Minimum Redundancy and Maximum Relevance
advertisement

Outline
Minimum Redundancy and
Maximum Relevance
Feature Selection and Its
Applications
!
!
!
!
Hanchuan Peng
!
Janelia Farm Research Campus,
Howard Hughes Medical Institute
!
What is mRMR feature selection
Applications in cancer classification
Applications in image pattern recognition
Theoretical basis of mRMR
Combinations with other methods
How to use mRMR programs
1
2
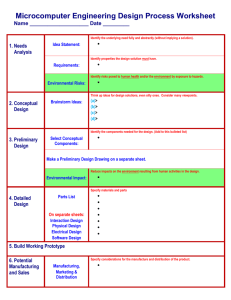
Feature Selection Problem
Redundancy in Features
S (selected variables, e.g. genes)
Data D
Target h
Variable pool
!={x1,…,x M}
!
A prediction example
Cancer
type h
! {Gene xi}
N Samples
G1
On
On
Baseline
Off
Baseline
On
Off
On
On
G2
On
Off
On
Off
Off
On
On
On
On
GM
On
Off
On
On
Off
On
Off
On
On
!
!
Prostate
Prostate
Liver
Liver
Lung
Lung
Lymphoma
Lymphoma
Lymphoma
Ding & Peng, CSB2003, JBCB2005
!
Ding & Peng, CSB2003, JBCB2005
!
WI =
" I (i, j )
!
Maximize Relevance:
!
V I = |S1 | " I (h, i )
i!S
VF =
1
| S | i! S
" F (i, h).
Minimize redundancy : Correlation c(i,j)
min Wc ,
I(i,j) is mutual information between features i and j
max V I ,
Maximize relevance : F-statistic: F(i,h)
max VF ,
1
| S | 2 i , j !S
S is the set of features.
!
4
mRMR: Continuous Variables
Minimize Redundancy:
min WI ,
Selected features are correlated;
Selected features cover narrow regions in space.
3
mRMR: Discrete Variables
!
Most used methods: select top-ranking
features based on mutual info., F-test, etc.,
without considering relationships among
features.
Problems:
Wc =
1
! | c(i, j ) |
|S |2 i , j
Relevance and redundancy can also be defined
using mutual info of hybrid variables (continuous
or categorical) (Peng, Long, & Ding, TPAMI, 2005)
h = target classes (e.g. types of different cancers, or annotations)
Ding & Peng, CSB2003, JBCB2005
5
Ding & Peng, CSB2003, JBCB2005
6
1
MRMR Selection Schemes
Combine Redundancy and Relevance
!
Additive combination
max(V ! W )
!
Multiplicative combination
max(V / W )
Ding & Peng, CSB2003, JBCB2005
Ding & Peng, CSB2003, JBCB2005
7
Most Related Methods
!
!
!
!
8
Outline
Most used feature selection methods: topranking features without considering
relationships among features.
Yu & Liu, 2003/2004. information gain,
essentially similar approach.
Wrapper: not filter approach, classifier-involved
and thus features do not generalize well.
PCA and ICA: Features are orthogonal or
independent, but not in the original feature
space.
!
!
!
!
!
!
What is mRMR feature selection
Applications in cancer classification
Applications in image pattern recognition
Theoretical basis of mRMR
Combinations with other methods
How to use mRMR programs
9
Microarray
Gene
Expression
Data Sets for
Cancer
Classification
10
Lymphoma Results
Ding & Peng, CSB2003, JBCB2005
11
Ding & Peng, CSB2003, JBCB2005
12
2
Do mRMR Features Better Cover the
Data Distribution Space and Thus
Perform Well on Different Classifiers?
What is the Role of
Redundancy Reduction?
Relevance
Redundancy
LOOCV Error
Average LOOCV errors of three different classifiers, NB, SVM, and
LDA on three multi-class datasets
(a) Relevance and (b) redundancy for MRMR features on discretized NCI dataset.
(c) The respective LOOCV errors obtained using the Naïve Bayes classifier
Ding & Peng, CSB2003, JBCB2005
LOOCV testing results (#error) for binarized NCI and
Lymphoma data using SVM classifier
Child Leukemia data (7 classes, 215 training samples, 112 testing samples)
testing errors. M is the number of features used in classification
Ding & Peng, CSB2003, JBCB2005
15
Comparison with Other Work
16
Outline
!
!
!
!
!
!
Ding & Peng, CSB2003, JBCB2005
14
What is the Relationship of mRMR
Features and Various Data
Discretization Schemes?
Do mRMR Features Generalize Well
on Unseen Data?
Ding & Peng, CSB2003, JBCB2005
Ding & Peng, CSB2003, JBCB2005
13
17
What is mRMR feature selection
Applications in cancer classification
Applications in image pattern recognition
Theoretical basis of mRMR
Combinations with other methods
How to use mRMR programs
18
3
Automatic Annotation of in situ Gene
Expression Patterns in Fly Embryos
!
!
!
!
Wavelet-embryo Features
Minimal redundant wavelet-embryo features were used to train classifiers.
456 genes, 80 ontological annotation terms, 6 developmental ranges.
Consistent automatic annotations with those manually generated in BDGP.
99.5% accuracy in predicting developmental stages of embryos by gene expression patterns.
re
co
es
nc
e
fid
on
nc
tio
c
i
ed
Pr
Short terms: ECNS: embryonic central nervous system; VNC: ventral
nerve cord; EH: embryonic hindgut; EM: embryonic midgut; EDE:
embryonic dorsal epidermis.
5-hours
5-years
Zhou & Peng, Bioinformatics, 2007
Peng et al, BMC Cell Bio, 2007
19
Annotation System
20
Tier 1 Accuracy: Developmental Stage Prediction
Wavelet-embryo features
Extract wavelet-embryo features
Tier 1
Select dev-stage-specific features
Assign dev-stage (classification)
Select annotation-Aspecific features
Select annotation-Bspecific features
…
Select annotation-Zspecific features
Predict annotation-A
(classification)
Predict annotation-B
(classification)
…
Predict annotation-Z
(classification)
mRMR feature selection
Tier 2
Classification
(LDA, SVM, etc.)
Overall accuracy >99%
Determine annotations based on probabilistic ranking
Peng et al, BMC Cell Bio, 2007
21
22
Tier 2 Accuracy: Specific Annotation Prediction
Outline
!
!
!
!
!
!
Zhou & Peng, Bioinformatics, 2007
23
What is mRMR feature selection
Applications in cancer classification
Applications in image pattern recognition
Theoretical basis of mRMR
Combinations with other methods
How to use mRMR programs
24
4
Mutual Information
Maximum Dependency Criterion
!
!
Statistical association
!
Definition
!
For two univariate variables x and y
I ( x; y ) = !! p ( x, y ) log
!
For multivariate variable Sm and the target h
Mutual information I(S,h)
I ( S m ; h) = !! p ( S m , h) log
not
S (selected variables)
Data D
p ( x, y )
dxdy
p( x) p( y )
Target h
p(x1, x 2 ,..., x m ,h)
dx1 Ldx m dh
p(x1 )L p(x m ) p(h)
p(Sm ,h)
p(Sm ,h)log
dSm dh
p(x1 )L p(x m ) p(h)
J(x1, x 2 ,..., x m ,h) = " L " p(x1,..., x m ,h)log
Variable pool
!={x1,…,x M}
Peng, Long, Ding, TPAMI 2005
p ( S m , h)
dS m dh
p ( S m ) p ( h)
= "L "
Peng, Long, Ding, TPAMI 2005
25
26
!
Maximum Dependency Feature
Selection is Combinatorial !
High-Dimensional Mutual Information
!
For multivariate variable Sm and the target h
!
p ( S m , h)
I ( S m ; h) = !! p ( S m , h) log
dS m dh
p ( S m ) p ( h)
!
p ( S m "1 , xm , h)
= !! p ( S m "1 , xm , h) log
dS m dh
p ( S m "1 , xm ) p (h)
= ! L ! p ( x1 , L , xm , h) log
!
Number of subspaces
!
p ( x1 , L , xm , h)
dx1 L dxm dh.
p ( x1 , L , xm ) p (h)
Total
Example:
2|!| or 2 M
For given number
of features |S|
&| ' |#
$$ | S | !!
%
"
Estimating high-dimensional I(S m,h) is difficult
!
!
An ill-posed problem to find inverse of large co-variance matrix
Insufficient number of samples
Peng, Long, Ding, TPAMI 2005
1000
1000
1000
1
2
3
103
0.5 x 106
~1.66 x 108
5000
3
~2.08 x 1010
#configurations
of selected
variables
Simplest case: the incremental search.
Peng, Long, Ding, TPAMI 2005
27
28
Upper & Lower Bounds of J(.)
Lower bound
Mutual information for multivariate variable S m
and the target h
J ( x1 , x2 ,..., xm ) ! 0.
p ( S m , h)
dS m dh
p ( S m ) p ( h)
when variables are maximally independent
Define:
p ( x1 , x 2 ,..., x m )
J ( x1 , x 2 ,..., x m ) = ! L ! p ( x1 ,..., x m ) log
dx1 L dx m
p ( x1 ) L p ( x m )
Upper bound
n
It can be proved:
J ( x1 , x2 ,..., xn ) $ min{! H ( xi ),
I ( S m , h ) = J ( h, S m ) ! J ( S m )
Peng, Long, Ding, TPAMI 2005
|S|
Heuristic search algorithms are necessary.
Factorize the Mutual Information
I ( S m ; h) = !! p ( S m , h) log
|!|
i =2
n
n
n "1
! H ( x ),L, ! H ( x ), ! H ( x )}
i
i =1,i # 2
i
i =1,i # n "1
i
i =1
when variables are maximally dependent
29
Peng, Long, Ding, TPAMI 2005
30
5
Factorize I(S m,h)
!
!
Advantages of mRMR
Relevance of S={x1, x2, …} and h, or RL(S,h)
Redundancy among variables {x1, x2, …}, or RD(S)
RL =
1
! I ( xi , h)
| S | xi "S
RD =
1
| S |2
! I (x , x
i
j
!
)
xi , x j "S
!
I ( S m , h) = J ( S m !1 , xm , h) ! J ( S m !1 , xm ).
!
!
For incremental search, max I(S,h) is “equivalent” to max
[RL(S,h) – RD(S)], i.e. combination of min-RedundancyMax-Relevance (mRMR).
!
!
Peng, Long, Ding, TPAMI 2005
Peng, Long, Ding, TPAMI 2005
!
!
!
!
In the pool ! find the variable x1 that has the largest
I(.,h). Exclude x1 from !.
Search x2 so that it maximizes I(.,h) – "I(.,x1)/|!|.
Iterate this process until an expected number of
variables have been obtained, or other constraints are
satisfied.
!
!
!
!
Complexity O(|S|*|!|)
!
Peng, Long, Ding, TPAMI 2005
32
Outline
Greedy search algorithm
!
mRMR is an optimal first-order approximation of I(.)
maximization
Relevance-only ranking only maximizes J(.)!
31
Search Algorithm of mRMR
!
Both relevance and redundancy estimation are lowdimensional problems (i.e. involving only 2 variables).
This is much easier than directly estimating
multivariate density or mutual information in the highdimensional space!
Faster speed
More reliable estimation
What is mRMR feature selection
Applications in cancer classification
Applications in image pattern recognition
Theoretical basis of mRMR
Comparison/Combinations with other
methods
How to use mRMR programs
33
Comparing Max-Dep and mRMR:
Complexity of Feature Selection
34
Comparing Max-Dep and mRMR: Accuracy
of Feature Selected in Classification
Time cost (seconds) for selecting individual features based on
mutual information estimation for continuous feature variables.
Leave-One-Out cross validation of feature classification
accuracies of mRMR and MaxDep
(Parallel experiments on a cluster of eight 3.06G Xeon CPUs
running Redhat Linux 9, with the Matlab implementation)
35
36
6
Use Wrappers to Refine Features
•
mRMR is a filter approach
• Fast
• Features might be redundant
• Independent of the classifier
•
Wrappers seek to minimize the number of errors directly
• Slow
• Features are less robust
• Dependent on classifier
• Better prediction accuracy
•
Use mRMR first to generate a short feature pool and
use wrappers to get a least redundant feature set with
better accuracy
Use Wrappers to Refine Features
Forward wrappers
(incremental selection)
Backward wrappers
(Decremental selection)
NCI data
37
mRMR Website
Outline
!
!
!
!
!
!
38
http://research.janelia.org/peng/proj/mRMR
What is mRMR feature selection
Applications in cancer classification
Applications in image pattern recognition
Theoretical basis of mRMR
Combinations with other methods
How to use mRMR programs
39
Available Datasets
Available Versions
!
!
!
All source codes. Can be embedded in your
programs easily.
!
C/C++ version
!
!
!
Matlab version
!
!
For Linux/Unix/Mac, simple command line
executable.
!
Online version
!
40
Upload the data sets (csv format: comma
separated values) and get the results right away.
!
41
NCI data (9 cancers, discretized as 3-states)
Lung Cancer data (7 subtypes, discretized as
3-states)
Lymphoma data (9 cancer-subtypes,
discretized as 3-states)
Leukemia data (2 subtypes, discretized as 3states)
Colon Cancer data (2 subtypes, discretized as
3-states)
The continuous-value raw data should be
obtained from the original sources.
42
7
Citations
!
!
This paper presents a theory of mutual information based feature selection. Demonstrates the
relationship of four selection schemes: maximum dependency, mRMR, maximum relevance, and
minimal redundancy. Also gives the combination scheme of "mRMR + wrapper" selection and
mutual information estimation of continuous/hybrid variables.
!
!
!
[JBCB05] Chris Ding, and Hanchuan Peng, "Minimum redundancy feature selection from
microarray gene expression data," Journal of Bioinformatics and Computational Biology,
Vol. 3, No. 2, pp.185-205, 2005.
!
!
!
This paper presents a comprehensive suite of experimental results of mRMR for microarray gene
selection on many different conditions. It is an extended version of the CSB03 paper.
[CSB03] Chris Ding, and Hanchuan Peng, "Minimum redundancy feature selection from
microarray gene expression data," Proc. 2nd IEEE Computational Systems Bioinformatics
Conference (CSB 2003), pp.523-528, Stanford, CA, Aug, 2003.
!
!
Conclusions
[TPAMI05] Hanchuan Peng, Fuhui Long, and Chris Ding, "Feature selection based on
mutual information: criteria of max-dependency, max-relevance, and min-redundancy,"
IEEE Transactions on Pattern Analysis and Machine Intelligence, Vol. 27, No. 8, pp.12261238, 2005.
The Max-Dependency feature selection can be efficiently
implemented as the mRMR algorithm.
Significantly outperforms the widely used max-relevance
selection method: mRMR features cover a broader
feature space with less features.
mRMR is very efficient and useful for gene selection and
many other applications. The programs are ready!
mRMR website:
http://research.janelia.org/peng/proj/mRMR
This paper presents the first set of mRMR results and different definitions of
relevance/redundancy terms.
[Bioinfo07] Jie Zhou, and Hanchuan Peng, "Automatic recognition and annotation of
gene expression patterns of fly embryos," Bioinformatics, Vol. 23, No. 5, pp. 589-596,
2007.
!
One application of mRMR in selecting good wavelet image features.
43
44
Acknowledgment
Collaborators:
Chris Ding
Fuhui Long
Jie Zhou
45
8