase
are added
DNA
inserted
Active lacZ
gene produces
blue colonies
Inactive lacZ
gene produces
white colonies
Transform
3
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
Plasmid Library
DNA fragments
from source DNA
DN A inserted
into plasmid vector
Transformation
Each cell contains a
single fragment. All cells
together are the library.
4
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
exons
introns
1
1
2
2
3
3
4
4
Eukaryotic DNA template
Transcription
5´ cap
3´ poly- A tail
Primary RNA transcript
Introns are cut out,
and coding regions are
spliced together.
3´ poly- A tail
5´ cap
Mature RNA transcript
Isolation of mRNA
Addition of reverse
transcriptase
Reverse
transcriptase
Reverse
transcriptase
utilizes mRNA
to create cDNA.
Addition of mRNAdegrading enzymes
mRNA–cDNA hybrid
Degraded
mRNA
DNA polymerase
Double-stranded cDNA
with no introns
5
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
5. A comparison with the original plate
identifies the colony containing the gene.
Filter paper
Film
1. Colonies of plasmid
containing bacteria, each
containing a single DNA
from the library, are grown
on agar.
4. The only sites on the
filter that will retain
probe DNA will contain
DNA complementary to
the probe. These
represent the sites of
colonies containing the
gene of interest.
2. A replica of the plate
is made by pressing
a piece of filter paper
against the agar and
bacterial colonies.
Some cells from
each colony adhere
to the filter.
3. The filter is washed with a solution to break the cells open
and denature the DNA, which sticks to the filter at the site
of each colony. The filter is incubated with a radioactively
labeled probe that can form hybrids with complementary
DNA in the gene of interest.
6
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
Test nucleic acids
Electrophoresis
1. Electrophoresis is
performed, using
radioactively labeled
markers as a size
guide in the first lane.
Radioactively
labeled markers
with specific sizes
Electrophoretic gel
2. The gel is covered
with a sheet of
nitrocellulose and
placed in a tray of
buffer on top of a
sponge. Alkaline
chemicals in the
buffer denature the
DNA into single
strands. The buffer
wicks its way up
through the gel and
nitrocellulose into a
stack of paper towels
placed on top of the
3. DNA in the gel is
transferred, or
“blotted,” onto the
nitrocellulose.
Stack of paper towels
Nitrocellulose filter
Gel
Buffer
Sponge
Nitrocellulose
paper now
contains nucleic
acid “print”
Gel
Radioactive
probe (singlestranded DNA)
4. Nitrocellulose with
bound DNA is
incubated with
radioactively labeled
nucleic acids and is
then rinsed.
Sealed
container
—AATGG—
—TTACC—
DNA fragments
within bands
5. Photographic film is
laid over the filter and
is exposed only in
areas that contain
radioactivity
(autoradiography).
Bands on the film
represent DNA in the
gel that is
complementary to the
probe sequence.
Film
Hybridized nucleic acids
Size markers
7
© SSPL/The Image Works
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
Original Sequence
of Restriction Sites
(no mutations)
Point Mutations
Change the
Sequence of
Restriction Sites
Sequence
Repetitions Can
Occur Between
Restriction Sites
Larger
fragments
restriction endonuclease
cutting sites
+
Single base-pair
change
Smaller
fragments
–
+
–
+
–
+
Sequence duplication
+
a. Three different
DNA duplexes
b. Cut DNA
c. Gel electrophoresis of
restriction fragments
8
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
Victim
Rapist’s semen
Suspect’s blood
Victim
Rapist’s semen
Suspect’s blood
Courtesy of Lifecodes Corp, Stamford CT
9
10
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
NH2
N
O
–O
N
CH2
O
P
N
5´
O
O–
4´
1´
3´
2´
H
H
11
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
Manual Enzymatic DNA Sequencing
Automated Enzymatic DNA Sequencing
Template
Template
DNA polymerase
DNA polymerase
5´
3´
T
A G C
C
A T
G
C
3´
T
Primer
Reaction
for ddG
Reaction
for ddC
Reaction
for ddA
Reaction
for ddT
A T
C G
5´
A T
C G
G
5´
A T
C G
G
5´
A T
C
5´
A T
C G
5´
A
5´
A T
5´
A T
5´
A T
C G
G
T
5´
A T
C G
G
T
C
C G
G
G
A
T
T
T
A C
G
A C
A
A C
G
T
T
C
A T
G
A
A T
5´
A T
C
5´
A T
C G
5´
A T
C G
G
5´
A T
C G
G T
5´
A T
C G
G T
A
5´
A T
C G
G T
A
C
5´
A T
C G
G T
A
C
G
5´
A T
C G
G T
A
C
G T
3´
T
G
C
A
T
G
G
C
T
A 5´
T
G
Laser
Photo detector
reads colors
A
T
G
G
5´
C
+
a.
A
5´
C
Shorter
segments
C
5´
3´
–
Longer
segments
A G C
Primer
5´
G
5´
A
A
T
C
G
G
T
A
C
G
T
3´
T
A
5´
b.
12
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
Adapter
DNA fragment
Dense primer lawn
in flow cell
DNA
Adapter
Adapters
Flow cell
a.
1 cm
b.
Bridge
amplification
with unlabeled
dNTPs
Free end
binds to
primer
c.
Denature
doublestranded
molecules
Attached
Free
terminus
Clusters
35 cycles
of bridge
amplification
f.
T
A
T
Attached
e.
d.
G
C
Fragments
become
doublestranded
N
G
NH2
C
O
A
A
–O
N
O
O
O–
T
4´
1´
3´
G
2´
OH
A
Image capture for each
round of synthesis
g.
N
CH2
5´
C
First round of
synthesis with
labeled dNTPs
P
Reversible terminator
h.
13
b: © 2007, Illumina Inc. All rights reserved
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
DNA segment
to be amplified
5´
3´
3´
5´
PCR
machine
1. Sample is first heated
to denature DNA.
DNA is denatured
into single strands
5´
3´
3´
5´
2. DNA is cooled to a
lower temperature
to allow annealing
of primers.
5´
3´
Primers anneal to DNA
3´
5´
3. DNA is heated to
72°C, the optimal
temperature for Taq
DNA polymerase to
extend primers.
5´
3´
3´
5´
Taq DNA polymerase
3´
5´
3´
5´
3´
3´
5´
3´
5´
5´
3´
3´
5´
5´
3´
3´
5´
5´
3´
3´
5´
3´
5´
3´
5´
5´
3´
Cycle 2:
4 copies
Cycle 3:
8 copies
5´
5´
3´
3´
5´
5´
3´
5´
3´
5´
3´
3´
5´
3´
3´
5´
3´
5´
3´
5´
5´
5´
3´
3´
5´
14
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
Yeast nucleus
Transcriptionactivating domain
Yeast cell
Gal4 protein
DNA
DNAbinding
domain
DNAbinding
domain
RNA polymerase
Bait vector
Prey vector
Inserted DNA
Inserted DNA
Fusion
proteins
Prey protein
Bait protein
Reporter gene
15
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
neo
neo
Embryonic stem (ES) cells with
knocked out gene
Gene to be knocked out
neo
1. Using recombinant DN A techniques, the gene
encoding resistance to neomycin (neo) is inserted
into the gene of interest, disrupting it. The neo gene
also confers resistance to the drug G418, which kills
mouse cells. This construct is then introduced into
ES cells.
2. In some ES cells, the construct will recombine
with the chromosomal copy of the gene to be
knocked out. This replaces the chromosomal
copy with the neo disrupted construct. This is
the equivalent to a double crossover event in
a genetic cross.
ES cells
containing
neo
G418-containing
medium
Surrogate mouse
Blastocyst
Dead cells without
knocked out gene
3. The ES cells are placed on G418containing medium. The G418 selects
cells that have had a replacement event,
and now contain a copy of the knocked
out gene.
4. The ES cells containing the knocked out
gene are injected into a blastocyst stage
embryo and then implanted into a female
to complete development.
Heterozygous
mouse carrying
the knockout gene
Homozygous
mouse for the
knockout gene
5. Offspring will contain one chromosome with
the gene of interest knocked out. Genetic
crosses can then produce mice homozygous
for the knocked out gene to assess the
phenotype. This can range from lethality to
no visible effect depending on the gene.
16
17
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
2. Herpes simplex
gene is isolated.
1. DNA is extracted.
3. Vaccinia DNA
is extracted and
cleaved.
Herpes simplex virus
Human immune
response
6. Antibodies directed
against herpes simplex
viral coat are made.
Gene specifying herpes
simplex surface protein
Harmless vaccinia
(cowpox) virus
4. Fragment containing
surface gene combines
with cleaved vaccinia DNA.
5. Harmless engineered
virus (the vaccine) with
surface like herpes
simplex is injected into
the human body .
18
19
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
Gene of
interest
Plasmid
Agrobacterium
Plant nucleus
1. Plasmid is
removed and cut open
with restriction
endonuclease.
2. A gene of interest is
isolated from the DN A
of another organism
and inserted into the
plasmid. The plasmid
is put back into the
Agrobacterium.
3. When used to infect plant cells,
Agrobacterium duplicates part
of the plasmid and transfers the
new gene into a chromosome of
the plant cell.
4. The plant cell divides, and each
daughter cell receives the new
gene. These cultured cells can
be used to grow a new plant
with the introduced gene.
20
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
SCIENTIFIC THINKING
Hypothesis: Petunias can acquire tolerance to the herbicide glyphosate by overexpressing EPSP synthase
Prediction: Transgenic petunia plants with a chimeric EPSP synthase gene with strong promoter will be glyphosate tolerant
Test:
1. Use restriction enzymes and ligase to “paste” the cauliflower mosaic virus promoter (35S) to the EPSP synthase gene and insert the
construct in Ti plasmids.
2. Transform Agrobacterium with the recombinant plasmid.
3. Infect petunia cells and regenerate plants. Regenerate uninfected plants as controls.
4. Challenge plants with glyphosate.
Agrobacterium
EPSP
synthase
35S
Glyphosate
Transformed,
regenerated
petunia plant
Ti plasmid
Cultured petunia cells
Non-tolerant
petunia
Tolerant
petunia
Result: Glyphosate kills control plants, but not transgenic plants.
Conclusion: Additional EPSP synthase provides glyphosate tolerance.
FurtherExperiments: The transgenic plants are tolerant, but not resistant (note bleaching at shoot tip). How could you determine if additional
copies of the gene would increase tolerance? Can you think of any downsides to expressing too much EPSP synthase in petunia?
(right): © Rob Horsch, Monsanto Company
21
Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display.
Daffodil
phytoene
synthase
gene (psy)
Bacterial
carotene
desaturase
gene (crtI )
Daffodil
lycopene
b-cyclase
gene (lcy)
Genes introduced
into rice genome
Rice
Rice
chromosome
chromosome
Expression
in endosperm
GGPP
psy
psy
crtI
crtI
lcy
Phytoene
synthase
Carotene
desaturase
β-Cyclase
Phytoene
Lycopene
in 1897,
β-Carotene
(Provitamin A)
22
23
24
