Hyun Soo Kim , AD Trahan , MD Lynch Department of
advertisement

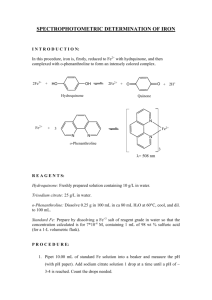
Citrate Synthase Metabolic Switch 1 Kim , Duke, Visible Thinking April 22, 2014 ABSTRACT The effectiveness of a CRISPR interference system as a metabolic switch for TCA flux control in E.coli was evaluated. The goal of controlling TCA (Tricarboxylic acid cycle) flux would be to reduce carbon dioxide formation and thereby increase yield to other desired products, such as those derived from malonyl-CoA. One key enzyme involved in controlling flux through the TCA cycle is the first step in the cycle, citrate synthase (CS). CS catalyzes the conversion of acetyl-CoA and oxaloacetate into citrate—the starting molecule in the TCA cycle. A CRISPR interference system was used to repress CS by repressing transcription of citrate synthase gene :gltA. Growth analysis showed that dCas9 induction causes a significant growth defect. Also, presence of gltA sgRNA led to a further growth defect. A commercially available CS enzyme assay kit was used to measure CS levels of different strains with different induction conditions. The enzyme assay protocol was optimized using various reactant concentrations. Enzyme assay results showed that base strain BWapldf and +sgRNA strains had unrepressed, high level of citrate synthase activity while AB1623 (gltA mutant) and negative control showed close to zero enzyme activity as expected. However, unrepressed activity in +sgRNA/dCas9 strain and repressed activity in +dCas9 strain was unexpected. Incompatibility of citrate synthase with “BugBugster” lysis buffer was suspected as the cause of the unexpected data and was experimentally confirmed. Alternative method for cell lysis is currently being implemented. 1 Trahan , 1 Lynch Hyun Soo A.D. M.D. 1Department of Biomedical Engineering, Duke University, Durham, NC EXPERIMENTAL RESULTS Growth Studies in Shake Flasks Design and Cloning of CRISPR System Procedure: • Design plasmid for insertion into E.coli • pSMART-HC-Kan vector, constitutive promoter, sgRNA sequence • yibD promoter [10] and phlD gene [11] • Series of PCR amplification and Gibson Assembly • Transformed into E.coli (DH5α and BWapldf) and sequenced Results: • Cloning steps were confirmed using gel electrophoresis at each stage • Three point mutations were found in the terminator • It was confirmed that the mutated sequence was still recognized as an effective terminator CONCLUSIONS Procedure: • 5 strains: BWapldf, BWapldf+sgRNA, BWapldf+dCas9, BWapldf+sgRNA+dCas9, AB1623 • Base strain BWapldf from work by Jian was an E.coli BW25113 mutant with reduced mixed acid fermentation (deletion of ackA-pta, poxB, ldhA, adhE, pflB genes; hence “BWapldf”) [12] • Negative control strain AB1623 contains mutation in the gltA gene (gltA5)--expected to have no citrate synthase activity [13]. Results: • Figure 4 shows dCas9 induction reduces the growth rate • [A] Strains with dCas9 induction had slower growth rate • [B] Earlier induction had slower growth rate Figure 3. Plasmid Construct Design • All three tests shows statistical significance (p-value ≈ 0.01, 0.0002, and 0.002). INTRODUCTION AND PURPOSE Malonyl-CoA derived products such as malonic acid and phloroglucinol have multiple potential uses as pharmaceuticals, industrial chemicals, and explosives [1, 2]. Traditionally, these chemicals are produced from petroleum and cause great harm to the environment. Preliminary work has shown the possibility of the biobased production of malonyl-CoA derivatives [4]. This approach has shown to work for the fermentation of polyketides 1,3,6,8tetrahydroxynaphthalene (THN) and 3-hydroxypropionic acid (3-HP). However, initial results for the fermentation of malonate from malonyl-CoA has been shown only at low levels [5]. Significant improvements are necessary to optimize the production process. A possible reason for the low production level of malonate is the unnecessary carbon consumption through the TCA cycle (tricarboxyl acid cycle) also known as the Krebs cycle. Procedure: • Protocol included in the citrate synthase enzyme assay kit (from Sigma-Aldrich, CS0720) • Cell pellets obtained from shake-flask experiment samples were lysed using BugBuster® Initial Results: • Only the positive control (purified citrate synthase) showed increasing absorbance. • All strains showed no citrate synthase activity (no change in absorbance; Figure 6A). Goal: Repress citrate synthase synthesis (gltA) in E.coli using a CRISPR interference approach. D E F G Optimization (Figure 6A): • Changes to sample volume and concentrations of reagents and buffers based on [8]. • Run time: 215 minutes • 30,000 MWCO centrifugal filter unit to prevent high background absorbance • 0.1 M potassium (KCl) addd [8]. Final Results (Figure 6B): • Confirmed that BWapldf and +sgRNA both had unrepressed enzyme activity • Confirmed that negative control and AB1623 had close-to-zero citrate synthase activity • However, +sgRNA/dCas9 had unrepressed activity while +dCas9 showed repressed activity • Unexpected and inconsistent results • Suspected BugBuster® was inhibiting activity Table 1. Post-Optimization Citrate Synthase Enzyme Assay Results BWapldf +sgRNA +dCas9 +sgRNA/dCas9 AB1623 High High High Low Low Theoretical (Unrepressed) (≈Unrepressed) (≈Unrepressed) (Repressed) (Absent) High Low High Low High Observed 0.023 units/mg 0.029 units/mg 0.005 units/mg 0.022 units/mg 0.003 units/mg Expected? Yes Yes No No Yes Figure 5. Specific Growth Rate of Key System Strains Confirmation of Incompatibility with BUGBUGSTER® Citrate Synthase Enzyme Assay Optimization Temperature sensitive mutant fabI (fabIts) stops producing fatty Acyl-ACP upon temperature shift, thereby relieving the feedback inhibition on ACCase that converts acetyl-CoA to malonyl-CoA. 2) A gene of choice can be used to produce malonyl-CoA derivative products. 3) Inhibition of citrate synthase (gltA) will reduce the flux of acetylCoA into TCA cycle and increase its flux through malonyl-CoA pathway. C Procedure: • Test citrate synthase compatibility with one or more steps in the experimental procedures. • The positive control protein was added to four tubes at different steps • 1) not added, 2) pre-lysis, 3) post-lysis/pre-filter, 4) post-filter. ` ` Add Citrate Synthase Centrifugal Filter Cell Lysis with BugBuster® ` Add Citrate Synthase Citrate Synthase Enzyme Assay (30,000 MWCO) Add Citrate Synthase ` Cell Pellet Samples Figure 7. Experimental Setup to Test Incompatibility Results (Figure 6C): • The earlier purified citrate synthase was added, the lower its enzymatic activity (increase in absorbance at 412nm) was. • Longer contact with BugBuster® led to lower citrate synthase activity. Figure 2. CRISPR Interference CRISPR interference approach. A targeting or guide RNA (sgRNA) that targets the gltA gene is produced, in addition to a catalytically inactive caspase 9 (dCas9) . When both enzymes are induced (“turned on”) the dCas9 and sgRNA bind to the gltA gene and silence transcription. FUTURE DIRECTIONS Growth and Induction Characterization Test effect of sgRNA anddCas9 at t=3HR induction • Follow-up experiment using a strain that only contains dCas9 with induction at t=3HR to test whether the growth defect for induction at t=3HR is due to dCas9 only or dCas9 and sgRNA together. Determine Magnitude of Citrate Synthase Inactivation Other methods could be used to quantify citrate synthase expression • Measure the gltA mRNA level using cDNA synthesis by qPCR. • Measure citrate synthase protein level by western-blot, SDS-page, or ELISA. • Targeted citrate synthase protein degradation (proteolysis) [14] • Tagging citrate synthase with a mutant ssrA tag that targets native E.coli protease ClpXP REFERENCES 1. "Intermediate Pharmaceutical Ingredients - Flopropione". Univar Canada. Retrieved 24 April 2009. 2. "Synthesis of trinitrophloroglucinol". The United States Patent and Trademark Office. 1984. Retrieved 24 April 2009. 3. Achkar J, Xian M, Zhao H, Frost JW (2005) Biosynthesis of phloroglucinol. J Am Chem Soc 127(15):5332–5333. 4. Mercogliano, C.P., Watson, F.D., Lipscomb, T.E.W., Liao, H.H. and Lynch, M.D., Compositions and Methods Regarding Direct NADH Utilization to Produce 3Hydroxypropionic Acid, Derived Chemicals and Further Derived Products, in PCT/US2012/056159. March 28, 2013, OPX Biotechnologies, Inc.: USA. 5. Corporation, R., Official Methods of Analysis: method #986.13. AOAC International, 17th edition, 2000. 6. Rao G, Lee JK, Zhao H (2013) Directed evolution of phloroglucinol synthase PhlD with increased stability for phloroglucinol production. Appl Microbiol Biotechnol 97(13):5861-7. 7. Lei, Q.S., Larson, M.H., Gilbert, L.A., Doudna, J.A., Weissman, J.S., Arkin, A.P. and Wendell, L.A., Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell, 2013. 152: p. 10. 8. Srere, P.A., Escherichia coli citrate synthase: Purification and the effect of potassium on some properties. Biochemistry., 8(11), 4497-4503 (1969). 9. Graham, J.M., and Rickwood, D. (eds.), Subcellular Fractionation: A Practical Approach, Oxford Univ. Press Inc. (New York, NY: 1997) p.135. 10.Lynch, M., Lipscomb, T.E.W., Trahan, A.D., Singh, A. and Wolter, T., Microbial Production of Chemical Products and Related Compositions, Methods and Systems, in PCT/US2012/030209. September 27, 2012, OPX Biotechnologies, Inc.: USA. 11.Frost, J.W., Green Synthesis of Phloroglucinol: Exploiting Pseudomonas Fluorescence and Scale-Up. Draths Corporation 2010 Annual Report. 12.Jian J., Zhang SQ., Shi ZY., Wang W., Chen GQ., Wu Q., Production of polyhydroalkanoates by Escherichia coli mutants with defected mixed acid fermentation pathways. Appl Microbiol Biotechnol 2010, 87:2257-2256. 13.Ashworth, J.M., H.L. Kornberg, D.L. Nothmann 1965. Location of the structural gene for citrate synthase on the chromosome of Escherichia coli K12. J.Mol.Biol. 11:654-657 14.McGinness, K.E., Baker, T.A. and Sauer, R.T., Engineering controllable protein degradation. Mol Cell, 2006. 22(5): p. 701-7. ACKNOWLEDGEMENTS Approach: 1) Design an sgRNA sequence to target the gltA gene. 2) Evaluate the level of gltA repression via: a. measuring the effect of gltA repression on growth. b. measure the amount of CS enzyme via enzyme assay Test for Incompatibility The current lysis procedure is causing results to be inconclusive • Longer contact with BugBuster® lysis buffer led to lower citrate synthase activity. • A different cell lysis procedure must be used to overcome this incompatibility with BugBuster®. Citrate Synthase Enzyme Assay Method Development Researchers should avoid using BugBuster® to maintain maximal citrate synthase activity • Mechanical rather than chemical lysis would be the preferred method for generating whole cell lysates for the strains used in this study Figure 4. Growth and Induction Characterization of System Strains A. Growth Curves vs. Different E.coli Strains; B. Growth Curves vs. Different Induction Conditions Figure 1. Metabolic Pathways B Optimization of the Citrate Synthase Enzyme Assay Protocol Confirmed that lysate clarification using a 30,000 MWCO centrifugal filter effective • Filter removed reactants contained in the cell lysate, reducing background absorbance Confirmed that potassium is an activator of citrate synthase (data not shown) • Potassium ions increased the citrate synthase activity, as shown in previous studies. Characterization of Citrate Synthase Enzyme Activity Citrate synthase activity was not inhibited as expected • BWapldf, +sgRNA, and +sgRNA/dCas9 showed citrate synthase activity uninhibited • +dCas9, AB1623, and the negative control showed no enzyme activity • Hypothesis Testing (t-tests, Figure 5) 1: Does dCas9 induced at t=0 cause growth defect? (A vs. C) 2: Does sgRNA, combined with dCas9 induced at t=0, cause further growth defect? (C vs. E) 3: Does the combination of sgRNA/dCas9 induced at t=3HR cause growth defect? (A vs. F) A Shake Flask Growth Studies Growth defects were observed with system induction • dCas9 induction at t=0 causes a growth defect • Addition of sgRNA to strains with dCas9 induction at t=0 causes further growth defect • sgRNA and dCas9 combined with dCas9 induction at t=3HR also causes growth defect • This research was supported by grants from: 1. Defense Advanced Research Projects Agency (DARPA) 2. Duke University Undergraduate Research Support (URS) Figure 6. Citrate Synthase Enzyme Assay Development (A: Before Protocol Optimization; B: After Protocol Optimization, C: Test for BugBuster® Incompatibility) Contact: Ashley D. Trahan▐ Duke University▐ Biomedical Engineering▐ LSRC Room A020▐ Durham, NC 27705 ▐ 1-919-684-0235 ▐ Email: adt20@duke.edu • Special thanks to all members of the Lynch Lab, McCafferty Lab and Toone Lab