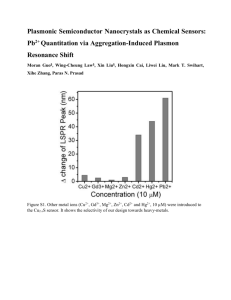
Zhanjiang (John) Liu
advertisement

Zhanjiang (John) Liu Associate Provost and Associate Vice President for Research Professor in the School of Fisheries, Aquaculture and Aquatic Sciences 202 Samford Hall, Auburn University, Auburn, AL 36849 USA Tel: (334)-844-4784 (office), (334)-246-0330 (home), FAX: 334-844-2937 E-mail: liuzhan@auburn.edu EXECUTIVE ACHIEVEMENT SUMMARY Career Development College education: Northwest A&F University, 1981 Master Education: University of Minnesota College of Agriculture, 1985 Doctoral education: University of Minnesota College of Biological Sciences, 1989 Postdoc: University of Minnesota Medical School, 1990-1993 R&D Director: National Biosciences, Inc., Minnesota, 1993-1994 Assistant Professor: Auburn University, 1995-1999 Associate Professor: Auburn University, 2000-2003 Professor: Auburn University, 2004-present Director, Aquatic Genomics Unit, Auburn University: 1999-2002 China Programs Coordinator: 2004-2007 Associate Dean for Research, College of Agriculture, Auburn University: 2007-2013 Associate Provost & Associate Vice President for Research, Auburn University: 2013present Scholarly Research Achievements Over 75 grants and contracts totaling over $47 million dollars; Published over 290 journal articles and book chapters; co-authored one patent; Published 3 books: “Aquaculture Genome Technologies”, “Next Generation Sequencing and Whole Genome Selection in Aquaculture”, “Functional Genomics in Aquaculture” and the 4th one “Bioinformatics in Aquaculture” is in press. Over 11,000 scientific citations with an H-index of 54, and i10-Index of 157; Research milestones include: 1990--Construction of the first “all-fish” expression vector that is still widely used in the world today; 1992--Production of transgenic northern pike; 1999--Application of DNA marker technologies in aquaculture; 2003--Construction of the catfish genetic linkage map; 2007--Generation of large (over 63,000) catfish BAC-end sequence resources; 2007--Development of high density microarray technology in catfish; 2007--Construction of the physical map of the catfish genome; 2009--Construction of gene-based genetic linkage map; 2010--Generation of over 500,000 EST resources in catfish by Sanger sequencing; 1 2010--Sequenced the catfish genome; 2011--Genome-wide discovery of 8.4 million catfish SNPs; 2012--Construction of the second-generation linkage map of the catfish genome and integration of genetic linkage and physical maps; 2012--Transcriptome sequencing and assembly of over 14,000 full-length cDNAs; 2012--Discovery and annotation of a unique set of 25,144 protein-encoding genes; 2014--Development of the catfish 250 SNP array technology 2014--Assembly of the catfish genome. Significant professional services and recognition by peers include: o AAAS Fellow o USDA recognition of significant contribution to aquaculture o Distinguished Research Award by Alabama Catfish Producers in recognition of outstanding research contributions and dedication to Alabama’s catfish industry. o Fulbright Specialist in Fish Genomics o Creative Research and Scholarship Award at Auburn University; o Academic Master for Aquaculture under “111” Project of China. o Alumni Professor at Auburn University; o Over 100 invited lectures including keynote and plenary lectures in over 20 countries; o Editor for Marine Biotechnology, Academic Editor for PLoS One, Associate Editor for BMC Genomics, Associate Editor for BMC Genetics, and on editorial board for various journals; o Adjunct or affiliate professor, honorary director or honorary dean for almost 20 universities and institutes in the world; o Congressional exhibition of research achievements; o Hou Ji Scholar, Northwest A&F University, China; o Deep Blue Scholar, Dalian Ocean University; o Board of Trustees, as an Eminent International Scholar, Guangdong Ocean University; o Eminent Professor, Shanghai Ocean University. Teaching, Advising and Training Achievements Developed two and taught three courses, with excellent student evaluations; Served on over 100 graduate committees; Served as major professor for more than 50 graduate students including over 40 Ph.D. and 10 M.S. students. Trained over 50 postdoctoral fellows or scholars. EDUCATION Ph.D. 1989 M.S. 1986 Molecular and Cellular Biology, University of Minnesota, St. Paul, MN Plant Pathology, University of Minnesota, St. Paul, MN 2 B.S. 1981 Plant Pathology and Entomology, with highest honor, Northwestern Agricultural University, Shaanxi Province, China ACADEMIC POSITIONS Associate Provost and Associate Vice President for Research Associate Dean for Research and Professor, College of Agriculture, and Assistant Director, Alabama Agricultural Experiment Station (AAES), Auburn University Alumni Professor and Director, Department of Fisheries and Allied Aquacultures and the Program of Cell and Molecular Biosciences Professor and China Programs Coordinator, College of Agriculture, Auburn University Associate Professor and Director, Department of Fisheries and Allied Aquacultures and the Program of Cell and Molecular Biosciences, Aquatic Genomics Unit, Auburn University Assistant Professor, Department of Fisheries and Allied Aquacultures, Auburn University Director, R&D, National Biosciences, Inc., Plymouth, Minnesota Research Professor, Institute of Human Genetics, University of Minnesota Postdoctoral Fellow, USDA-ARS Cereal Disease Laboratory, St. Paul, MN Graduate Research Assistant and Teaching Assistant, Department of Genetics and Cell Biology, University of Minnesota Graduate Research Assistant, Department of Plant Pathology, University of Minnesota Researcher, Plant Pathology, Beijing Agricultural University Research Assistant, Northwestern Agricultural University, China County Agent for Agricultural Extension, Yanchuan, Shaanxi 2013-Present 2007-2013 2002-2007 2004-2007 1999-2002 1995-1999 1994-1995 1991-1994 1990-1991 1986-1989 1983-1985 1982-1983 1978-1981 1974-1977 HONORS AND AWARDS Dean’s Grantsmanship Award, College of Agriculture Fulbright Specialist Grant Award, US Department of State Dean’s Grantsmanship Award, College of Agriculture Super Professor, FacultyRow Academy of Fellows, College of Agriculture, Auburn University Hou Ji Scholar, Northwest A&F University, Yanglin, China Honorary Dean, College of Marine Science and Engineering, Qingdao Agricultural University, China Guest Professor, Dalian Ocean University, China Academy of Inventors, Auburn University USDA Recognition for significant contribution to "National Animal Genome Program", USDA CSREES. 2015 2015 2014 2012 2012 2011-present 2012-present 2012-present 2011 2009 3 Honorary Director, Applied Genomics Institute, the Chinese Academy of Fisheries Sciences Second place award for most cited paper of the year, International Society of Animal Genetics. Guest professor, Northwest A&F University, China Guest professor, Liaoning Fisheries Research Academy, China Fellow, American Association for the Advancement of Science (AAAS). Creative Research & Scholarship Award, the highest research award at Auburn University. Alumni Professorship, Auburn University, in recognition of “outstanding contributions in research and teaching”. Guest professor, Ocean University of China Academic Master for Aquaculture under “111” Project of China. Director’s Outstanding Senior Research Award, Alabama Agricultural Experiment Station. Guest professor, Shanxi Agricultural University Guest professor, Heilongjiang Fisheries Research Institute Guest professor, Shanghai E-Institute Golden Summit Award from the Governor, Sichuan Province, China in recognition of “enthusiastic support to economic construction and social progress in Sichuan Province, and prominent contributions to international exchange and friendly cooperation between Sichuan and foreign countries.” Honorary Director, Key Laboratory of Aquaculture Genetics and Ecology, Ministry of Agriculture, Shanghai Fisheries University “Catfish Genome Research” exhibited on Capitol Hill, sponsored by NASULGC. Research featured in the AU Football season program and Auburn Tigers Magazine. Distinguished Research Award by Alabama Catfish Producers in recognition of outstanding research contributions and dedication to Alabama's catfish industry. Research featured on the cover of Biotechnology. Outstanding Student Leader Award, University of Minnesota, for service in the Association of International Student Associations. Presidential Award for Outstanding Student Leadership Competitive Graduate Fellowship, University of Minnesota. Certification for vital role in promoting friendship between people of the US and China, US-China Peoples’ Friendship Association of Minnesota. Graduate Scholarship, the Educational Council of China. Top Student List, Northwestern Agricultural University. Governor's Award for Model Student, Shaanxi Province, China. Governor’s Student Leadership Award, Shannxi Province. 2009-present 2008 2008-present 2007-present Since 2007 2007 2002-2007 2007-present 2007-present 2006 2005-present 2005-present 2004-present 2003 2003-present 2000 2000 1999 1990 1987 1986 1986 1985 1983 1981 1981 1981 4 President's Award for Model Student, Northwestern Agricultural University. Governor's Award for Model Student, Shaanxi Province. 1978-1981 1980 ADMINISTRATIVE/LEADERSHIP EXPERIENCE Associate Provost and Associate Vice President for Research, Auburn University: 2013present The responsibility of this position is to provide leadership for strategic planning and implementation for overall research enterprise at Auburn University that include, but not limited to, growing the research enterprise; fostering and facilitating interdisciplinary collaborations; enhancing discoveries, innovations, and scholarship; enhancing technology development and transfer for economic development; supporting a state-of-the-art research infrastructure; and ensuring research integrity and compliance. In addition to the institutional roles for research leadership, I also have direct responsibilities in 1) serving as the liaison among the Office of the Vice President for Research, the Provost Office, and the University’s colleges/schools, Auburn University Montgomery, Alabama Agricultural Experiment Station and Alabama Cooperative Extension System, for research, scholarship, and other sponsored program development; 2) Coordinating activities among the associate deans for research, Faculty Research Committee, Research Compliance and related committees, and other appropriate University and Senate committees, the University Research Council, and similar entities such as research task forces/councils/committees associated with implementing the strategic research goals of Auburn University; 3) Coordinating Human Resource matters (annual employee goal setting, performance reviews, hiring, improvement plans, reclassification, etc., for the Office of the Vice President for Research and Economic Development (VPRED); 4) Providing budgetary oversight for all units of OVPRED; 5) Providing leadership for Auburn Speaks, for This Is Research” student and faculty symposia, and for IGP; 6) facilitating for AU federal research agenda; 7) Engaging Research Advisory Board, especially the Academic Committee; and 8) Promoting and enhancing faculty recognition and awards programs; and 9) Carry out routine functions of the Provost Office related to research such as Review and approve start up packages, review and approve Professional Improvement Leave packages, review and approve SEC Faculty Achievement Award packages, develop and implement processes for cluster hire proposals, review and make recommendations for cluster hire proposals, review and approve Scholarship Incentive Program (SIP) packages, review and approve SURA Award packages; Provide leadership for web page development, maintenance, and update for OVPRED; and Oversight of all the offices related to internal functions: OSP, PDFS, Compliance, OTT, RA, University Veterinarian, Research Security, Business/HR, and Undergraduate Research, and responsible for the budget planning and operations of the Office of Vice President for Research and Economic Development (annual budget of ~$11M). Associate Dean for Research and Assistant Director, College of Agriculture and Alabama Agricultural Experiment Station (AAES): 2007-2013 Primary responsibilities include providing leadership for the College of Agriculture and AAES for research and graduate education; managing the administrative aspects of research for the college and AAES; developing and implementing research policies, ensuring compliance of 5 federal and state laws and regulations relevant to research in public and land grant universities, providing strong advocacy and leadership for development, implementation and funding of research opportunities within the College and AAES including interacting with US congressional offices and Alabama State legislative offices; enhancing collaborative research of faculty; managing AAES competitive grants programs; facilitating research efforts of the faculty through timely notification of funding opportunities and deadlines; and provide data and reports related to research programs to various groups and constituents, internal and external; leading and coordinating the preparation of five-year plans of work required by USDA-CSREES (now USDA-NIFA) from Alabama’s three land-grant universities and prepare annual reports of progress on these research programs; coordinating the preparation and approval of federally required Hatch and Multi-State Research funded projects submitted by faculty; working with Associate Deans of Research in AAES colleges and department heads/chairs, director, Outlying Units and faculty in identifying these needs for the college and other AAES units; serving as the interface between the college and the Office of Vice President for Research; and representing the Dean and Director at various meetings as requested. External duties include working with various commodity groups, Alabama Farmers Federation, state legislature, state agencies, and federal government, participation and contribution to the Southern Association of Agricultural Experiment Station Directors, serving as administrative advisor for multistate project(s); provide advice to the Dean on personnel decisions such as approval of faculty and staff hiring, budget development and management, tenure and promotion reviews, and annual faculty reviews. International/China Programs Coordinator, College of Agriculture: 2004-2007 The College of Agriculture at Auburn University plays a leading role in international research collaborations, academic exchanges, and outreach activities and interactions. A significant part of such activities involves China. To enhance such international activities, I was appointed to serve as the International/China Programs Coordinator during 2004-2007. My major responsibilities included leadership for the development of collaborative relationships, execution of collaborative and/or cooperative agreements, and coordination of various exchange programs such as Maymester teaching programs, study abroad, and joint/dual degree programs, and hosting various visitors and delegations. Director of Aquatic Genomics Unit and Executive, Cell and Molecular Biosciences Program: 2000-2007 I was one of the founding members of the inter-departmental CMB program involving over 70 faculty members. Genomics was one of the three major components of CMB. I was the founding member of the Aquatic Genomics Unit and have served as its Director since 2000. Leadership as the Coordinator of the USDA National Animal Genome Program, NRSP-8 Aquaculture: 2000-present The USDA National Animal Genome Program was established as a National Research Program. Its leadership and coordination was conducted through NRSP-8. I led the effort of aquaculture genome and brought this then “falling behind group” successfully to NRSP-8 as an official program, and obtained research support funds (annually $65,000). Active leadership was provided to over 400 scientists all over the world in the area of aquaculture genetics. 6 PROFESSIONAL SERVICE EXPERIENCE Grant Review Panel Experience Genome BC, Canada USDA AFRI Review Panel National Cheng Kung University Aim for the Top University Project review USDA AFRI Review Panel Overseas Expert Review Panel, Chinese Academy of Sciences Integrative Organismal Systems, National Science Foundation Grant Review, Portuguese Foundation for Science and Technology (Fundação para a Ciência e a Tecnologia) Grant Review Panel, USDA NIFA Translational Genomics Grant Review Panel, NSF SBIR “Food Safety” Program. Grant Review Panel, USDA “Critical Issues in Emerging Plant and Animal Diseases" program. South Africa’s National Research Foundation Research Evaluations. Natural Sciences and Engineering Research Council (NSERC) of Canada grant review. Grant Review Panel, USDA NRI Whole Genome-based Selection. Grant Review Panel, NSF SBIR Phase II Program. Grant Review Panel, Natural Science Foundation of China. Grant Review Panel, USDA NRI Animal Genome Program. Grant Review Panel, Natural Science Foundation of China. Grant Panel Manager, USDA NRI Porcine Genome Sequencing Panel. USDA Special Grants Program for Tropical and Subtropical Agriculture. Review Panel, USDA ARS Scientific Quality Program Review, NP 106Biology, Genetics, and Husbandry Program. NIH/CSR study section, Immunology. Aquaculture Chairperson, National Animal Genome Project NRSP-8. Grant Review Panel, Wisconsin Sea Grant. Grant Review Panel, USDA NRI Animal Genomes and Genetic Mechanisms Program Grant Review Panel, USDA NRI Animal Growth and Nutrition Program. External Expert Review Panel, Genoma Espana and Genome Canada. Grant Review Panel, USDA NRI Animal Growth and Nutrition Program. Grant Review Panel, USDA SBIR Aquaculture Program. Grant Review Panel, Maryland Sea Grant. 2015 2015 2014 2014 20132016 2013 2012 2012 2010 2009 2008 2008 2008 2007 2007 2006 2006 2005 2005 2004 2004 20022003 2003 2003 2003 2003 2002 2002 2001 7 Canadian AquaNet Research Projects review. Review Panel, USDA ARS Aquaculture Program. Grant Review Panel, Maryland Sea Grant. USDA ARS review, Catfish Genetics Research Unit Five Year Research Plan. Grant Review Panel, NOAA Sea Grant, Aquaculture Endocrinology and Molecular Genetics. 2001 2000 1999 1998 1997 Journal Editorial Experience 2004-present Co-Editor-in-Chief, Marine Biotechnology. 2010-present Academic Editor, PLoS One 2010- present Associate Editor, BMC Genomics 2009- present Associate editor, BMC Genetics. 2014-present Editorial Board, Frontiers of Agricultural Science and Engineering 2015-2020 Editorial Board, Journal of Fishery Sciences of China 2004-2014 Editorial Board, Aquaculture. 2007-present Editorial Board, Animal Biotechnology 2008-present Editorial Board, Reviews in Aquaculture. 2007-present Editorial Board, Chinese Journal of Oceanology and Limnology. 2014-present Editorial Board, Frontiers of Agricultural Science and Engineering 2002-2004 Editorial Board, Marine Biotechnology. Also served as reviewer for over 40 professional journals including Acta Zoologica Sinica, Animal Biotechnology, Animal Genetics, Aquaculture, Aquaculture Research, Aquatic Living Resources, Biochemical Genetics, Biochemical Systematics and Ecology, BMC Genomics, BMC Evolutionary Biology, BMC Molecular Biology, Cell Research, Comparative Biochemistry and Physiology, Developmental and Comparative Immunology, Developmental Dynamics, Diseases of Aquatic Organisms, DNA Sequence, DNA and Cell Biology, Electrophoresis, Fish Physiology and Biochemistry, Food Microbiology, Gene, General and Comparative Endocrinology, Genetics, Genome Research, Genomics, Global Science Books, In Vitro Cellular and Developmental Biology, International Journal of Biological Sciences, Journal of Animal Science, Journal of Comparative Physiology, Journal of Experimental Zoology, Journal of Fish Biology, Journal of Heredity, Journal of Molecular Endocrinology, Journal of Nutrition, Journal of the World Aquaculture Society, Mammalian Genome, Marine Biotechnology, Micron, Molecular and General Genetics, Molecular Genetics and Genomics, Molecular Ecology, Molecular Marine Biology and Biotechnology, Nature, Nucleic Acids Research, The FASEB Journal, and Transactions of the American Fisheries Society. National & International Program Leadership and Advisory Panels APLU ESCOP Science & Technology Committee. Academic Committee, Qingdao Marine Sciences and Technology National Laboratory USDA National Aquaculture Genome Coordinator. Advisory Panel, Oceans and Human Health Initiative, NOAA, 2008-2013 2015-2020 2003- present 2006-2012 8 Department of Commerce. Administrative Advisor for NRSP-7, "A National Agricultural Program for Minor Use Animal Drugs”. Administrative Advisor for Multistate Project SERA 17, Minimizing Agricultural Phosphorus Run-off Losses for Protection of the Water Resource. Administrative Advisor, Multi-State Project S-1042, Modeling for TMDL Development, and Watershed Based Planning, Management and Assessment. Southern Regional Aquaculture Center Board of Directors. Advisory Council, National Egg Processing Center. Academic Committee, Key Laboratory of Marine Genetics and Breeding, the Ministry of Education of China Consultant, U.S. Veterinary Immune Reagent Network. National Coordinator of the Catfish Species Gene Mapping Efforts. National Aquaculture Gene Mapping Executive Committee. Expert Advisory Panel, Ministry of Education, Ministry of Agriculture, and the Ministry of Science and Technology of China. Expert Panelist, Conference on Aquaculture Genetics in the Third Millennium, Bangkok, Thailand, organized by the United Nations. 2009- 2013 2009- 2013 2008- 2013 2010-2013 2008-2013 2011-present 2005-2009 1997-2003 1997-2003 1999 2000 Conference Organizer and Session Chair Organizer, USDA Aquaculture Genomics, Genetics and Breeding Workshop Scientific Committee, ISGA XII, Santiago de Compostela, Spain Session Chair, Functional Genomics: Fish, International Conference of Fish and Shellfish Immunology, Vigo, Spain Organizing Committee, The Third Conference of Genomics in Aquaculture (GIA) 2013, Bodo, Norway Session Chair, Hi-Techs in Aquaculture, World Aquaculture Society Aquaculture 2012, Prague, Czech, September 1-5, 2012. Session Chair, Genomics, World Aquaculture Society Aquaculture America 2011, New Orleans, Louisiana February 28-March 3, 2011. Session Co-Chair, Finfish Genetics, World Aquaculture Society Aquaculture 2011, Natal, Brazil, June 6 - 10, 2011. Organizing Committee, International Plant & Animal Genome Conference. Co-Chairperson, International Plant & Animal Genome Conference XVIII, January 8-14, 2009, San Diego, USA. Session Chair, Aquaculture genetics, World Aquaculture Society, Veracruz, Mexico. Scientific Committee, International Symposium of Genetics in Aquaculture (ISGA 2009), June 22-26, 2009. Bangkok, Thailand. Session Chair, Molecular immune response mechanisms of fish, 5th 2016 2015 2013 2013 2012 2011 2011 2009-2012 2009 2009 2009 2008 9 World Fisheries Congress, October 20-24, Yokohama, Japan. Vice Chair, Gordon Conference for Marine Genomics, BioInformatics and Climate. Chairman, International Conference on Healthy Aquaculture, May 2427, Qingdao, China. Session Chair for Molecular aspects and biotechnology, and Session Chair for Genomics, US-Israel Aquaculture Genetics Workshop. International Advisory Committee, 13th International Congress on Genes, Gene Families and Isozymes and 2005’ Forum on Fishery Science and Technology, Sept. 17-21, 2005. Shanghai, China. Expert Panel, Workshop on Genetic Improvement and Selective Breeding for the Hybrid Striped Bass Industry. Stuttgart, Arkansas. Expert Consultation on Strategies and Plans for Dissemination of Improved Fish Breeds, NAGRI, Pathumthani, Thailand. International Co-Chair, Aquatic Genomics Session, World Aquaculture Society Annual Conference, Beijing, China. Organizer, National Aquaculture Genomics Workshop 2002 in San Diego, CA. Program Review, Kuwait Science Foundation, "Developing and Application of DNA Fingerprinting in Zobaidy (Pampus Aregenteus) in the Northern Arabian Gulf|", Project Code: 2004-1207-10. 2008 2008 2006 2005 2002 2002 2002 2001-2002 2010 Tenure and Promotion Reviews I have evaluated P&T packages for many Auburn University faculty members and faculty members or scientists from various universities and government agencies including: Brigham Young University Israel Oceanographic & limnological Research Ltd. Memorial University of New Foundland, Canada Rutgers University Texas A&M University University of Delaware University of Hawaii University of Idaho University of Malaysia Terengganu University of Victoria, canada University of Washington USDA-ARS Virginia Institute of Marine Science Virginia Polytech and State University Washington State University Auburn University Committee Services Auburn University RESTORE Initiative Working Group 2012-present 10 Auburn University Council for Energy, Environment, and Economic Research (CEEER) Luce Professor of Forestry Selection Committee, School of Forestry & Wildlife Sciences College Facilitator, Department Head Review for Entomology and Plant Pathology College Facilitator, Department Head Review for Agricultural Economics and Rural Sociology College Facilitator, Department Head Review for Animal Sciences Search Committee for Chief Financial Officer, AAES and College of Agriculturre Endowed Professors Selection Committee, School of Forestry & Wildlife Sciences Promotion and Tenure Committee, Chair every other year Search Committee Chair for Director, AU Food Systems Initiative Auburn University Research Council Budget Committee, College of Agriculture Graduate School Deans Liaison Council AU Intramural Grant Review Limited Submission Committee Chair, Research Committee, College of Agriculture Space Grant Review Panel. Advisor for Department Head BioGrant Review Committee, Auburn University. Competitive Research Grant Committee, Auburn University. Research/Hatch Project Review Committee. Research Grant-in-Aid Grant Committee. Creative Research & Scholarship Award Selection Committee, Auburn University. Director’s Research Award Committee, AAES. Institutional Biosafety Committee, Auburn University. Radiological Safety Committee, Auburn University. Institutional Biosafety Committee, Auburn University. Interim Chair, Institutional Biosafety Committee, Auburn University. Radiological Safety Committee, Auburn University. Instrumentation Committee, Auburn University. “Core Team” member for the AU Food Safety Initiative Chair, Discipline group of “Genetics, Health, Nutrition, Processing”, Department of Fisheries and Allied Aquacultures. Founding Committee, Inter-college Cell and Molecular Biosciences Program 2012-2014 2012 2012 2012 2012 2012 2011 2010-present 2009 2008-present 2008-present 2009-present 2008-present 2008-present 2008 2005-2008 2005-2008 2001-2004 1997-present 2002-2005 2007-present 2007 1997-1999 1998-2000 2000-2003 2003 2001-2004 2000-2004 2010 1998-present 11 Executive Committee, Cell and Molecular Biosciences Program Planning Committee, Department of Fisheries and Allied Aquacultures. Cell and Molecular Biosciences Program-Initiating Group. Strategic Planning Committee Chair, P & T Committee, College of Agriculture Search Committee member for Associate Dean for Research and Graduate Studies, College of Sciences and Mathematics Search Committee Chair for Director, AU Food Safety Initiative Search Committee Chair, Aquaculture Genomics Faculty Position. Board of Directors, National Poultry Technology Center. Search Committee Chair, Microbial Genomics faculty position. Chair, Search Committee, research assistant. Search Committee for research associate Acquisition #30490. Title Change Committee (to research professor from research associate), University of Minnesota. 1999-2006 1997-present 1998 2010-present 2010-present 2010 2010 2008 2008-present 2001 1996 1996 1993-1994 PROFESSIONAL SOCIETY AFFILIATIONS American Association for the Advancement of Science (AAAS). World Aquaculture Society. American Fisheries Society. Physiology Section, American Fisheries Society. Genetics Section, American Fisheries Society. Honorary Member, American Institute of Biological Sciences. American Phytopathological Society. 2007-present 2004-present 1998-2003 1998-2003 1998-2003 1997-1998 1983-1985 TEACHING, ADVISING AND TRAINING EXPERIENCE Classroom Teaching FISH7660/CMBL7660, Molecular Genetics and Biotechnology, taught 1996-2008 FISH7970, Special problems of molecular genetics, taught 1995-2013 FISH7653, Fish Genetics and Genetic Improvement, taught 1996-2000 Student Advising Served as major professor for over 40 Ph.D and 10 master students 12 Served as committee member for 45 students including 25 Ph.D. and 20 master students Trained over 50 postdoctoral fellows and visiting scientists Most of my former students are now university faculty members Served as reader for 7 international Ph.D. students SCHOLARLY AND CREATIVE RESEARCH EXPERIENCE Published Books 1. Liu ZJ. Editor, Aquaculture Genome Technologies, pp.551, Blackwell Publishing, 2007, Ames, IA. ISBN: 978-0-8138-0203-9. This book has been translated into Chinese language by Bao et al. and published by the Chinese Chemical Publishing, 2011. 2. Liu ZJ. Editor, Next Generation Sequencing & Whole Genome Selection in Aquaculture, pp.221, Wiley and Blackwell Publishing, 2011, Ames, IA. ISBN: 978-0-8138-0637-2 3. Saroglia M and Liu ZJ, editors, Functional Genomics in Aquaculture, pp. 407, Wiley and Blackwell Publishing, 2012, Ames, IA. ISBN 978-0-4709-6008-0. This book has been translated into Chinese language by Zhang et al. and published by the Scientific Press of China, 2015. 4. Liu ZJ. Editor, Bioinformatics in Aquaculture, Wiley and Blackwell Publishing, 2016, Ames, IA. In press. Patents Rex Dunham, Zhanjiang Liu, and Gregory Warr. US Patent 7,183,079 “Compositions and Methods for Enhancing Disease Resistance in Fish”, Awarded on February 27, 2007. Journal Articles and Book Chapters 1. Liu S and Liu ZJ. 2016. Chapter 1 Introduction to Linux and command line tools for bioinformatics. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 2. Gao S, Yuan Z, Li N, Zhang J and Liu ZJ. 2016. Chapter 2 Determining sequence identities: BLAST, Phylogenetic analysis, and syntenic analyses. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 3. Li N, Wang X and Liu ZJ. 2016. Chapter 3 Assembly of genomic sequences generated from next generation sequencing. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 4. Wang R, Bao L, Liu S and Liu ZJ. 2016. Chapter 4 Genome annotation: determination of the coding potential of the genome. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 5. Bao L and Liu ZJ. 2016. Chapter 5 Analysis of repetitive elements in the genome. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 13 6. Wang R and Liu ZJ. 2016. Chapter 6 Analysis of duplicated genes and multi-gene families. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 7. Yao J, Jiang C, Li C, Zeng Q and Liu ZJ. 2016. Chapter 8 Assembly of RNA-Seq short reads into transcriptome sequences. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 8. Li C, Zeng Q, Jiang C, Yao J and Liu ZJ. 2016. Chapter 9 Analysis of differentially expressed genes and co-expressed genes using RNA-Seq datasets. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 9. Zhou T, Yao J and Liu ZJ. 2016. Chapter 10. Gene ontology, enrichment analysis and pathway analysis. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 10. Yao J, Wang R and Liu ZJ. 2016. Chapter 11 Genetic analysis using RNA-Seq: Bulk segregant RNA-Seq (BSR-Seq). In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 11. Wang R, Bao L, Liu S and Liu ZJ. 2016. Chapter 12 Analysis of long non-coding RNAs (lncRNAs). In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 12. Liu S and Liu ZJ. 2016. Chapter 13 Analysis of microRNAs and their target genes. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 13. Li Y, Chen A and Liu ZJ. 2016. Chapter 14 Analysis of allele-specific expression. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 14. Wang S, Jiang Y and Liu ZJ. 2016. Chapter 16 Bioinformatic mining of microsatellite markers from genomic and transcriptomic sequences. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 15. Zeng Q, Sun L, Fu Q, Liu S and Liu ZJ. 2016. Chapter 17 SNP identification from next generation sequencing datasets. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 16. Liu S, Zeng Q, Wang X and Liu ZJ. 2016. Chapter 18 Development of SNP arrays and genotyping analysis. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 17. Li Y, Liu S, Wang R, Qin Z and Liu ZJ. 2016. Chapter 20 Bioinformatic considerations and approaches for high-density linkage mapping in aquaculture. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 18. Geng X, Zhi D and Liu ZJ. 2016. Chapter 23 Genome-wide association studies of performance traits. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 14 19. Liu S, Zeng P and Liu ZJ. 2016. Chapter 24 Gene set analysis of SNP data from genomewide association studies. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 20. Yuan Z, Yang Y, Liu S and Liu ZJ. 2016. Chapter 26 NCBI resources useful for informatics issues in aquaculture. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 21. Jin Y, Tan S, Yao J and Liu ZJ. 2016. Chapter 27 Resources and Bioinformatic Tools in Ensembl. In: Bioinformatics in Aquaculture (edited by Liu ZJ.), Blackwell Publishing, Ames, IA. In press. 22. Zhou X, Cui J, Liu S, Kong D, Sun H, Gu C, Wang H, Qiu X, Chang Y, Liu ZJ, Wang X. 2016. Comparative transcriptome analysis of papilla and skin in the sea cucumber, Apostichopus japonicas. PeerJ, in press. 23. Li Z, Yao J, Xie Y, Geng X, Liu ZJ. 2016. Phosphoinositide 3-kinase family in channel catfish and their regulated expression after bacterial infection. Fish Shellfish Immunology 49: 364-373. 24. Fu Q, Li Y, Yang Y, Li C, Yao J, Zeng Q, Qin Z, Li Dao, Liu ZJ. 2016. Septin genes in channel catfish (Ictalurus punctatus) and their involvement in disease defense responses. Fish and Shellfish Immunology 49: 110-121. 25. Liu ZJ, Liu S, Yao J, Bao L, Zhang J, Li Y, Jiang C, Sun L, Zhang Y, Zhou T, Zeng Q, Fu Q, Gao S, Li N, Wang R, Koren S, Jiang Y, Zimin A, Xu P, Phillippy AM, Geng X, Song L, Sun F, Li C, Wang X, Chen A, Jin Y, Yuan Z, Yang Y, Tan S, Peatman E, Lu J, Qin Z, Dunham R, Li Z, Sonstegard T, Feng J, Danzmann RG, Schroeder S, Scheffler B, Duke MV, Ballard L, Kucuktas H, Kaltenboeck L, Liu H, Armbruster J, Xie Y, Kirby ML, Tian Y, Flanagan ME, Mu W, Waldbieser GC. 2016. The channel catfish genome sequence and insights into evolution of scale formation in teleosts. Nature Communications, under revision. 26. Chen A, Wang, R, Liu S, Peatman E, Sun L, Bao L, Jiang, C, Li C, Li Y, Zeng Q, Liu ZJ. 2016. Ribosomal protein genes are highly enriched among genes with allele-specific expression in the interspecific F1 hybrid catfish. Molecular Genetics and Genomics, in press. DOI: 10.1007/s00438-015-1162-z 27. Jin Y, Liu S, Yuan Z, Yang Y, Tan S, Liu ZJ. 2016. Catfish genomic studies: progress and perspectives. In: Genomics in Aquaculture, edited by Simon MacKenzie & Sissel Jentoft. In press. 28. Song L, Li C, Xie Y, Liu S, Zhang J, Yao J, Jiang C, Li Y, Liu ZJ. 2016. Genome-wide identification of Hsp70 genes in channel catfish and their regulated expression after bacterial infection. Fish and Shellfish Immunology 49: 154-162. 29. Su B, Shang M, Grewe P, Patil JG, Peatman E, Perera D, Cheng Q, Li C, Weng C, Li P, Liu ZJ and Dunham R. 2015. Suppression and restoration of primordial germ cell marker gene expression in channel catfish, Ictalurus punctatus, using knock-down constructs regulated by copper transport protein gene promoters: potential for reversible transgenic sterilization. Theriogenology 84: 1499-1512. 15 30. Jiang C, Zhang J, Yao J, Liu S, Li Y, Song L, Li C, Wang X, Liu ZJ. 2015. Complement regulatory protein genes in channel catfish and their involvement in disease defense response. Developmental and Comparative Immunology 53: 33-41. 31. Wang H, Liu S, Cui J, Li C, Hu Y, Zhou W, Chang Y, Qiu X, Liu ZJ, Wang Xiuli. 2015. Identification and characterization of microRNAs from longitudinal muscle and respiratory tree in sea cucumber (Apostichopus japonicus) using high-throughput sequencing. PLoS One 10(8): e0134899. doi:10.1371/journal.pone.0134899. 32. Xie, Y, Song L, Weng Z, Liu S, Liu ZJ. 2015. Hsp90, Hsp60 and sHsp families of heat shock protein genes in channel catfish and their expression after bacterial infections. Fish and Shellfish Immunology 44: 642-651. 33. Tian Y, Yao J, Liu S, Jiang C, Zhang J, Li Y, Liu ZJ. 2015. Identification and expression analysis of 26 oncogenes of the receptor tyrosine kinase family in channel catfish after bacterial infection and hypoxic stress. Comparative Biochemistry and Physiology Part D, Genomics and Proteomics 14: 16-25. 34. Liu S, Li Y, Qin Z, Geng X, Bao L, Kaltenboeck L, Kucuktas H, Dunham R, Liu ZJ. 2015. High-density SNP-based genetic mapping of interspecific hybrid catfish reveals intrinsic genomic incompatibility between Channel catfish and Blue catfish. Animal Genetics 47: 8190. doi: 10.1111/age.12372. 35. Geng X, Sha J, Liu S, Bao L, Zhang J, Wang R, Yao J, Li C, Feng J, Sun F, Sun L, Jiang C, Dunham R, Zhi D, Liu ZJ. 2015. A genome-wide association study in catfish reveals the presence of functional hubs of related genes within QTLs for columnaris disease resistance. BMC Genomics 16: 196. DOI 10.1186/s12864-015-1409-4. 36. Sun L, Liu S, Bao L, Li Y, Feng J, Liu ZJ. 2015. Claudin multigene family in channel catfish and their expression profiles in response to bacterial infection and hypoxia as revealed by meta-analysis of RNA-Seq datasets. Comparative Biochemistry and Physiology, Part D, Genomics and Proteomics 13: 60-69. 37. Li Y, Liu S, Qin Z, Yao J, Jiang C, Song L, Dunham R, Liu ZJ. 2015. The serpin superfamily in channel catfish: Identification, phylogenetic analysis and expression profiling in mucosal tissues after bacterial infections. Developmental and Comparative Immunology 49:267-277. 38. Li Y, Liu S, Qin Z, Waldbieser G, Wang R, Sun L, Bao L, Danzmann R, Dunham R, Liu ZJ. 2015. Construction of a high-density high-resolution genetic map and its integration with BAC-based physical map in channel catfish. DNA Research 22:39-52. doi:10.1093/dnares/dsu038 39. Feng J, Liu S, Wang R, Zhang J, Wang X, Kaltenboeck L, Li J, Liu ZJ. 2015. Molecular characterization, phylogenetic analysis and expression profiling of myoglobin and cytoglobin genes in response to heat stress in channel catfish. Journal of Fish Biology 86: 592-604. 40. Song L, Zhang J, Li C, Yao J, Jiang C, Li Y, Liu S, Liu ZJ. 2015. Genome-wide identification of Hsp40 genes in channel catfish and their regulated expression after bacterial infection. PLoS One 9(12): e115752. doi:10.1371/journal.pone. 16 41. Mu W, Yao J, Zhang J, Liu S, Wen H, Feng J, Liu ZJ. 2015. Expression of tumor suppressor genes in channel catfish after bacterial infections. Developmental and Comparative Immunology 48: 171-177. 42. Yao J, Mu W, Liu S, Zhang J, Wen H, Liu ZJ. 2015. Identification, phylogeny and expression analysis of suppressors of cytokine signaling in channel catfish. Molecular Immunology 64:276-284. 43. Su B, Shang M, Grewe P, Patil JG, Peatman E, Perera D, Cheng Q, Li C, Weng C, Li P, Liu ZJ; Dunham R. 2015. Reversible transgenic sterilization in channel catfish, Ictalurus punctatus, using copper transport protein gene promoters and copper sulfate as a repressor. PLoS One, submitted. 44. Liu ZJ. 2014. Editorial, GIA 2013: Genomics in Aquaculture 2013 symposium. Marine Genomics, 18: 1-2. doi:10.1016/j.margen.2014.10.005 45. Wang H, Liu S, Cui J, Li C, Qiu X, Chang Y, Liu ZJ, Wang X. 2014. Characterization and expression analysis of microRNAs in the tube foot of sea cucumber Apostichopus japonicas. PLoS One 9(11):e111820. 46. Liu S, Zhang J, Yao J, and Liu ZJ. 2014. The complete mitochondrial genome of the armored catfish, Hypostomus plecostomus (Siluriformes: Loricariidae). Mitochondria DNA 20: 1-2. 47. Liu S, Yao J, Zhang J, and Liu ZJ. 2014. Next generation sequencing yields the complete mitochondrial genome of the striped raphael catfish, Platydoras armatulus (Siluriformes: Doradidae). Mitochondria DNA, in press. 48. Sun L, Liu S, Wang R, Jiang Y, Zhang Y, Zhang J, Bao L, Kaltenboeck L, Dunham R, Waldbieser G, Liu ZJ. 2014. Identification and analysis of genome-wide SNPs provide insight into signatures of selection and domestication in catfish. PLoS One 9(10): e109666. 49. Argue B, Kuhlers D, Liu ZJ, Dunham RA. 2014. Growth of channel catfish (Ictalurus punctatus), blue catfish (I. furcatus), and their F1, F2, F3, and F1 reciprocal backcross hybrids in earthen ponds. Journal of Animal Science 92(10):4297-305. 50. Xu Peng, Xiaofeng Zhang, Xumin Wang, Jiongtang Li, Guiming Liu, Youyi Kuang, Jian Xu, Xianhu Zheng, Lufeng Ren, Guoliang Wang, Yan Zhang, Linhe Huo, Zixia Zhao, Dingchen Cao, Cuiyun Lu, Chao Li, Yi Zhou, Zhanjiang Liu, Zhonghua Fan, Guangle Shan, Xingang Li, Shuangxiu Wu, Lipu Song, Guangyuan Hou, Yanliang Jiang, Zsigmond Jeney, Dan Yu, Li Wang, Changjun Shao, Lai Song, Jing Sun, Peifeng Ji, Jian Wang, Qiang Li, Liming Xu, Fanyue Sun, Jianxin Feng, Chenghui Wang, Shaolin Wang, Baosen Wang, Yan Li, Yaping Zhu, Wei Xue, Lan Zhao, Jintu Wang, Ying Gu, Weihua Lv, Kejing Wu, Jingfa Xiao, Jiayan Wu, Zhang Zhang, Jun Yu, Xiaowen Sun. 2014. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nature Genetics 6(11):1212-1219. doi: 10.1038/ng.3098. 51. Sun L, Liu S, Wang R, Li C, Zhang J, Liu ZJ. 2014. Pathogen recognition receptors in channel catfish: IV. Identification, phylogeny and expression analysis of peptidoglycan recognition proteins. Developmental and Comparative Immunology 46:291-299. 17 52. Jiang Y, Xu P, Liu ZJ. 2014. Generation of physical map contig-specific sequences. Frontiers in Genetics 5: 243. 53. Wong L, Kucuktas H; Arai T; Dobson S; Supriharti D; Liu ZJ. 2014. Assessment of Population Genetic Structure of Redeye Bass (Micropterus coosae): Implications for Fish Stock Management. PLoS One, submitted. 54. Dunham R, Taylor JF, Rise M, Liu ZJ, 2014. Development of strategies for integrated breeding, genetics and applied genomics for genetic improvement of aquatic organisms. Aquaculture 420-421:S121-S123. 55. Zhang J, Yao J, Wang R, Zhang Y, Liu S, Sun L, Jiang Y, Feng J, Liu N, Nelson D, Waldbieser G, Liu ZJ. 2014. The cytochrome P450 genes of channel catfish: their involvement in disease defense responses as revealed by meta-analysis of RNA-Seq datasets. Biochimica et Biophysica Acta-General Subjects 1840:2813-2828. 56. Yao J, Li C, Zhang J, Liu S, Feng J, Wang R, Li Y, Jiang C, Song L, Chen A, Liu ZJ. 2014. Expression of nitric oxide synthase (NOS) genes in channel catfish is highly regulated and time dependent after bacterial challenges. Developmental and Comparative Immunology 45:74-86. 57. Hutson AM, Liu ZJ., Kucuktas H, Umali-Maceina G, Su B, and Dunham RA. 2014. QTL map for growth and morphometric traits using a channel catfish x blue catfish interspecific hybrid system. Journal of Animal Science 92:1850-1865. 58. Wang R, Zhang Y, Liu S, Li C, Sun L, Bao L, Feng J, Liu ZJ. 2014. Analysis of 52 Rab GTPases from channel catfish and their involvement in immune responses after bacterial infections. Developmental and Comparative Immunology 45:21-34. 59. Wang H, Liu S, Cui J, Li C, Qiu X, Chang Y, Liu ZJ, Wang X. 2014. Characterization and expression analysis of microRNAs in the tube foot of sea cucumber Apostichopus japonicus. PLoS ONE, in review. 60. Liu S, Sun L, Li Y, Sun F, Jiang Y, Zhang Y, Zhang J, Feng J, Kaltenboeck L, Kucuktas H, and Liu ZJ. 2014. Development of the catfish 250K SNP array for genome-wide association studies. BMC Research Notes 7:135. DOI: 10.1186/1756-0500-7-135. 61. Geng X, Feng J, Liu S, Wang Y, 2, Arias C, Liu ZJ. 2014. Transcriptional regulation of hypoxia inducible factors alpha (HIF-alpha) and their inhibiting factor (FIH-1) of channel catfish (Ictalurus punctatus) under hypoxia. Comparative Biochemistry and Physiology, Part B, Biochemistry and Molecular Biology 169:38-50. 62. Cui J, Liu S, Zhang B, Wang H, Sun H, Song S, Qiu X, Liu Y, Wang X, Jiang Z, Liu ZJ. 2014. Transciptome analysis of the gill and swimbladder of Takifugu rubripes by RNASeq. PLoS ONE 9:e85505. doi:10.1371/journal.pone.0085505 63. Cui J, Wang H, Liu S, Zhu L, Qiu X, Jiang Z, Wang X, Liu ZJ. 2014. SNPs discovery from transcriptome of the swimbladder of Takifugu rubripes. PLoS ONE 9: e92502. doi:10.1371/journal.pone.0092502 64. Feng J, Liu S, Wang X, Kaltenboeck L, Kucuktas H, Li J, Liu ZJ 2014. Hemoglobin genes are differentially regulated under heat stress conditions between sensitive and tolerant 18 catfish in different tissues. Comparative Biochemistry and Physiology, Part D, Genomics and Proteomics 9:11-22. 65. Wong LL, peatman E, Kelly L, Kucuktas H, Na-Nakorn U, and Liu ZJ. 2014. Catfish species identification using Lab-on-Chip PCR-RFLP. Journal of Aquatic Food Product Technology 23:2-13. 66. Wang R, Sun L, Bao L, Zhang J, Jiang Y, Yao J, Song L, Feng J, Liu S, Liu ZJ. 2013. Bulk segregant RNA-seq reveals expression and positional candidate genes and allele-specific expression for disease resistance against enteric septicemia of catfish. BMC Genomics 14: 929. DOI: 10.1186/1471-2164-14-929 67. Jiang Y, Gao X, Liu S, Zhang Y, Liu H, Sun F, Bao L, Waldbieser G, Liu ZJ. 2013. Whole genome comparative analysis of channel catfish (Ictalurus punctatus) with four model fish species. BMC Genomics 14:780. DOI: 10.1186/1471-2164-14-780. This paper was highlighted in The Scientist http://www.thescientist.com//?articles.view/articleNo/38334/title/Genome-Digest/ 68. Sookruksawong S, Sun F, Liu ZJ, Tassanakajon A. 2013. RNA-Seq analysis reveals genes associated with resistance to Taura syndrome virus (TSV) in the Pacific white shrimp Litopenaeus vannamei. Developmental and Comparative Immunology 41:523-533. doi: 10.1016/j.dci.2013.07.020. 69. Sun F, Liu S, Gao X, Jiang Y, Perera D, Wang X, Li C, Sun L, Zhang J, Kaltenboeck L, Dunham R, Liu ZJ. 2013. Male-biased genes in catfish as revealed by RNA-Seq analysis of the testis transcriptome. PLoS One 8(7):e68452. doi:10.1371/journal.pone.0068452. 70. Sahu D, Panda SP, Panda S, Das P, Meher PK, Hazra RK, Peatman E, Liu ZJ, Eknath AE, and Nandi S. 2013. Identification of reproduction-related genes and SSR-markers through expressed sequence tags analysis of a monsoon breeding carp rohu, Labeo rohita (Hamilton). Gene 524:1-14. 71. Wang R, Feng J, Li C, Liu S, Zhang Y, Liu ZJ. 2013. Four lysozymes (one c-type and three g-type) in catfish are drastically but differentially induced after bacterial infection. Fish and Shellfish Immunology 35:136-145. 72. Liu S, Wang X, Sun F, Zhang J, Feng J, Liu H, Rajendran KV, Sun L, Zhang Y, Jiang Y, Kaltenboeck L., Kucuktas H, and Liu ZJ. 2013. RNA-Seq reveals expression signatures of genes involved in oxygen transport, protein synthesis, folding and degradation in response to heat stress in catfish. Physiological Genomics 45:462-476. 73. Niu D, Wang L, Sun F, Feng B, Jin K, Liu ZJ, Li J. 2013. Development of molecular resources for an intertidal clam, Sinonovacula constricta using 454 transcriptome sequencing. PLoS One 8: e67456. 74. Zhang J, Liu S, Rajendran KV, Sun L, Zhang Y, Sun F, Kucuktas H, Liu H, Liu ZJ. 2013. Pathogen recognition receptors in channel catfish: Ш. phylogeny and expression analysis of Toll-like receptors. Developmental and Comparative Immunology 40: 185-194. 75. Sun F, Ninwichian P, Liu SK, Liu, ZJ. 2013. Identification and application of sex-specific markers in catfish. In: Sex determination in fishes, ed. Songlin Chen, pp. 131-139, Academic Press, Beijing. 19 76. Zhang Y, Liu S, Lu J, Jiang Y, Gao X, Ninwichian P, Li C, Waldbieser G, Liu ZJ. 2013. Comparative genomic analysis of catfish linkage group 8 reveals two homologous chromosomes in zebrafish with extensive inter-chromosomal rearrangements. BMC Genomics, BMC Genomics 14:387, DOI: 10.1186/1471-2164-14-387. 77. Liu S, Li Q, Liu ZJ. 2013. Genome-wide identification, characterization and phylogenetic analysis of 50 catfish ATP-binding cassette (ABC) transporter genes. PLoS ONE 8(5):e63895. doi:10.1371/journal.pone.0063895. 78. Jiang Y, Ninwichian P, Liu S, Zhang J, Liu H, Kucuktas H, Liu ZJ. 2013. Generation of physical contig-specific sequences useful for whole genome sequence assembly and scaffolding. PloS one 8 (10), e78872. 79. Preziosa E, Liu S, Gao X, Liu H, Kucuktas H, Terova G, and Liu ZJ. 2013. Effect of Nutrient Restriction and Re-feeding on Calpain Family Genes in Skeletal Muscle of Channel Catfish (Ictalurus punctatus). PLoS One 8:e59404. 80. Sarropoulou E, Fernandes J, Liu ZJ. 2012. Editorial, GIA 2011: Genomics in Aquaculture 2011 symposium. Marine Biotechnology 14(5): 1-2. 81. Sun F, Peatman E, Li C, Liu S, Jiang Y, Zhou Z, Liu ZJ. 2012. Transcriptomic signatures of attachment, NF-κB suppression and IFN stimulation in the catfish gill following columnaris bacterial infection. Developmental and Comparative Immunology 38: 169-180. 82. Wang R, Li C, Stoeckel J, Moyer G, Liu ZJ, and Peatman E. 2012. Rapid development of molecular resources for a freshwater mussel, Villosa lienosa (Bivalvia: Unionidae) using a RNA-seq-based approach. Freshwater Science (formerly Journal of the North American Benthological Society) 31: 695-708. 83. Ninwichian P, Peatman E, Liu H, Kucuktas H, Somridhivej B, Liu S, Li P, Jiang Y, Sha Z, Kaltenboeck M, Abernathy J, Wang W, Chen F, Lee Y, Wong L, Wang S, Lu J, Liu ZJ. 2012. Second generation genetic linkage map and its integration with the BAC-based physical map in channel catfish. G3: Genes, Genomes, Genetics 2: 1233-1241. 84. Lu J, Peatman E, Tang H, Lewis J, Liu ZJ. 2012. Profiling of gene duplication patterns of teleost genomes: Evidence for rapid lineage-specific genome expansion mediated by recent tandem duplications. BMC Genomics 13:246. 85. Liu S, Zhang Y, Zhou Z, Waldbieser G, Sun F, Lu J, Zhang J, Jiang Y, Zhang H, Wang X, Rajendran KV, Kucuktas H, Peatman E, Liu ZJ. 2012. Efficient assembly and annotation of the transcriptome of catfish by RNA-Seq analysis of a doubled haploid homozygote. BMC Genomics 13:595. doi:10.1186/1471-2164-13-595 86. Ninwichian P, Peatman E, Perera D, Liu S, Kucuktas H, Dunham R, and Liu Z.J. 2012. Identification of a sex-linked marker for channel catfish. Animal Genetics 43:476-477. doi:10.1111/j.1365-2052.2011.02276.x 87. Li C, Zhang Y, Wang R, Lu J, Nandi S, Mohanty S, Terhune J, Liu ZJ, Peatman E. 2012. RNA-Seq analysis of mucosal immune responses reveals signatures of intestinal barrier disruption and pathogen entry following Edwardsiella ictaluri infection in channel catfish, Ictalurus punctatus. Fish & Shellfish Immunology 32:816-827. 20 88. Rajendran KV, Zhang J, Liu S, Kucuktas H, Wang X, Liu H, Wood T, Terhune J, Peatman E, Liu ZJ. 2012. Pathogen recognition receptors in channel catfish: II. Identification, phylogeny and expression of retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), Developmental and Comparative Immunology 37:381-389. 89. Browdy, C.L., Hulata, G., Liu, Z., Allan, G.L., Sommerville, C., Passos de Andrade, T., Pereira, R., Yarish, C., Shpigel, M., Chopin, T., Robinson, S., Avnimelech, Y. & Lovatelli, A. 2012. Novel and emerging technologies: can they contribute to improving aquaculture sustainability? In R.P. Subasinghe, J.R. Arthur, D.M. Bartley, S.S. De Silva, M. Halwart, N. Hishamunda, C.V. Mohan & P. Sorgeloos, eds. Farming the Waters for People and Food. Proceedings of the Global Conference on Aquaculture 2010, Phuket, Thailand. 22–25 September 2010. pp. 149–191. FAO, Rome and NACA, Bangkok. 90. Zhang H, Peatman E, Liu H, Feng T, Chen L, Liu ZJ. 2012. Molecular characterization of three L-type lectin genes from channel catfish, Ictalurus punctatus and their responses to Edwardsiella ictaluri challenge. Fish and Shellfish Immunology 32:598-608. doi: 10.1016/j.fsi.2011.12.009. 91. Rajendran KV, Zhang J, Liu Shikai, Kucuktas H, Wang X, Liu H, Sha Z, Terhune J, Peatman E, Liu ZJ. 2012. Pathogen recognition receptors in channel catfish: I. Identification, phylogeny and expression of NOD-like receptors. Developmental and Comparative Immunology 37:77-86. 92. Liu, S, Zhang Y, Sun F, Jiang Y, Wang R, Li C, Zhang J, Liu ZJ. 2012. Approaches for Functional Genomics in aquaculture. pp. 1-40. In: Functional Genomics in Aquaculture, Wiley and Blackwell Publishing, Ames, IA. 93. Zhang J, Jiang Y, Wang R, Li C, Zhang Y, Sun F, Liu, S, Liu ZJ. 2012. Chapter 2, Genomic resources for aquaculture functional genomics. pp. 41-77. In: Functional Genomics in Aquaculture, Wiley and Blackwell Publishing, Ames, IA. 94. Zhou Z, Liu H, Liu S, Sun F, Peatman E, Kucuktas H, Kaltenboeck L, Feng T, Zhang H, Niu D, Lu J, Waldbieser G, Liu ZJ. 2012. Alternative complement pathway of channel catfish (Ictalurus punctatus): molecular characterization, mapping and expression analysis of factors Bf/C2 and Df. Fish and Shellfish Immunology 32:186-195. doi:10.1016/j.fsi.2011.11.012. 95. Zhang H, Peatman E, Liu H, Niu D, Feng T, Kucuktas H, Waldbieser G, Chen L, Liu ZJ. 2012. Characterization of mannose-binding lectin from channel catfish (Ictalurus punctatus). Research in Veterinary Science 92:408-413. 96. Jiang Y, Lu J, Peatman E, Kucuktas H, Liu S, Wang S, Sun F, Liu ZJ. 2011. A Pilot study for channel catfish whole genome sequencing and de novo assembly. BMC Genomics 12:629 doi:10.1186/1471-2164-12-629. 97. Feng T, Zhang H, Liu H, Zhou Z, Niu D, Wong L, Kucuktas H, Liu X, Peatman E, Liu ZJ. 2011. Molecular characterization and expression analysis of the channel catfish cathepsin D genes. Fish and Shellfish Immunology 31:164-169. 98. Niu D, 1, Peatman E, Liu H, Lu J, Kucuktas H, Liu S, Sun F, Zhang H, Feng T, Zhou Z, Terhune J, Waldbieser G, Li J, Liu ZJ. 2011. Microfibrillar-associated protein 4 (MFAP4) 21 genes in catfish play a novel role in innate immune responses. Comparative Immunology 35:568-579. Developmental and 99. Abernathy J, Xu D-H, Peatman E, Kucuktas H, Klesius P, Liu ZJ. 2011. Gene expression profiling of a fish parasite Ichthyophthirius multifiliis: insights into development and senescence-associated avirulence. Comparative Biochemistry and Physiology, Part D, Genomics and Proteomics, 6: 382–392. 100. Wong L, Peatman E, Kucuktas H, Zhou C, Na-Nakorn U, and Liu ZJ. 2011. DNA Barcoding of Catfish: Species Authentication and Phylogenetic Assessment. PLoS One 6: e17812. 101. Liu H, Peatman E, Wang W, Abernathy J, Liu S, Kucuktas H, Terhune J, Xu D-H, Klesius P, Liu ZJ. 2011. Molecular responses of ceruloplasmin to Edwardsiella ictaluri infection and iron overload in channel catfish (Ictalurus punctatus). Fish and Shellfish Immunology 30: 992-997. 102. Liu H, Peatman E, Wang W, Abernathy J, Liu S, Kucuktas H, Lu J, Xu D-H, Klesius P, Waldbieser G, Liu ZJ. 2011. Molecular responses of calreticulin genes to iron overload and disease infection in channel catfish (Ictalurus punctatus). Developmental and Comparative Immunology 35: 267-272. 103. Liu S, Zhou Z, Lu J, Sun F, Wang S, Liu H, Jiang Y, Kucuktas H, Kaltenboeck L, Peatman E, Liu Z.J. 2011. Generation of genome-scale gene-associated SNPs in catfish for the construction of a high-density SNP array. BMC Genomics 12:53. 104. Bao B, Ke Z, Xing J, Peatman E, Liu ZJ, Xie C, Xu B, Gai J, Gong X, Yang G, Jiang Y, Tang W, and Ren D. 2011. Proliferating cells in suborbital tissue drive eye migration in flatfish. Developmental Biology 351: 200-207. 105. Liu, Z.J. 2011. Aquaculture Genomics: A Case Study with Catfish. In: Comprehensive Biotechnology (2nd Edition), Article 293 ‘Aquaculture Genomics’, pp.371-380. 106. Liu ZJ. 2011. Development of genomic resources in support of sequencing, assembly, and annotation of the catfish genome. Comparative Biochemistry and Physiology, Part D, Genomics and Proteomics 6:11-17. doi:10.1016/j.cbd.2010.03.001. 107. Xu D-H, Klesius P, Peatman E, and Liu ZJ. 2011. Susceptibility of channel catfish, blue catfish and channel × blue catfish hybrid to Ichthyophthirius multifiliis. Aquaculture 311:2530. 108. Lu J, Peatman E, Yang Q, Wang S, Hu Z, Kucuktas H, Liu ZJ. 2011. Catfish Genome Database cBARBEL: A informatic platform for genome biology of ictalurid catfish. Nucleic Acids Research 39 (Database Issue): D815-21, doi:10.1093/nar/gkq765. 109. Liu W, Li H, Bao X, Gao X, Li Y, He C, and Liu ZJ. 2010. Genetic differentiation between natural and hatchery stocks of Japanese scallop (Mizuhopecten yessoensis) as revealed by AFLP analysis. International Journal of Molecular Sciences 11: 3933-3941. doi:10.3390/ijms11103933. 110. Liu ZJ. 2010. Genomic variations, marker technologies and their usefulness for genomebased selection. pp. 3-20. In: Next Generation Sequencing, SNPs, and Whole Genome Selection, Wiley and Blackwell Publishing, Ames, IA. 22 111. Lu J and Liu ZJ. 2010. Copy number variations (CNV) and whole genome selection. pp. 21-34. In: Next Generation Sequencing, SNPs, and Whole Genome Selection, Wiley and Blackwell Publishing, Ames, IA. 112. Kucuktas H and Liu ZJ. 2010. Construction of libraries for next generation sequencing. pp. 57-68. In: Next Generation Sequencing, SNPs, and Whole Genome Selection, Wiley and Blackwell Publishing, Ames, IA. 113. Wang S and Liu ZJ. 2010. SNP discovery through EST data mining. pp. 91-108. In: Next Generation Sequencing, SNPs, and Whole Genome Selection, Wiley and Blackwell Publishing, Ames, IA. 114. Wang S, Liu H, and Liu ZJ. 2010. SNP quality assessment. pp. 109-122. In: Next Generation Sequencing, SNPs, and Whole Genome Selection, Wiley and Blackwell Publishing, Ames, IA. 115. Wang S, Liu H, Bushek D, Ford S, Peatman E, Kucuktas H, Quilang J, Li P, Wallace R, Wang Y, Guo X, and Liu ZJ. 2010. Microarray analysis of gene expression in eastern oyster (Crassostrea virginica) reveals a novel combination of antimicrobial and oxidative stress host responses after dermo (Perkinsus marinus) challenge. Fish and Shellfish Immunology 29: 921-929. 116. Xu B, Wang S, Jiang Y, Yang L, Li P, Xie C, Xing J, Ke Z, Li J, Gai J, Yang G, Bao B, and Liu ZJ. 2010. Generation and analysis of ESTs from the grass carp, Ctenopharyngodon idellus. Animal Biotechnology 21: 1-9. 117. Chen F, Lee Y, Jiang Y, Wang S, Peatman E, Abernathy J, Liu H, Liu S, Kucuktas H, Ke C, Liu ZJ. 2010. Identification and characterization of full-length cDNAs in catfish (Ictalurus spp.). PLoS One 12, e11546. 118. Lu J, Peatman E, Wang W, Yang Q, Abernathy J, Wang S, Kucuktas H, Liu ZJ. 2010. Alternative splicing in teleost fish genomes: Same-species and cross-species analysis and comparisons. Molecular Genetics and Genomics 283:531-539. 119. Liu ZJ, Peatman E. 2010. Whole Genome Sequencing of Aquaculture Species: Identifying the Genotypes behind Production Phenotypes. Global Aquaculture Advocate, March/April 2010:68-70. 120. Liu H, Takano T, Peatman E, Abernathy J, Wang S, Sha Z, Kucuktas H, Liu ZJ. 2010. Molecular characterization and gene expression of the channel catfish ferritin H subunit after bacterial infection and iron treatment. Journal of Experimental Zoology, Part A: Ecological Genetics and Physiology 313:359-368. 121. Jiang Y, Abernathy J, Peatman E, Liu H, Wang S, Xu D-H, Kucuktas H, Klesius P, Liu Z.J. 2010. Identification and characterization of matrix metalloproteinase-13 sequence structure and expression during embryogenesis and infection in channel catfish (Ictalurus punctatus). Developmental and Comparative Immunology 34:590-597. 122. Sha Z, Wang S, Zhuang Z, Wang Q, Wang Q, Li P, Ding H, Wang N, Liu ZJ, Chen S. 2010. Generation and analysis of 10,000 ESTs from the half smooth tongue sole, Cynoglossus semilaevis and identification of microsatellite and SNP markers. Journal of Fish Biology 76:1190–1204. 23 123. Wang S, Abernathy J, Waldbieser G, Lindquist E, Richardson P, Lucas S, Wang M, Li P, Thimmapuram J, Liu L, Vullaganti D, Kucuktas H, Murdock C, Small B, Wilson M, Liu H, Jiang Y, Lee Y, Chen F, Lu J, Wang W, Peatman E, Xu P, Somridhivej B, Baoprasertkul P, Quilang J, Sha Z, Bao B, Wang Y, Wang Q, Takano T, Nandi S, Liu S, Wong L, Kaltenboeck L, Quiniou S, Bengten E, Miller N, Trant J, Rokhsar D, Liu ZJ., and Catfish Genome Consortium. 2010. Assembly of 500,000 inter-specific catfish expressed sequence tags and large scale gene-associated marker development for whole genome association studies. Genome Biology 11 (1):R8. 124. Wang S, Zhang L, Hu J, Bao Z, Liu ZJ. 2010. Molecular and cellular evidence for biased mitotic gene conversion in hybrid scallop. BMC Evolutionary Biology 10:6. doi:10.1186/1471-2148-10-6. 125. Abernathy J, Peatman E, Liu ZJ. 2010. Basic Aquaculture Genetics, SRAC Publication No. 5001, 1-14. 126. Liu H, Tomokazu T, Abernathy J, Wang S, Sha Z, Terhune J, Kucuktas H, Jiang Y, Liu ZJ. 2010. Structure and expression of transferrin gene of channel catfish, Ictalurus punctatus. Fish and Shellfish Immunology 28:159-166. 127. Abernathy J, Lu J, Liu H, Kucuktas H, Liu ZJ. 2009. Molecular characterization of complement factor I reveals constitutive expression in channel catfish. Fish & Shellfish Immunology 27:529–534. 128. Liu H, Jiang Y, Wang S, Ninwichian P, Somridhivej B, Xu P, Abernathy J, Kucuktas H, Liu ZJ. 2009. Comparative analysis of catfish BAC end sequences with the zebrafish genome. BMC Genomics 10:592. 129. Kucuktas H, Wang S, Li P, He C, Xu P, Sha Z, Liu H, Jiang Y, Baoprasertkul P, Somridhivej B, Wang Y, Abernathy J, Guo X, Liu L, Muir W, Liu ZJ. 2009. Construction of genetic linkage maps and comparative genome analysis of catfish using gene-associated markers. Genetics 181:1649-1660. 130. Xu D, Panangala V, van Santen V, Dybvig K, Abernathy J, Klesius P, Liu ZJ, and Russo R. 2009. Molecular characterization of an immobilization antigen gene of the fish-parasite protozoan Ichthiophthirius multifiliis strain ARS-6. Aquaculture Research 40:1884-1892. 131. Sha Z, Abernathy JW, Wang S, Li P, Kucuktas H, Liu H, Peatman E, and Liu ZJ. 2009. NOD-like subfamily of the nucleotide-binding domain and leucine rich repeat containing family receptors and their expression in channel catfish. Dev. Comp. Immunology 31:991999. 132. Abernathy JW, Xu D-H, Li P, Klesius P, Liu ZJ. 2009. Transcriptomic profiling of Ichthyophthirius multifiliis reveals polyadenylation of the large subunit ribosomal RNA. Comparative Biochemistry and Physiology, Part D, Genomics and Proteomics 4:179-186. 133. Liu, Z.J. 2009. Genome-based technologies useful for aquaculture research and genetic improvement of aquaculture species. pp. 3-54. In: New Technologies in Aquaculture (eds. Gavin Burnell and Geoff Allan) Woodhead Publishing, London. 134. Rise M, Liu ZJ, Douglas SE, Brown LL, Nash JHE, and McFall-Ngai MJ. 2009. Chapter 4, Aquaculture-related Applications of DNA Microarray Technology. Pp. 63-101, In: 24 Molecular Research in Aquaculture (eds., Overturf and Small). Wiley-Blackwell Publishing, Ames, IA. 135. Liu, Z.J. 2008. Aquaculture genomics---progress in identifying species genetics continues. Global Aquaculture Advocate, November/December 2008:75-78. 136. Wang S, Sha Z, Sonstegard TS, Liu H, Xu P, Somridhivej B, Peatman E, Kucuktas H, Liu ZJ. 2008. Quality assessment parameters for EST-derived SNPs from catfish. BMC Genomics 9:450, doi:10.1186/1471-2164-9-450. 137. Lamkom T, Kucuktas H, Liu Z.J, Li P, Na-Nakorn U, Klinbunya S, Hutson A, Chaimongkol A, Ballenger J, Umali G, and Dunham R. 2008. Microsatellite Variation among Domesticated Populations of Channel (Ictalurus punctatus) and Blue Catfish (I. furcatus). Kasetsart Fisheries Bulletin 32:37-47. 138. Liu ZJ, Li RW, Waldbieser G. 2008. Utilization of microarray technology for functional genomics in ictalurid catfish. Journal of Fish Biology 72(9): 2377-2390. 139. Takano T, Sha Z, Peatman E, Terhune J, Liu H, Kucuktas H, Li P, Edholm E-S, Wilson M, Liu ZJ. 2008. The two channel catfish intelectin genes exhibit highly differential patterns of tissue expression and regulation after infection with Edwardsiella ictaluri. Developmental and Comparative Immunology 32: 693-705. 140. Sha Z, Xu P, Takano T, Liu H, Terhune J, Liu ZJ. 2008. The warm temperature acclimation protein Wap65 as an immune response gene: Its duplicates are differentially regulated by temperature and bacterial infections. Molecular Immunology 45: 1458-1469. 141. Somridhivej B, Wang S., Sha Z., Liu H., Quilang J., Xu P, Li P, Liu ZJ. 2008. Characterization, polymorphism assessment, and database construction for microsatellites from BAC end sequences of catfish: a resource for integration of linkage and physical maps. Aquaculture 275: 76-80. 142. Peatman E, Terhune J, Baoprasertkul P, Xu P, Nandi S, Wang S, Somridhivej B, Kucuktas H, Li P, Dunham R, Liu ZJ. 2008. Microarray analysis of gene expression in the blue catfish liver reveals early activation of the MHC class I pathway after infection with Edwardsiella ictaluri. Molecular Immunology 45: 553-566. 143. He C., Ge L, Zhou Z, X Mu Y, Liu W, Gao X, and Liu ZJ. 2008. Sequence and organization of the complete mitochondrial genome of spotted halibut (Verasper variegatus) and barfin flounder (Verasper moseri). DNA Sequence 19:246-255. 144. Liu, Z.J. 2007. Genomic resources and their informatic analysis for comparative genomics in catfish. Aquaculture 272: S286-S286. 145. Wang S, Zhang L, Zhan A, Wang X, Liu ZJ, Hu J, and Bao Z. 2007. Patterns of concerted evolution of rDNA family in natural population of Zhikong scallop, Chlamys farreri. Journal of Molecular Evolution 65:660-667. 146. Peatman E., and Liu ZJ. 2007. Evolution of CC chemokines in teleost fish: a case study in gene duplication and implications for immune diversity. Immunogenetics 59: 613-623. 25 147. Wang W., Chen L., Yang P., Hou Lin, He C., Gu Z., Liu ZJ. 2007. Assessing genetic diversity of populations of topmouth culter (Culter alburnus) in China using AFLP markers. Biochemical Systematics and Ecology 35: 662-669. 148. Quilang J., Wang S, Li P, Abernathy J, Peatman E, Wang, Y., Wang, L., Shi, Y., Wallace R, Guo, X., and Liu ZJ. 2007. Generation and analysis of ESTs from the eastern oyster, Crassostrea virginica Gmelin and identification of microsatellite and SNP markers. BMC Genomics 8: 157. 149. Li P, Peatman E, Wang S, Feng J, He C, Baoprasertkul P, Xu P, Kucuktas H, Nandi S, Somridhivej B, Serapion J, Simmons M, Turan C, Liu L, Muir W, Dunham R, Brady Y, Grizzle J, Liu ZJ. 2007. Towards the ictalurid catfish transcriptome: generation and analysis of 31,215 catfish ESTs. BMC Genomics 8: 177. 150. Xu P, Wang S, Liu L, Thorsen J, Kucuktas H, Liu ZJ. 2007. A BAC-based physical map of the channel catfish genome. Genomics 90:380-388. 151. Wang S, Xu P, Thorsen, J., Zhu, B., de Jong, P., Waldbieser, G., Kucuktas, H., Liu ZJ. 2007. Characterization of a BAC library from channel catfish Ictalurus punctatus: indications of high rates of chromosomal reshuffling among teleost genomes. Marine Biotechnology 9:701-711. 152. Abernathy J, Xu P, Li P, Xu, D., Kucuktas H, Klesius, P., Arias, C., and Liu ZJ. 2007. Generation and analysis of expressed sequence tags from the ciliate protozoan parasite Ichthyophthirius multifiliis. BMC Genomics 8:176. 153. Peatman E, Baoprasertkul P, Terhune J, Xu P, Nandi S, Kucuktas H, Li P, Wang S, Somridhivej B, Dunham R, Liu, Z.J.. 2007. Expression analysis of the acute phase response in channel catfish (Ictalurus punctatus) after infection with a Gram negative bacterium. Developmental and Comparative Immunology 31:1183-1196. 154. Baoprasertkul P, Xu P, Peatman E, Kucuktas H, Liu ZJ. 2007. Divergent Toll-like receptors in catfish (Ictalurus punctatus): TLR5S, TLR20, TLR21. Fish Shellfish Immunology 23:1218-1230. 155. Bao B, Li J, Wang G, Liu ZJ. 2007. Characterization of chemokine gene SCYA126 and its alternative splicing in channel catfish (Ictalurus punctatus). Journal of Fishery Sciences of China 14:1-7. 156. Nandi S, Peatman E, Xu P, Wang S, Li P, and Liu ZJ. 2007. Repeat structure of the catfish genome: A genomic and transcriptomic assessment of Tc1-like transposon elements in channel catfish (Ictalurus punctatus). Genetica 131:81-90 157. Jenny MJ, Chapman RW, Mancia A, Chen YA, McKillen DJ, Trent H, Lang P, Escoubas J-M, Bachere E, Boulo V, Liu ZJ, Gross PS, Cunningham C, Cupit PM, Guo X, Tanguy A, De Guise S, Almeida JS, Warr GW. 2007. Characterization of a cDNA Microarray for Crassostrea virginica and C. gigas. Marine Biotechnology 9: 577-591. 158. Baoprasertkul, B, Peatman E, Abernathy J, Liu ZJ. 2007. Structural characterization and expression analysis of Toll-like receptor 2 gene from catfish. Fish and Shellfish Immunology 22:418-426. 26 159. Liu ZJ. 2008. Chapter 3, Catfish. Pp. 85-100. In “The Genome: A Series on Genome Mapping, Molecular Breeding & Genomics of Economic Species” (C. Kole, Kocher eds.) Vol. 6, Oxford & IBH and Science Pub. Inc. 160. Liu ZJ. 2007. Chapter 1 Concept of genomes and genomics. In: Aquaculture Genome Technologies (edited by Liu ZJ.), pp. 1-10, Blackwell Publishing, Ames, IA. 161. Liu ZJ. 2007. Chapter 2 Marking the genome: Restriction fragment length polymorphism (RFLP). In: Aquaculture Genome Technologies (edited by Liu ZJ.), pp. 11-20, Blackwell Publishing, Ames, IA. 162. Liu ZJ. 2007. Chapter 3 Random amplified polymorphic DNA (RAPD). In: Aquaculture Genome Technologies (edited by Liu ZJ.), pp. 21-28, Blackwell Publishing, Ames, IA. 163. Liu ZJ. 2007. Chapter 4 Amplified fragment length polymorphism (AFLP). In: Aquaculture Genome Technologies (edited by Liu ZJ.), pp. 29-42, Blackwell Publishing, Ames, IA. 164. Liu ZJ. 2007. Chapter 5 Microsatellite markers and assessment of marker utility. In: Aquaculture Genome Technologies (edited by Liu ZJ.), pp. 43-58, Blackwell Publishing, Ames, IA. 165. Liu ZJ. 2007. Chapter 6 Single nucleotide polymorphism (SNP). In: Aquaculture Genome Technologies (edited by Liu ZJ.), pp. 59-72, Blackwell Publishing, Ames, IA. 166. Liu ZJ. 2007. Chapter 7 Allozyme and mitochondrial DNA markers. In: Aquaculture Genome Technologies (edited by Liu ZJ.), pp. 72-86, Blackwell Publishing, Ames, IA. 167. Xu P, Wang S, and Liu ZJ. 2007. Chapter 15 Physical characterization through BAC end sequencing. In: Aquaculture Genome Technologies (edited by Liu ZJ.), pp. 263-276, Blackwell Publishing, Ames, IA. 168. Liu ZJ. 2007. Chapter 16 Genomescape: characterizing the repeat structure of the genome. In: Aquaculture Genome Technologies (edited by Liu ZJ.), pp. 277-290, Blackwell Publishing, Ames, IA. 169. Liu ZJ. 2007. Chapter 20 Transcriptome characterization through the analysis of expressed sequence tags. In: Aquaculture Genome Technologies (edited by Liu ZJ.), pp. 301-356, Blackwell Publishing, Ames, IA. 170. Peatman E, and Liu ZJ. 2007. Chapter 21 Microarray fundamentals: Basic principles and application in aquaculture. In: Aquaculture Genome Technologies (edited by Liu ZJ.), pp. 357-370, Blackwell Publishing, Ames, IA. 171. Liu ZJ. 2007. Chapter 25 DNA sequencing technologies. In: Aquaculture Genome Technologies (edited by Liu ZJ.), pp. 465-476, Blackwell Publishing, Ames, IA. 172. Liu ZJ. 2007. Chapter 26 Sequencing the genome. In: Aquaculture Genome Technologies (edited by Liu ZJ.), pp. 477-490, Blackwell Publishing, Ames, IA. 173. Liu ZJ. 2007. Fish genomics and genetic analytical technologies, with examples of their potential applications in management of fish genetic resources. Policy paper for FAO’s Commission on Genetic Resources for Food and Agriculture. In: Workshop on Status and 27 Trends in Aquatic Genetic Resources---A basis for international policy, FAO Fisheries Proceedings 5 (eds. Devin Bartley, Brian Harvey, Roger Pullin), FAO, Rome, pp. 145-179. 174. Bao B, Wang G, Li J, Liu ZJ. 2006. Homologue analysis of chemokine gene SCYA107 between Ictalurus punctatus and Ictalurus furcatus. Journal of Shanghai Fisheries University 15:385-389. 175. Baoprasertkul P, Peatman E, Somridhivej B, Liu ZJ. 2006. Toll-like receptor 3 and TICAM genes in catfish: species-specific expression profiles following infection with Edwardsiella ictaluri. Immunogenetics 58:817-830. 176. Zhang Y, Chen S, Liu Y, Sha Z, and Liu ZJ. 2006. Major histocompatibility complex class II B allele polymorphism and its association with resistance/susceptibility to Vibrio Anguillarum in Japanese flounder Paralichthys olivaceus. Marine Biotechnology 8:600-610. 177. Dunham R, Liu ZJ. 2006. Transgenic fish-where we are and where do we go? Israel Journal of Aquaculture-Bamidgeh 58:297-319. 178. Liu ZJ. 2006. Transcriptome characterization through the generation and analysis of expressed sequence tags: Factors to consider for a successful EST project. Israel Journal of Aquaculture-Bamidgeh 58:328-341. 179. Xu P, Wang S, Liu L, Peatman E, Somridhivej B, Thimmapuram J, Gong G, Liu ZJ. 2006. Channel catfish BAC-end sequences for marker development and assessment of syntenic conservation with other fish species. Animal Genetics 37:321-326. 180. Hou L, Bi X, Zou X, He C, Yang L, Qu R, Liu ZJ. 2006. Molecular systematics of bisexual Artemia populations. Aquaculture Research 37:671-680. 181. Arias CR, Abernathy J, and Liu ZJ. 2006. Combined use of 16S ribosomal DNA and automated ribosomal intergenic spacer analysis (ARISA) to study the bacterial community in catfish ponds. Letters in Applied Microbiology 43:287-292. 182. Peatman E, and Liu ZJ. 2006. CC chemokines in zebrafish: evidence for extensive intrachromosomal gene duplications. Genomics 88:381-385. 183. Liu ZJ, Peatman E. 2006. Chemokines in fish: a rapidly expanding repertoire. pp. 121144. In Focus on Immunology Research, (Editor, Barbara A. Veskler), Nova Science Publishers, Inc., New York. 184. Simmons M, Mickett K, Kucuktas H, Li P, Dunham R, Liu ZJ. Comparison of domestic and wild catfish populations provide no evidence for genetic impact. 2006. Aquaculture 252:133-146. 185. Bao B, Peatman E, Xu P, Baoprasertkul B, Wang G, Liu ZJ. 2006. Characterization of 23 CC chemokine genes and analysis of their expression in channel catfish (Ictalurus punctatus). Developmental and Comparative Immunology 30:783-796. 186. Peatman E, Bao B, Xu P, Baoprasertkul P, Liu ZJ. 2006. Catfish CC chemokines: genomic clustering, duplications, and expression after bacterial infection with Edwardsiella ictaluri. Molecular Genetics and Genomics 275:297-309. 187. Wang Q, Wang Y, Xu P, Liu ZJ. 2006. NK-lysin of channel catfish: gene triplication, sequence variation, and expression analysis. Molecular Immunology 43:1676-1686. 28 188. Wang Y, Wang Q, Baoprasertkul P, Peatman E, Liu ZJ. 2006. Genomic organization, gene duplication, and expression analysis of interleukin-1 in channel catfish (Ictalurus punctatus). Molecular Immunology 43:1653-1664. 189. Wang Q, Bao B, Wang Y, Peatman E, and Liu ZJ. 2006. Characterization of a NK-lysin antimicrobial peptide gene from channel catfish. Fish and Shellfish Immunology 20:419426. 190. Bao B, Peatman E, Xu P, Li P, Zeng H, He C, and Liu ZJ. 2006. The catfish liverexpressed antimicrobial peptide 2 (LEAP-2) gene is expressed in a wide range of tissues and developmentally regulated. Molecular Immunology 43:367-377. 191. Liu ZJ. 2005. Genetic Analysis-Amplified Fragment Length Polymorphism (AFLP), Chapter 19, In: Stock Identification Methods, Steve Cadrin, Keven D. Friedland, John Waldman, (eds.), Elsevier Press, New York, pp.389-411. 192. Bao B, Yang G, Liu ZJ, Li S, Wang Z, Ren D. 2005. Isolation of Sfrs3 gene and its differential expression during metamorphosis involving eye migration of Japanese flounder Paralichthys olivaceus. Biochimica et Biophysica Acta 1725:64-70. 193. Peatman E, Bao B, Baoprasertkul, P, and Liu ZJ. 2005. In silico identification and expression analysis of 12 novel CC chemokines in catfish. Immunogenetics 57:409-419. 194. Bao B, Peatman E, Li P, He C, and Liu ZJ. 2005. Catfish hepcidin gene is expressed in a wide range of tissues and exhibits tissue-specific upregulation after bacterial infection. Developmental and Comparative Immunology 29:939-950. 195. Xu P, Bao B, He Q, Peatman E, He C, Liu ZJ. 2005. Characterization and expression analysis of bactericidal permeability-increasing protein (BPI) antimicrobial peptide gene from channel catfish Ictalurus punctatus. Developmental and Comparative Immunology 29:865-878. 196. Baoprasertkul P, He C, Peatman E, Zhang S, Li P, Liu ZJ. 2005. Constitutive expression of three novel catfish CXC chemokines: homeostatic chemokines in teleost fish. Molecular Immunology 42:1355-1366. 197. Chen L, He C, Baoprasertkul, P, Xu P, Li P, Serapion, J, Waldbieser G, Wolters, W, Liu ZJ. 2005. Analysis of a catfish gene resembling interleukin-8: cDNA cloning, gene structure, and expression after infection with Edwardsiella ictaluri. Developmental and Comparative Immunology 29:135-142. 198. Liu YG, Chen SL, Li BF, Wang ZJ, Liu ZJ. 2005. Analysis of genetic variation in selected stocks of hatchery flounder, Paralichthys olivaceus, using AFLP markers. Biochemical Systematics and Ecology 33:993-1005. 199. Liu ZJ, Cordes JF. 2004. Erratum to DNA marker technologies and their applications in aquaculture genetics. Aquaculture 242:735-736. 200. Ligeon C, Jolly C, Argue B, Phelps R, Liu ZJ, Yant R, Benfrey J, Crews J, Gagalac F, and Dunham R. 2004. Economics of production of channel catfish, Ictalurus punctatus, female x blue catfish, I. furcatus, male hybrid eggs and fry. Journal of Aquaculture Economics and Management 8:253-268. 29 201. Ligeon C, Dunham R, Jolly CJ, Argue B, Liu ZJ, Yant R, Benfrey J, Gagalac F, and Dunham R. 2004. Economics of production of channel catfish, Ictalurus punctatus, female x blue catfish, I. furcatus, male hybrid fingerlings and foodfish. Journal of Aquaculture Economics and Management 8:269-280. 202. Serapion J, Waldbieser GC, Wolters W, Liu ZJ. 2004. Development of Type I markers in channel catfish through intron sequencing. Animal Genetics 35:463-466. 203. Liu ZJ, Cordes J. 2004. DNA marker technologies and their applications in aquaculture genetics. Aquaculture 238:1-37. 204. He C, Peatman E, Baoprasertkul P, Kucuktas H, and Liu ZJ. 2004. Multiple CC chemokines in channel catfish and blue catfish as revealed by analysis of expressed sequence tags. Immunogenetics 56:379-387. 205. Pridgeon JW, Liu ZJ, and Liu N. 2004. Identification of Mariner Elements from House Flies (Musca domestica) and German Cockroaches (Blattella germanica). Insect Molecular Biology 13:443-447. 206. Chen L, Jiang H, Zhou Z, Li K, Li K, Deng GY, and Liu ZJ. 2004. Purification of vitellin from the ovary of Chinese mitten-handed crab, Eriocheir sinensis, and development of an anti-vitellin ELISA. Comparative Biochemistry and Physiology B Biochemistry and Molecular Biology 138:305-311. 207. Karsi A, Waldbieser G, Small B, Liu ZJ, and Wolters W. 2004. Molecular cloning of proopiomelanocortin cDNA and multi-tissue mRNA expression in channel catfish. General and Comparative Endocrinology 137:312-321. 208. Serapion J, Kucuktas H, Feng J, Liu ZJ. 2004. Bioinformatic Mining of Type I Microsatellites from Expressed Sequence Tags of Channel Catfish (Ictalurus punctatus). Marine Biotechnology 6:364-377. 209. Peatman E, Wei X, Feng J, Liu L, Kucuktas H, Li L, He C, Rouse D, Wallace R, Dunham R, Liu ZJ. 2004. Development of Expressed Sequence Tags (ESTs) from Eastern Oyster (Crassostrea virginica): Lessons Learned from Previous Efforts. Marine Biotechnology 6: S491-496. 210. Kocabas A, Dunham R, Liu ZJ. 2004. Alterations in gene expression in the brain of white catfish (Ameirus catus) in response to cold acclimation. Marine Biotechnology 6:S431-438. 211. Baoprasertkul P, Peatman E, He C, Kucuktas H, Li P, Chen L, Simmons M, and Liu ZJ. 2004. Sequence analysis and expression of a CXC chemokine in resistant and susceptible catfish after infection of Edwardsiella ictaluri. Developmental and Comparative Immunology 28:769-780. 212. He C, Chen L, Simmons M, Li P, Kim S, and Liu ZJ. 2003. Putative SNP discovery in interspecific hybrids of catfish by comparative EST analysis. Animal Genetics 34:445-448. 213. Liu ZJ, and Jensen JW. 2003. Aquaculture has great expectations of the genome revolution. Pp. 50-52. In: Celebration of the 50 Years of DNA, A Commemorative Book for the discovery of the DNA structure by Francis Crick and James Watson (eds. T. John, Foreword by Tony Blair), Cambridge. 30 214. Argue B, Liu ZJ, and Dunham RA. 2003. Dress-out and fillet yields of channel catfish, Ictalurus punctatus, blue catfish, I. furcatus, and their F1, F2 and backcross hybrids. Aquaculture 228:81-90. 215. Liu ZJ. 2003. A review of catfish genomics: progress and perspectives. Comparative and Functional Genomics 4:259-265. 216. Mickett K, Morton C, Feng J, Li P, Simmons M, Dunham RA, Cao D, and Liu ZJ. 2003. Assessing genetic diversity of domestic populations of channel catfish (Ictalurus punctatus) in Alabama using AFLP markers. Aquaculture 228:91-105. 217. Liu ZJ, Karsi A, Li P, Cao D, and Dunham R. 2003. An AFLP-based genetic linkage map of channel catfish (Ictalurus punctatus) constructed by using an interspecific hybrid resource family. Genetics 165:687-694. 218. Patterson A, Karsi A, Feng J, and Liu ZJ. 2003. Translational Machinery of Channel Catfish: II. Complementary DNA and Expression of the Complete Set of 47 60S Ribosomal Proteins. Gene 305:151-160. 219. Dunham R, and Liu ZJ. 2003. Gene mapping, isolation and genetic improvement in catfish. In N.Shimizu, T Aoki, I. Hirono and F. Takashima (eds.) Aquatic Genomics: Steps Toward a Great Future. Springer-Verlag, New York pp. 45-60. 220. Dunham R, Kucuktas H, Liu ZJ, Keefer L, Spencer M. 2002. Biochemical Genetics of Brook Trout in Georgia: Management Implications. Proc. Ann. South. Assoc. Fish Wildl. 55:63-80. 221. Kocabas A, Li P, Cao D, Karsi A, He C, Patterson A, Ju Z, Dunham R, and Liu ZJ. 2002. Expression profile of the channel catfish spleen: analysis of genes involved in immune functions. Marine Biotechnology 4:526-536. 222. Ju Z, Dunham R, and Liu ZJ. 2002. Differential gene expression in the brain of channel catfish (Ictalurus punctatus) in response to cold acclimation. Molecular Genetics and Genomics 268:87-95. 223. Karsi A, Patterson A, Feng J, and Liu ZJ. 2002. Translational machinery of channel catfish: I. A transcriptomic approach to the analysis of 32 40S ribosomal protein genes and their expression. Gene 291:177-186. 224. Karsi A, Cao D, Li P, Patterson A, Kocabas A, Feng J, Ju Z, Mickett K, and Liu ZJ. 2002. Transcriptome analysis of channel catfish (Ictalurus punctatus): Initial analysis of gene expression and microsatellite-containing cDNAs in the skin. Gene 285:157-168. 225. Kocabas A, Kucuktas H, Dunham RA, and Liu ZJ. 2002. Molecular characterization and differential expression of the myostatin gene in channel catfish (Ictalurus punctatus). Biochimica et Biophysica Acta 1575:99-107. 226. Kucuktas H, Wagner BK, Shopen, R, Gibson M, Dunham RA, and Liu ZJ. 2002. Genetic analysis of Ozark hellbenders (Cryptobranchus alleganiensis bishopi) utilizing RAPD markers. Proc. Ann. Conf. SEAFWA 55:126-137. 227. Liu ZJ, Li P, Kocabas A, Ju Z, Karsi A, Cao D, Patterson A. 2001. Microsatellitecontaining genes from the channel catfish brain: evidence of trinucleotide repeat expansion 31 in the coding region of nucleotide excision repair gene RAD23B, Biochemical and Biophysical Research Communications 289:317-324. 228. Liu ZJ, Kim S, Karsi A. 2001. Channel catfish follicle stimulating hormone and luteinizing hormone: cDNA cloning and their expression during ovulation. Marine Biotechnology 3:590-599. 229. Liu ZJ, Feng J. 2001. Gene mapping and marker-assisted selection in channel catfish (Ictalurus punctatus). Acta Agriculturae Boreali-Occidentalis Sinica 10:110-114. 230. Liu ZJ, Kim S, Kucuktas H, and Karsi A. 2001. Multiple isoforms and an unusual cathodic isoform of creatine kinase from channel catfish (Ictalurus punctatus). Gene 275:207-215. 231. Cao D, Kocabas A, Ju Z, Karsi A, Li P, Patterson A, and Liu ZJ. 2001. Transcriptome of channel catfish (Ictalurus punctatus): initial analysis of genes and expression profiles from the head kidney. Animal Genetics 32:169-188. 232. Karsi A, Moav B, Hackett P, Liu ZJ. 2001. Effects of insert size on transposition efficiency of the Sleeping Beauty transposon in mouse cells. Marine Biotechnology 3:241245. 233. Altinok I, Grizzle J, and Liu ZJ. 2001. Detection of Yersinia ruckeri in rainbow trout blood by use of polymerase chain reaction. Disease of Aquatic Organisms 44:29-34. 234. Dunham RA, Majumdar K, Hallerman E, Hulata G, Gjedrem T, Mair G, Bartley D, Gupta M, Liu ZJ, Pongthana N, Rothlisberg P, and Horstweeg-Schwark G. 2001. Review of the status of aquaculture genetics. In: R.P. Subasinghe, P. Bueno, M.J. Phillips, C. Hough, S.E. McGladdery, and J.R. Arthur, eds. Aquaculture in the Third Millenium. Technical Proceedings of the Conference on Aquaculture in the Third Millenium Pp. 137-166. NACA, Bangkok and FAO, Rome. 235. Liu ZJ. 2001. Gene mapping, marker-assisted selection, gene cloning, genetic engineering, and integrated genetic improvement programs at Auburn University. In: Fish Genetics Research in Member Countries and Institutions of the International Network on Genetics in Aquaculture, ed. M.V. Gupta &B.O. Acosta, ICLARM Conf. Proc. 64:109-118. 236. Dunham R, Majumdar K, Hallerman E, Hulata G, Gjedrem T, Mair G, Bartley D, Gupta M, and Liu ZJ. 2000. Aquaculture Genetics in the Third Millennium. In: Book of Synopses, International Conference on Aquaculture in the Third Millennium, pp.139-144. 237. Ju Z, Karsi A, Kocabas A, Patterson A, Li P, Cao D, Dunham R, and Liu ZJ. 2000. Transcriptome analysis of channel catfish (Ictalurus punctatus): genes and expression profile from the brain. Gene 261:373-382. 238. Kim S, Karsi A, Dunham R, Liu ZJ. 2000. The skeletal muscle -actin gene of channel catfish (Ictalurus punctatus) and its association with piscine-specific SINE elements. Gene 252:173-181. 239. Dunham RA, Lambert DM, Argue B J, Ligeon C, Yant R, and Liu ZJ. 2000. Comparison of manual stripping and pen spawning for production of female channel catfish x blue catfish hybrids and aquarium spawning of channel catfish. North American Journal of Aquaculture 62:260-265. 32 240. Liu ZJ. 1999. Gene Mapping and marker-assisted selection in channel catfish (Ictalurus punctatus). Proceedings of Sino-US Symposium on Agricultural Biotechnology. pp. 101112. 241. Tan G, Karsi A, Li P, Kim S, Zheng X, Kucuktas H, Argue BJ, Dunham RA, and Liu ZJ. 1999. Polymorphic microsatellite markers in Ictalurus punctatus and related catfish species. Molecular Ecology 8:1758-1760. 242. Liu ZJ, Tan G, Li P, and Dunham RA. 1999. Transcribed dinucleotide microsatellites and their associated genes from channel catfish, Ictalurus punctatus. Biochemical and Biophysical Research Communications 259:190-194. 243. Liu ZJ. 1999. Gene mapping, marker-assisted selection, gene cloning, genetic engineering, and integrated genetic improvement programs at Auburn University. The Fifth Conference of the International Network on Genetics in Aquaculture Proceedings, Kuala Lumpur, Malaysia. pp. 1-19. 244. Lambert DM, Argue BJ, Liu ZJ, Dunham RA. 1999. Effects of seasonal variations, thyroid and steroid hormones, and carp pituitary extract on the artificial production of channel catfish (Ictalurus punctatus) X blue catfish (I. furcatus) hybrid embryos. Journal of the world Aquaculture Society 30:80-89. 245. Liu ZJ, Karsi A, and Dunham RA. 1999. Development of polymorphic EST markers suitable for genetic linkage mapping of catfish. Marine Biotechnology 1:437-447. 246. Liu ZJ, Tan G, Kucuktas H, Li P, Karsi A, Yant DR, and Dunham RA. 1999. High levels of conservation at microsatellite loci among Ictalurid catfishes. J. Heredity 90:307312. 247. Liu ZJ, Li P, Kucuktas H, and Dunham RA. 1999. Characterization of nonautonomous Tc1-like transposable elements of channel catfish (Ictalurus punctatus). Fish Physiology and Biochemistry 21:65-72. 248. Liu ZJ, Li P, Argue BJ, and Dunham RA. 1999. Random amplified polymorphic DNA markers: usefulness for gene mapping and analysis of genetic variation of catfish. Aquaculture 174:59-68. 249. Liu ZJ, Li P, Kucuktas H, Nichols A, Tan G, Zheng X, Argue BJ, Yant R, and Dunham RA. 1999. Development of amplified fragment length polymorphism (AFLP) markers suitable for genetic linkage mapping of catfish. Trans. Amer. Fish. Soc. 128:317-327. 250. Dunham RA., Liu ZJ, and Argue B. 1998. The effect of the absence or presence of channel catfish males on induced ovulation of channel catfish females for artificial fertilization with blue catfish sperm. The Progressive Fish-Culturist 60:297-300. 251. Karsi A, Li P, Dunham R, and Liu ZJ. 1998. Transcriptional activities in the pituitaries of channel catfish (Ictalurus punctatus) before and after induced ovulation as revealed by expressed sequence tag analysis. Journal of Molecular Endocrinology 21:121-129. 252. Waldbieser G, Liu ZJ, Wolters W, and Dunham R. 1998. Genetic linkage and QTL mapping of catfish. Proceedings of the National Aquaculture Species Gene Mapping Workshop, 31-38. 33 253. Liu ZJ, Li P, Argue B, and Dunham R. 1998. Inheritance of RAPD markers in channel catfish (Ictalurus punctatus), blue catfish (I. furcatus) and their F1, F2 and backcross hybrids. Animal Genetics 29:58-62. 254. Liu ZJ, Nichols A, Li P, and Dunham R. 1998. Inheritance and usefulness of AFLP markers in channel catfish (Ictalurus punctatus), blue catfish (I. furcatus) and their F1, F2 and backcross hybrids. Molecular and General Genetics 258:260-268. 255. Liu ZJ, Li P, and Dunham R. 1998. Characterization of an A/T-rich family of sequences from the channel catfish (Ictalurus punctatus). Molecular Marine Biology and Biotechnology 7:232-239. 256. Dunham RA, Liu ZJ, Wolters W, Waldbieser G, Tiersch TR. and Winn R. 1998. Summary of catfish mapping group. Proceedings of the National Aquaculture Species Gene Mapping Workshop. Northeastern Regional Aquaculture Center Publication No. NRAC 98001.pp. 66-67. 257. Liu ZJ, and Dunham R. 1997. Genetic linkage and QTL mapping of ictalurid catfish. Alabama Agricultural Experiment Station Circular Bulletin 321:1-19. 258. Liu ZJ, Li P, Argue B, and Dunham R. 1997. Gonadotropin subunit glycoprotein from channel catfish (Ictalurus punctatus) and its expression during hormone-induced ovulation. Molecular Marine Biology & Biotechnology 6:217-227. 259. Liu ZJ. 1996. Heterostagger cloning: efficient and rapid cloning of PCR products. Nucleic Acids Research 24:2458-2459. 260. Ghai J, Ostrow R, Tolar J, McGlennen R, Lamke T, Tobolt D, Liu ZJ, and Faras A. 1996. The E5 gene product of rhesus papillomavirus is an activator of endogenous ras and phosphatidylinositol-3’-kinase in NIH 3T3 cells. Proceedings of the National Academy of Sciences USA. 93:12879-12884. 261. Liu ZJ. 1995. Faithful cloning of PCR products by heterostegger GC vector. NIH Journal 7:78. 262. Liu ZJ, Ostrow R, and Faras A. 1995. The expression levels of HPV-16 E7 determines the immortalization or transformation phenotypes of BRK cells. Virology 207:260-270. 263. Ostrow R, Coughlin S, McGlennen R, Liu ZJ, Zelterman D, and Faras A. 1994. Topical CTC-96 accelerates wart growth in rabbits infected with cottontail rabbit papillomavirus. Antiviral Research 24:27-35. 264. Liu ZJ, Ostrow R, McGlennen R, and Faras A. 1994. The E6 gene of human papillomavirus type 16 is Sufficient for transformation of baby rat kidney cells in cotransfection with activated H-ras. Virology 201:388-396. 265. Bushnell W, Liu ZJ. 1994. Incompatibibility conditioned by the Mla gene in powdery mildew of barley: Timing of the effect of cordycepin on hypersensitive cell death. Physiological and Molecular Plant Pathology 44:389-402. 266. Liu ZJ, and Hackett PB. 1993. Reverse Cloning: rapid generation of subclones for DNA sequencing. Methods in Enzymology (Ray Wu ed.) 218:47-58. 34 267. Liu ZJ, Szabo L, and Bushnell W. 1993. Molecular cloning and analysis of abundant and stage-specific mRNAs from Puccinia graminis. Molecular Plant-Microbe Interactions 6:8491. 268. Ostrow RS, Liu ZJ, Schneider JF, McGlennen RC, Forslund K, and Faras AJ. 1993. The products of the E5, E6, or E7 open reading frames of RhPV 1 can individually transform NIH 3T3 cells or in cotransfections with activated ras can transform primary rodent epithelial cells. Virology 196:861-867. 269. Gross M, Schneider J, Moav N, Alvarez C, Myster S, Liu ZJ, Hallerman E, Hackett P, Guise K, Faras A, and Kapuscinski A. 1992. Molecular analysis and growth evaluation of northern pike (Esox Iucius) microinjected with growth hormone genes. Aquaculture 103:253-273. 270. Moav B, Liu ZJ, Faras A, Guise K, Kapuscinski A, and Hackett PB. 1992. Regulation of expression of transgenes in developing fish. Transgenic Research 2:153-161. 271. Moav B, Liu ZJ, and Hackett P. 1992. Selection of promoters for gene transfer into fish. Molecular Marine Biology & Biotechnology 1:338-345. 272. Bushnell W, Clark TA, Liu ZJ, Smith AG, Szabo LJ, and Zeyen RJ. 1992. Host response gene expression in powdery mildew of barley. Vortr. Pflanzenzuchtg. 24:48-50. 273. Liu ZJ, Moav B, Faras A, Guise K, Kapuscinski A, and Hackett P. 1991. Importance of CArG boxes in regulation of the carp -actin gene expression. Gene 108:211-216. 274. Moav B, and Liu ZJ, Moav N, Faras A, Guise K, Kapuscinski A, and Hackett P. 1991. Expression of heterologous genes in transgenic fish. In: Transgenic Fish (Hew C., ed.) World Scientific Pub. Co. 121-140. 275. Hallerman E, Schneider J, Gross M, Liu ZJ, Yoon SJ, He L, Hackett P, Faras A, Kapuscinski A, and Guise K. 1990. Gene expression promoted by the RSV long terminal repeat element in transgenic goldfish. Animal Biotechnology 1:79-93. 276. Yoon SJ, Hallerman E, Gross M, Liu ZJ, Schneider J, Faras A, Hackett P, Kapuscinski A, and Guise K. 1990. Transfer of the gene for neomycin resistance into goldfish, Carassius auratus. Aquaculture 85:21-33. 277. Liu ZJ, Zhu Z, Roberg K, Faras A, Guise K, Kapuscinski A, and Hackett P. 1990. Isolation and characterization of the -actin gene of carp (Cyprinus carpio). DNA Sequence 1:125-136. 278. Liu ZJ, Moav B, Faras A, Guise K, Kapuscinski A, and Hackett P. 1990. Functional analysis of elements affecting expression of the -actin gene of carp. Molecular and Cellular Biology 10:3432-3440. 279. Liu ZJ, Moav B, Faras A, Guise K, Kapuscinski A, and Hackett P. 1990. Development of expression vectors for transgenic fish. Nature Biotechnology 8:1268-1272. 280. Yoon SJ, Liu ZJ, Kapuscinski A, Hackett P, Faras A, and Guise K. 1989. Successful gene transfer in fish. In: UCLA Symposia on Molecular and Cellular Biology and Gene Transfer and Gene Therapy, New series, Vol. 87:29-34. 35 281. Yoon SJ, Liu ZJ, Kapuscinski A, Hackett P, Faras A, and Guise K. 1990. Successful gene transfer in fish. 16th US-Japan Natural Resources Panel on Aquaculture Symposium. NOAA Technical Report NMFS 92:39-44. 282. Liu ZJ, and Hackett P. 1989. Sequence variation of the Rous sarcoma virus PrA src gene. Nucleic Acids Research 17:3986. 283. Liu ZJ, Zhu Z, Roberg K, Faras A, Guise K, Kapuscinski A, and Hackett P. 1989. The actin gene of carp (Ctenopharyngodon idella). Nucleic Acids Research 17:5850. 284. Liu ZJ, and Hackett P. 1989. Rapid generation of subclones for DNA sequencing using the reverse cloning procedure. BioTechniques 7:722-728. 285. Bushnell WR, Mendgen K, and Liu ZJ. 1987. Accumulation of potentiometric and other dyes in haustoria of Erysiphe graminis in living host cells. Physiological and Molecular Plant Pathology 31:237-250. 286. Liu ZJ, Bushnell WR, and Brambl R. 1987. Potentiometric cyanine dyes are sensitive probes for mitochondria in intact plant cells: kinetin enhances mitochondrial fluorescence. Plant Physiology 84:1385-1390. 287. Bushnell WR, Liu ZJ, and Mendgen K. 1986. Cyanine potentiometric probes for mitochondria of Erysiphe graminis. Phytopathol. 76:1123. 288. Liu ZJ, and Bushnell WR. 1986. Effects of cytokinins on fungus development and host response in powdery mildew of barley. Physiological Plant Pathology 29:41-52. 289. Li J, Bai X, and Liu ZJ. 1982. A preliminary taxonomy of powdery mildew in Shaanxi Province. Northwestern Agricultural University Academic Journal 20:70-76. Translated books The Genetic Resource Studies of Silver Carp, Bighead, and Grass Carp in the Yangtze River, Zhu River, and Heilong River. Edited by Li Sifa, Wu Lizhao, Wang Qiang, Chou Qianru,Chen Yongle, Published by Shanghai Science and Technology Press, Translated by Z. John Liu. Pp. 1-228. Published Abstracts/Presentations 1. Xiaozhu Wang, Chen Jiang, Shikai Liu, Xin Geng, Tao Zhou, Ning Li, Jun Yao, Lisui Bao, Yun Li, Qifan Zeng, Sen Gao, Yulin Jin, Yujia Yang, Rex Dunham and Zhanjiang Liu. A Genome-Wide Association Study for Low Oxygen Tolerance in Catfish Using the 250K SNP Array. January 9-14, 2016. International Plant and Animal Genome XXIV, San Diego, CA, USA. 2. Tao Zhou, Shikai Liu, Xin Geng, Chen Jiang, Yulin Jin, Xiaozhu Wang, Ning Li, Suxu Tan, Lisui Bao, Jun Yao, Yu Zhang, Jiaren Zhang, Luyang Sun, Jianbin Feng, Ludmilla Kaltenboeck, Zhanjiang Liu. Candidate genes for ESC Disease Resistance of Catfish as Revealed by a Genome Wide Association Study. January 9-13, 2016, International Plant and Animal Genome XXIV, San Diego, CA, USA. 36 3. Qiang Fu, Qifan Zeng, Yun Li, Chen Jiang, Sen Gao, Tao Zhou, Jun Yao, Shikai Liu, Zhanjiang Liu. Comparative transcriptome analysis of the swimbladder reveals expression signatures in response to hypoxia in channel catfish, Ictalurus punctatus. January 9-13, 2016, International Plant and Animal Genome XXIV, San Diego, CA, USA. 4. Shikai Liu, Yun Li, Zhenkui Qin, Xin Geng, Lisui Bao, Ludmilla Kaltenboeck, Huseyin Kucuktas, Rex Dunham, Zhanjiang Liu. High-density genetic linkage mapping in hybrid catfish revealed genomic incompatibility between channel catfish and blue catfish. Jan 913, 2016, Plant & Animal Genome XXIV, San Diego, CA, USA. 5. Shikai Liu, Yun Li, Chen Jiang, Qifan Zeng, Lisui Bao, Xin Geng, Jun Yao, Zhaohong Wong, Zhanjiang Liu. The catfish MicroRNAome: identification, annotation and expression profiling in response to bacterial infections and hypoxia stress. Jan 9-13, 2016, Plant & Animal Genome XXIV, San Diego, CA, USA. 6. Xin Geng, Shikai Liu, Jun Yao, Lisui Bao, Jiaren Zhang, Chao Li, Ruijia Wang, Xiaozhu Wang, Jin Sha, Peng Zeng, Degui Zhi, Zhanjiang Liu. Mapping of genetic regions for head size in catfish: the involved GTPase pathway is evolutionarily conserved for skull morphometric traits. Jan 9-13, 2016, Plant & Animal Genome XXIV, San Diego, CA, USA. 7. Sen Gao, Yao Jun, Zhanjiang Liu. Integrative and Comparative Analysis of Skin Transcriptome of Various Fish Species. January 9-13, 2016, International Plant and Animal Genome XXIV, San Diego, CA, USA 8. Qifan Zeng, Qiang Fu, Shikai Liu, Yun Li, Zhanjiang Liu. Development of ultra-high density SNP genotyping array with 690K markers for catfish. January 9-13, 2016, International Plant and Animal Genome XXIV, San Diego, CA, USA. 9. Xin Geng, Jin Sha, Shikai Liu, Lisui Bao, Jiaren Zhang, Ruijia Wang, Jun Yao, Chao Li, Jianbin Feng, Fanyue Sun, Luyang Sun, Chen Jiang, Yu Zhang, Ailu Chen, Rex Dunham, Degui Zhi, Zhanjiang Liu. GWAS in catfish reveals the presence of functional hubs of related genes within major QTLs for columnaris disease resistance. Jan 10-14, 2015, Plant & Animal Genome XXIII, San Diego, CA, USA. W048/P0243. 10. Qifan Zeng, Shikai Liu, Jun Yao, Yu Zhang, Baofeng Su, Rex Dunham, Zhanjiang Liu. RNA-Seq Analysis Unraveled Genes and Pathways Involved in Testicular Differentiation in Channel Catfish. January 10-14, 2015, International Plant and Animal Genome XXIII, San Diego, CA, USA, P0241. 11. Jun Yao, Jiaren Zhang, Shikai Liu, Qifan Zeng, Luyang Sun, Baofeng Su, Chen Jiang, Yun Li, Rex Dunham, Zhanjiang Liu. Transcriptomic Signatures in Common Carp Skin during Scale Regeneration. January 10-14, 2015, International Plant and Animal Genome XXIII, San Diego, CA, USA, P0245. 12. Chen Jiang, Jiaren Zhang, Jun Yao, Shikai Liu, Yun Li, Lin Song, Chao Li, Zhanjiang Liu. Characterization of Complement Regulatory Genes in Channel Catfish and Their Involvement in Disease Defense Response. January 10-14, 2015, International Plant and Animal Genome XXIII, San Diego, CA, USA, P0242. 13. Waldbieser G., Liu, Z.J. Production of a genome sequence assembly for channel catfish, Ictalurus punctatus. January 9-14, 2015, Plant and Animal Genome XXI, San Diego. 14. Study of performance traits: Genomics made it easy, Invited Lecture, National Chengkong University, Taiwan, June 8, 2014. 15. Liu, Z.J. 2014. Genomic approaches to the understanding of disease resistance and low oxygen tolerance. Next Generation Genome View of Plants, Animals and Microbes, March 5-7, Singapore 37 16. 17. 18. 19. 20. 21. 22. 23. 24. 25. 26. 27. 28. Liu, Z.J. 2013. Genomic approaches to the understanding of disease resistance and innate immunity in catfish, Keynote speaker, the First International Conference of Fish and Shell Fish Immunology, June 25028, 2013, Vigo, Spain. Fanyue Sun, Yu Zhang, Shikai Liu, and Zhanjiang Liu. Towards the identification of sexdetermining genes in channel catfish. January 12-16, 2013, Plant and Animal Genome XXI, San Diego. Jianbin Feng, Shikai Liu, Yu zhang, Jiaren Zhang, Ludmilla Kaltenboeck, Huseyin Kucuktas, Zhanjiang Liu. Transcriptome and expression profiling of response to acute hypoxia in the gill of adult catfish. January 12-16, 2013, Plant and Animal Genome XXI, San Diego. Luyang Sun, Shikai Liu, Yu Zhang, Jiaren Zhang, Jianbin Feng, Ludmilla Kaltenboeck, Geoff Waldbieser, Huseyin Kucuktas, Zhanjiang (John) Liu. Whole genome SNP discovery and analysis of catfish genetic diversity. January 12-16, 2013, Plant and Animal Genome XXI, San Diego. Shikai Liu, Zhanjiang (John) Liu. Genome-wide identification and evolutionary analysis of the ATP-binding cassette (ABC) transporter superfamily in catfish. January 12-16, 2013, Plant and Animal Genome XXI, San Diego. Shikai Liu, Luyang Sun, Fanyue Sun, Yanliang Jiang, Yu Zhang, Jiaren Zhang, Jianbin Feng, Ludmilla Kaltenboeck, Huseyin Kucuktas, Zhanjiang (John) Liu. Development of the catfish 250K SNP array. January 12-16, 2013, Plant and Animal Genome XXI, San Diego. Yanliang Jiang, Yu Zhang, Parichart Ninwichian, Geoff Waldbieser, and Zhanjiang Liu. Genome-wide comparative mapping of channel catfish with zebrafish. January 12-16, 2013, Plant and Animal Genome XXI, San Diego. Geoff Waldbieser, Michael Kertesz, Dmitry Pushkarev, Tim Blauwkamp, and Zhanjiang Liu. Production of Moleculo Long Reads for the blue catfish genome assembly. January 12-16, 2013, Plant and Animal Genome XXI, San Diego. Eric Peatman, Chao Li, Fanyue Sun, Yu Zhang, Ruijia Wang, Jianguo Lu, Zhanjiang Liu. RNA-seq analysis of mucosal immune responses in catfish following exposure to bacterial pathogens. June 25-29, 2012, The 11th International Symposium on Genetics in Aquaculture, Auburn, AL USA. Abstract p.87. Zhanjiang Liu. Challenges of genome era aquaculture genetics research. June 25-29, 2012, The 11th International Symposium on Genetics in Aquaculture, Auburn, AL USA. Abstract p.72. Chao Li, Yu Zhang, Ruijia Wang, Jianguo Lu, Samiran Nandi, Sriprakash Mohanty, Jeffery Terhune, Zhanjiang Liu, Eric Peatman. RNA-seq analysis of mucosal immune responses reveals signatures of intestinal barrier disruption and pathogen entry following Edwardsiella ictaluri infection in channel catfish, Ictalurus punctatus. January 14-18, 2012, Plant and Animal Genome XX, San Diego. Fanyue Sun, Chao Li, Shikai Liu, Yanliang Jiang, Zunchun Zhou, Huseyin Kucuktas, Eric Peatman, Zhanjiang Liu. RNA-Seq analysis of differentially expressed genes in channel catfish after Flavobacterium columnare challenge. January 14-18, 2012, Plant and Animal Genome XX, San Diego. Hao Zhang, Eric Peatman, Hong Liu, Tingting Feng, Liqiao Chen, Zhanjiang Liu. Molecular characterization and expression responds to Edwardsiella ictaluri challenge of three L-type lectin genes from channel catfish, Ictalurus punctatus. January 14-18, 2012, Plant and Animal Genome XX, San Diego. 38 29. 30. 31. 32. 33. 34. 35. 36. 37. 38. 39. 40. 41. 42. Jianguo Lu, Eric Peatman, Haibao Tang, Joshua Lewis, Huseyin Kucuktas, Zhanjiang Liu. Genome expansion in zebrafish is mediated by recent tandem duplications. January 14-18, 2012, Plant and Animal Genome XX, San Diego. Jiaren Zhang, Rajendran Kooloth Valappil, Shikai Liu, Huseyin Kucuktas, Zhenxia Sha, Luyang Sun, Jeffery Terhune, Eric Peatman, Xiuli Wang, Hong Liu, Hao Zhang, Zhanjiang Liu. Pathogen recognition receptors (PRRs) in channel catfish, Ictalurus punctatus: Identification, phylogeny and expression of NLRs, RLRs and TLRs. January 14-18, 2012, Plant and Animal Genome XX, San Diego. Ruijia Wang, Chao Li, James Stoeckel, Gregory Moyer, Zhanjiang Liu, Eric Peatman. Rapid development of molecular resources for a freshwater mussel, Villosa lienosa (Bivalvia: Unionidae) using a RNA-seq-based approach. January 14-18, 2012, Plant and Animal Genome XX, San Diego. Shikai Liu, Yu Zhang, Zunchun Zhou, Geoff Waldbieser, Fanyue Sun, Jianguo Lu, Jiaren Zhang, Yanliang Jiang, Hao Zhang, Xiuli Wang, Rajendran Kooloth Valappil, Huseyin Kucuktas, Eric Peatman, Zhanjiang Liu. Comprehensive annotation of the transcriptome of channel catfish (Ictalurus punctatus) by paired-end RNA-Seq. January 14-18, 2012, Plant and Animal Genome XX, San Diego. Xiuli Wang, Shikai Liu, Fanyue Sun, Jiaren Zhang, Hong Liu, Rajendran Kooloth Valappil, Jianguo Lu, Luyang Sun, Hao Zhang, Yu Zhang, Yanliang Jiang, Eric Peatman, Ludmilla Kaltenboeck, Huseyin Kucuktas, Zhanjiang Liu. Identification of genes associated with heat tolerance in catfish exposed to chronic thermal stress using RNA-Seq. January 14-18, 2012, Plant and Animal Genome XX, San Diego. Yu Zhang, Jianguo Lu, Hao Zhang, Chao Li, Zhanjiang Liu. Comparative genomic analysis of catfish linkage group 28 with teleost species genomes. January 14-18, 2012, Plant and Animal Genome XX, San Diego. Li Lian Wong, Eric Peatman, Huseyin Kucuktas, Lenore Kelly,Chuanjiang Zhou, Uthairat Na-Nakorn, Zhanjiang Liu 2011. Catfish species authentication using DNA barcoding and lab-on-a-chip-based PCR-RFLP assays. World Aquaculture Society Conference. Liu, Z.J. Strategies for the application of whole genome sequences for improving production and performance traits in catfish. Keynote lecture, Functional Genomics Session, World Aquaculture Society Conference, Natal, Brazil, June 6-10, 2011. Liu, Z.J. Strategies for efficient assembly and annotation of the catfish whole genome sequence. Finfish Genetics Session, World Aquaculture Society Conference, Natal, Brazil, June 6-10, 2011. Liu, Z.J. Strategies for efficient assembly and annotation of the catfish whole genome sequence. January 15-19, 2011, Plant and Animal Genome XIX, San Diego. Zunchun Zhou, Hong Liu, Shikai Liu, Fanyue Sun, Eric Peatman, Huseyin Kucuktas, Ludmilla Kaltenboeck, Tingting Feng, Hao Zhang, Donghong Niu, Geoff Waldbieser, Zhanjiang Liu. Alternative Complement Pathway of Channel Catfish (Ictalurus punctatus): Molecular Characterization and Expression Analysis of Factors B and D. January 15-19, 2011, Plant and Animal Genome XIX, San Diego. Jianguo Lu, Eric Peatman, Qing Yang, Shaolin Wang, Zhiliang Hu, James Reecy, Huseyin Kucuktas and Zhanjiang Liu. Development of cBARBEL, a Catfish Genome Database. January 15-19, 2011, Plant and Animal Genome XIX, San Diego. Shikai Liu, Eric Peatman, Zunchun Zhou, Jianguo Lu, Shaolin Wang, Hong Liu, Yanliang Jiang, Fanyue Sun, Huseyin Kucuktas, Zhanjiang Liu. Genome-scale gene-associated SNPs in catfish for the construction of a high-density SNP array. January 15-19, 2011, Plant and Animal Genome XIX, San Diego. 39 43. 44. 45. 46. 47. 48. 49. 50. 51. 52. 53. 54. Donghong Niu, Eric Peatman, Hong Liu, Jianguo Lu, Huseyin Kucuktas, Shikai Liu, Fanyue Sun, Hao Zhang, Tingting Feng, Cova Arias, Jeffery Terhune, Geoff Waldbieser, Jiale Li, Zhanjiang Liu. Microfibril-associated protein 4 (MFAP4) genes in catfish play a novel role in innate immune responses. January 15-19, 2011, Plant and Animal Genome XIX, San Diego. Tingting Feng, Eric Peatman, Hong Liu, Shaolin Wang, Zunchun Zhou, Hao Zhang, Donghong Niu, Lilian Wong, Xiaolin Liu, Zhanjing Liu. Discovery and mapping of SNPs in immune-response genes of channel catfish, Ictalurus punctatus. January 15-19, 2011, Plant and Animal Genome XIX, San Diego. Jianguo Lu, Eric Peatman, Qing Yang, Haibao Tang, Huseyin Kucuktas, and Zhanjiang Liu. Genome-wide gene duplication in teleost fish: same-species and cross-species analysis and comparisons. January 15-19, 2011, Plant and Animal Genome XIX, San Diego. Liu, Z.J. Liu, Z.J. Strategies for efficient assembly and annotation of the catfish whole genome sequence. International Workshop on Marine and Aquaculture Genomics, November 16-17, 2010, Shenzhen, China. Conference Book page 13. Jianguo Lu, Eric Peatman, Qing Yang, Shaolin Wang, Zhiliang Hu, James Reecy, Huseyin Kucuktas and Zhanjiang Liu. Development an Informatic Platform for Genome Biology of Catfish. World Aquaculture Society 2011, February 28-March 4, 2011, New Orleans, LA. Shaolin Wang, Shikai Liu, Yanliang Jiang, Hong Liu, Jianguo Lu, Eric Peatman, Huseyin Kucuktas, Zhanjiang Liu. Identification And Criteria-Based Screening Of Putative SNP Markers For A High-Density SNP Chip For Catfish. P655, January 8-13, 2010, Plant and Animal Genome XVIII, San Diego. Yoona Lee, Fei Chen, Shaolin Wang, Yanliang Jiang, Hong Liu, Shikai Liu, Jason Abernathy, Eric J Peatman , Huseyin Kucuktas, Zhanjiang Liu. Identification And Characterization Of Catfish Full-Length cDNAs. P656, January 8-13, 2010, Plant and Animal Genome XVIII, San Diego. Jason W. Abernathy, De-Hai Xu, Eric Peatman, Huseyin Kucuktas, Phillip Klesius, Zhanjiang Liu. Microarray Analysis For Developmental And Pathogenic Gene Regulation In The Fish Protozoan Parasite Ichthyophthirius multifiliis. P654, January 8-13, 2010, Plant and Animal Genome XVIII, San Diego. Hong Liu, Wenqi Wang, Shikai Liu, Huseyin Kucuktas, Eric Peatman, Zhanjiang Liu. Structure And Expression Of Calreticulin And Ceruloplasmin Genes In Channel Catfish, Ictalurus punctatus. P638, January 8-13, 2010, Plant and Animal Genome XVIII, San Diego. Jianguo Lu, Eric Peatman, Wenqi Wang, Qing Yang, Jason Abernathy, Shaolin Wang, Zhanjiang Liu. Alternative Splicing in Teleost Fish Genomes: Cross-Species Analysis and Comparisons. W055, January 8-13, 2010, Plant and Animal Genome XVIII, San Diego. Hong Liu, Yanliang Jiang, Shaolin Wang, Zhanjiang Liu. Comparative genome analysis using BAC end sequences of catfish. January 9-14, 2009, Plant and Animal Genome XVII, San Diego. Huseyin Kucuktas, Shaolin Wang, Jason Abernathy, Parichart Ninwichian, Ping Li, Chongbo He, Peng Xu, Zhenxia Sha, Hong Liu, Yanliang Jiang, Puttharat Baoprasertkul, Benjaporn Somridhivej, Yaping Wang, Lei Liu, William Muir, Ximing Guo, Zhanjiang Liu. Construction of the genetic linkage and comparative maps of channel catfish (Ictalurus punctatus) using est-derived microsatellite markers. January 9-14, 2009, Plant and Animal Genome XVII, San Diego. 40 55. 56. 57. 58. 59. 60. 61. 62. 63. 64. Jason W. Abernathy, Jianguo Lu, Hong Liu, Zhenxia Sha, and Zhanjiang Liu. Complement control factor I in catfish immunity: molecular characterization and gene expression after bacterial challenge. January 9-14, 2009, Plant and Animal Genome XVII, San Diego. Yanliang Jiang, Jason W. Abernathy, Hong Liu, Shaolin Wang, Atra Chaimongkol, Jianguo Lu, and Zhanjiang Liu. Identification of matrix metalloproteinase-13 gene in channel catfish (Ictalurus punctatus) and expression through embryogenesis and pathology. January 9-14, 2009, Plant and Animal Genome XVII, San Diego. Hong Liu, Tomokazu Takano, Jason Abernathy, Shaolin Wang, Zhenxia Sha, Jeffery Terhune, Huseyin Kucuktas, Yanliang Jiang, Zhanjiang Liu. Molecular characterization and gene expression of the channel catfish ferritin heavy chain gene after bacterial infection and iron treatment. January 9-14, 2009, Plant and Animal Genome XVII, San Diego. Hong Liu, Tomokazu Takano, Jason Abernathy, Shaolin Wang, Zhenxia Sha, Jeffery Terhune, Huseyin Kucuktas, Yanliang Jiang, Zhanjiang Liu. Expression analysis of the transferrin gene in channel catfish suggests its importance in iron regulation with Gramnegative bacterial infection. January 9-14, 2009, Plant and Animal Genome XVII, San Diego. Zhenxia Sha, Jason W. Abernathy, Shaolin Wang, Ping Li, Huseyin Kucuktas, Hong Liu, and Zhanjiang Liu. The NOD-like subfamily of NLR receptor genes exhibit tissue-specific expression patterns in the channel catfish (Ictalurus punctatus). January 9-14, 2009, Plant and Animal Genome XVII, San Diego. Shaolin Wang, Jason Abernathy, Geoff Waldbieser, Erika Lindquist, Paul Richardson, S Lucas, M Wang, Ping Li, Jyothi Thimmapuram, Lei Liu, Deepika Vullaganti, Eric Peatman, Peng Xu, Christopher Murdock, Brian C. Small, Melanie Wilson, Huseyin Kucuktas, Benjaporn Somridhivej, Puttharat Baoprasertkul, Jonas Quilang, Hong Liu, Zhenxia Sha, Jianguo Lu, Baolong Bao, Yaping Wang, Qun Wang, Tomokazu Takano, Samiran Nandi, Sylvie Quiniou, Eva Bengten, Norman Miller, John Trant, Daniel Rokhsar, Zhanjiang Liu. JGI completed catfish EST sequencing project. January 9-14, 2009, Plant and Animal Genome XVII, San Diego. Shaolin Wang, Hong Liu, David Bushek, Susan E. Ford, Huseyin Kucuktas, Jonas Quilang, Ping Li, Richard Wallace, Yongping Wang, Ximing Guo, and Zhanjiang Liu. Microarray analysis of gene expression in eastern oysters (Crassostrea virginica) reveals activation of apoptosis inhibition after dermo (perkinsus marinus) challenge. January 9-14, 2009, Plant and Animal Genome XVII, San Diego. Shaolin Wang, Zhenxia Sha, Tad S. Sonstegard, Hong Liu, Peng Xu, Benjaporn Somridhivej1, Eric Peatman, Huseyin Kucuktas, Zhanjiang Liu. Quality assessment parameters for EST-derived SNPs from catfish. January 9-14, 2009, Plant and Animal Genome XVII, San Diego. David C. Glover, Dennis R. DeVries, Russell A. Wright, Alicia J. Norris, Troy M. Farmer, Huseyin Kucuktas, Zhanjiang Liu, and Rex A. Dunham. Understanding the slow-growth, high-condition paradox of largemouth bass in the mobile-Tensaw river delta, AL: Integrating bioenergitic modeling, life-history, and genetics. MISSISSIPPI-ALABAMA Bays and Bayous Symposium, October 28-29, 2008, Mississippi Coast Coliseum and Convention Center, Biloxi, Mississippi. Zhanjiang Liu. 2008. Healthy aquaculture: the genomic approaches. AU-OUC Joint Symposium, Healthy Aquaculture, Qingdao, China, May 25-27, 2008. 41 65. 66. 67. 68. 69. 70. 71. 72. 73. 74. 75. 76. 77. Zhanjiang Liu. 2008. Genomic approaches to the understanding of genes involved in resistance of catfish. The 5th World Fisheries Congress, Yokohama, Japan, October 20 to 24, 2008. Jason W. Abernathy, Jianguo Lu, Zhenxia Sha, and Zhanjiang (John) Liu. 2008. Complement control factor I in the channel catfish (Ictalurus punctatus) is conserved through teleost evolution. The Southeastern Ecology and Evolution Conference at Florida state University. Zhanjiang Liu. 2008. Status and trends of aquaculture genomics. World Aquaculture Society 2008, May 12-16, Pusan, Korea. Eric Peatman, Jason Abernathy, Jeffery Terhune, Puttharat Baoprasertkul, Peng Xu, Samiran Nandi, Shaolin Wang, Benjaporn Somridhivej, Huseyin Kucuktas, Ping Li, Rex Dunham, Zhanjiang Liu. 2008. Microarray analysis of gene expression in the blue catfish liver reveals early activation of the MHC class I pathway after infection with Edwardsiella ictaluri. Plant and Animal Genome XVI, San Diego. Jason W Abernathy, De-Hai Xu, Ping Li, Phillip Klesius, and Zhanjiang Liu. 2008. Posttranscriptional polyadenylation of the large subunit ribosomal RNA in Ichthyophthirius multifiliis. Plant and Animal Genome XVI, San Diego. Tomokazu Takano, Hong Liu, Zhenxia Sha, Eric Peatman, Jeffery Terhune, Huseyin Kucuktas, Ping Li, Eva-Stina Edholm, Melanie Wilson, Zhanjiang Liu. 2008. The two channel catfish intelectin genes exhibit highly differential patterns of tissue expression and regulation after infection with Edwardsiella ictaluri. Plant and Animal Genome XVI, San Diego. Jonas Quilang, Shaolin Wang, Ping Li, Jason Abernathy, Eric Peatman, Yongping Wang, Lingling Wang, Yaohua Shi, Richard Wallace, Ximing Guo, and Zhanjiang Liu. 2008. Generation and analysis of ESTs from the eastern oyster Crassostrea virginica and identification of microsatellite and SNP markers. Plant and Animal Genome XVI, San Diego. P613. Shaolin Wang, Peng Xu, Hong Liu, Lei Liu, Jim Thorsen, Huseyin Kucuktas, Zhanjiang Liu. 2008. A BAC-based physical map of the channel catfish genome. Plant and Animal Genome XVI, San Diego. Zhenxia Sha, Peng Xu, Tomokazu Takano, Hong Liu, Jeffery Terhune, Zhanjiang Liu. 2008. The warm temperature acclimation protein wap65 as an immune response gene: its duplicates are differentially regulated by temperature and bacterial infections. Zhanjiang Liu, Brad Argue, Renee Beam and Rex Dunham. 2008. Reproductive characteristics of F1, F2, and backcross channel-blue hybrid catfish. World Aquaculture Society Annual Conference, Orlando, FL. Zhanjiang Liu. 2007. "Status and challenges of aquaculture genomics: Making reactions work without reagents". Invited lecture for the Conference of “Convergence of Genomics and the Land Grant Mission: Emerging Trends in the Application of Genomics in Agricultural Research”, page 38-39, Purdue University, West Lafayette, Indiana, September 10-12, 2007. Eric Peatman, Puttharat Baoprasertkul, Jefferey Terhune, Peng Xu, Samiran Nandi, Huseyin Kucuktas, Ping Li, Shaolin Wang, Benjaporn Somridhivej, Thanathip Lamkom, Wonkyo Lee, Rex Dunham, Zhanjiang Liu. 2007. Global assessment of the acute phase response in catfish after infection with a Gram negative bacterium. Plant and Animal Genome XV, San Diego. Jason Abernathy, Peng Xu, Ping Li, Dehai Xu, Huseyin Kucuktas, Phillip Klesius, Covadonga Arias, and Zhanjiang Liu. 2007. A genomic resource for the analysis of 42 78. 79. 80. 81. 82. 83. 84. 85. 86. 87. 88. 89. developmental and virulent factors of the protozoan fish parasite Ichthyophthirius multifiliis using microarrays. Plant and Animal Genome XV, San Diego. Shaolin Wang, Peng Xu, Jim Thorsen, Baoli Zhu, pieter J de Jong, Geoff Waldbieser, Zhanjiang Liu. 2007. Characterization of a BAC library from channel catfish Ictalurus punctatus: indications of high rates of evolution among teleost genomes. Plant and Animal Genome XV, San Diego. Peng Xu, Shaolin Wang, Lei Liu, Jim Thorsen, Zhanjiang Liu 2007. A BAC-based physical map of the channel catfish genome. USDA CSREES Principal Investigator Meeting, San Diego. Eric Peatman, Puttharat Baoprasertkul, Jefferey Terhune, Peng Xu, Samiran Nandi, Huseyin Kucuktas, Ping Li, Shaolin Wang, Benjaporn Somridhivej, Thanathip Lamkom, Wonkyo Lee, Rex Dunham, Zhanjiang Liu. Differentially expressed genes of catfish after bacterial infection as assessed by a 28000 gene microarray. IMBC 2007, The 8th International Marine Biotechnology Conference, Eilat, Israel. Zhanjiang Liu. Status and trends in genomics and modern genetic technologies in aquaculture and fisheries. Abstracts of the CAS Conference on Marine Science and Technology, Qingdao, China, July 29-31, 2006, pp. 49-52. Zhanjiang Liu, Huseyin Kucuktas, Ping Li, Puttharat Baoprasertkul, Peng Xu, Eric Peatman, Shaolin Wang, Benjaporn Somridhivej, Jason Abernathy, Wonkyo Lee, Samiran Nandi. Genomic resources and their informatic analysis for comparative genomics in catfish. International Symposium For Genetics in Aquaculture IX, Montpellier, France June 25-30, 2006. Eric Peatman, Baolong Bao, Xu Peng, Puttharat Baoprasertkul, Shaolin Wang, and Zhanjiang Liu. Genomic organization, architecture, and expression of catfish CC chemokines. Sixth Congress of International Developmental and Comparative Immunology Society, Charleston, SC. July 1-6, 2006. Eric Peatman, Puttharat Baoprasertkul, Peng Xu, Baolong Bao, Qun Wang, Yaping Wang, Zhanjiang Liu. Genomic approaches for characterization of the teleost innate immune system. Sixth Congress of International Developmental and Comparative Immunology Society, Charleston, SC. July 1-6, 2006. Zhanjiang Liu, Huseyin Kucuktas, Ping Li, Puttharat Baoprasertkul, Peng Xu, Eric Peatman, Shaolin Wang, Benjaporn Somridhivej, Jason Abernathy, Wonkyo Lee, Samiran Nandi. Informatic analysis of genomic resources for comparative genomics in catfish. In BARD Workshop, Aquaculture Genetics, Status and Prospects, Eilat, Israel, February 2023, 2006, Program and Abstracts, p.14. Xu P, Wang S, Liu L, Liu ZJ. 2006. Generation of 25,000 BAC-end sequences for genome analysis of catfish. Plant and Animal Genome XIV, San Diego. Peatman E, Bao B, Xu P, Baoprasertkul P, Wang S, and Liu ZJ. 2006. Genomic organization, architecture, and expression of catfish CC chemokines. Plant and Animal Genome XIV, San Diego. Wang,Y., He C, Xu P, Wang, Q., Peatman E, Bao B, Kucuktas H, Arias, C., and Liu ZJ. 2006. Novel hypoxia-induced factors (HIF) and their inhibiting factor (FIH-1) from channel catfish (Ictalurus punctatus) and their expression under hypoxia. Plant and Animal Genome XIV, San Diego. Wang, Y., Wang, Q., Baoprasertkul P, Peatman E, Wang S, and Liu ZJ. 2006. Genomic organization, gene duplication, and expression analysis of interleukin-1 in channel catfish (Ictalurus punctatus). Plant and Animal Genome XIV, San Diego. 43 90. 91. 92. 93. 94. 95. 96. 97. 98. 99. 100. 101. 102. 103. Liu ZJ, Waldbieser, G., Wilson, M., Trant, J., Quiniou, S., Small, B., Murdock, C., Li , P. 2006. JGI to sequence 300,000 catfish ESTs. Plant and Animal Genome XIV, San Diego. Wang, Q., Wang, Y., Xu P, Li P, and Liu ZJ. 2006. NK-lysin of channel catfish: gene triplication, sequence variation, and expression analysis. Plant and Animal Genome XIV, San Diego. Peatman E, Xu P, Wang S, Abernathy J, Baoprasertkul P, Someridhivej, B., Nandi, S., Li P, Kucuktas H, Liu ZJ. 2006. Identification of catfish candidate genes for comparative genome analysis utilizing chromosome-specific genes of the Tetraodon Genome. Plant and Animal Genome XIV, San Diego. Baoprasertkul P, Abernathy J, Li P, Kucuktas H, and Liu ZJ. 2006. Expression of Tolllike receptors in resistant and susceptible catfish after infection with bacterial pathogen Edwardsiella ictaluri. Plant and Animal Genome XIV, San Diego. Liu ZJ. 2006. BAC-end sequences and candidate genes of catfish for comparative genome analysis utilizing chromosome-specific genes of the Tetraodon Genome. Plant and Animal Genome XIV, San Diego. Liu ZJ, Genomewise thinking: new approaches demanded by living in the genomics era. Fisheries Research Forum and 13th International Conference on Genes, Gene Families, and Isozymes. September 17-21, 2005, Shanghai, China. Liu ZJ, P. Baoprasertkul, B. Bao, E. Peatman, P. Xu, Q. Wang, L. Chen, H., Kucuktas, Y. Wang, P. Li, C. He, Q. He, S. Zhang, H. Zeng. Catfish Innate Immunity Components: Chemokines and Antimicrobial Peptides. International Marine Biotechnology Conference, Newfoundland, Canada. June 7-12, 2005. Liu ZJ, Eric Peatman, Ping Li, Puttharat Baoprasertkul, Chongbo He, Micah Simmons, Huseyin Kucuktas. Genomic studies of catfish using channel catfish x blue catfish interspecific hybrid system. World Aquaculture Society Aquaculture America 2005. New Orleans, January 17-20, 2005. Peng Xu, Baolong Bao, Shazhong Zhang, Eric Peatman, Chongbo He, Qun Wang, Yaping Wang, Zhanjiang Liu. 2005. Characterization And Expression Analysis Of Bactericidal Permeability-Increasing Protein (BPI) Antimicrobial Peptide Gene From Channel Catfish Ictalurus punctatus. Plant and Animal Genome XIII, San Diego, p610. Eric Peatman, Puttharat Baoprasertkul, Chongbo He, Ping Li, Huseyin Kucuktas, Zhanjiang Liu. 2005. Chemokine Diversity In Fish: Growing Complexity In The Teleost Innate Immune System. Plant and Animal Genome XIII, San Diego, W028. Baolong Bao, Eric Peatman, Qiang He, Shanzhong Zhang, Hang Zeng, Chongbo He, Ping Li, Zhanjiang Liu. 2005. The Catfish Liver-Expressed Antimicrobial Peptide 2 (LEAP-2) Gene Is Expressed In A Wide Range Of Tissues And Developmentally Regulated. Plant and Animal Genome XIII, San Diego, p590. Puttharat Baoprasertkul, Eric Peatman, Ping Li, Chongbo He, Huseyin Kucuktas and Zhanjiang Liu. 2005. Cloning, Characterization And Expression Of Three CXC Chemokines In Channel Catfish (Ictalurus punctatus). Plant and Animal Genome XIII, San Diego, p594. Cunningham C, Jenny MJ, Chapman R, Vasta GR, Liu J, Gomez-Chiarri M, Escoubas J-M, Bachere E, Almeida J, Guo X, & Warr G. 2005. The effect of Perkinsus marinus on the transcriptome of the Eastern oyster Crassostrea virginica. International Marine Biotechnology Conference, Canada. Alison Hutson*, Yoni Zohar, Eytan Abraham, Anang Kristanto, Joseph Ballenger, Michael Templeton, Gloria Umali, Deborah Beam, Fajin Wang, Carel Ligeon, Phillip Waters,Jr., Zhanjiang Liu, and Rex A. Dunham. Evaluation of LHRHa delivered via liquid injections 44 104. 105. 106. 107. 108. 109. 110. 111. 112. 113. 114. 115. 116. or implants for the production of channel-blue hybrid catfish embryos. WAS, New Orleans, 2005. Lundqvist, M., Jenny, M., Warr, G., Gross, P., Vasta, G., Robledo, J., Liu ZJ, Tompkins, J., Fang, G., Saski, C., Gomez-Chiarri, M., Escoubas, J-M., Bachere, E., Roche, P., Hedgecock, D., Chapman, R. 2004. Current state of oyster (Crasostrea) functional genomics resources. Plant and Animal Genome XII, San Diego, p735. Liu ZJ. 2004. Mapping the catfish genome: DNA marker technologies, linkage mapping, and functional analysis. AAAS Symposium: Aquaculture: Recent Advances in Fish Culture, Breeding and the Mitigation of Environmental Impact. Seattle. Z.J. Liu, P. Li, L. Liu, C. He, H. Kucuktas, J. Feng, L. Chen, E. Peatman, P. Baoprasertkul, M. Simmons, W. Muir, J. Grizzle, R. Dunham, Y. Brady 2004. 30,000 catfish ESTs: a case study for a set of quality standards established for the analysis of ESTs. Plant and Animal Genome XII, San Diego, W22. Baoprasertkul P, E. Peatman, C. He, H. Kucuktas, P. Li, L. Chen, M. Simmons, and Z.J. Liu 2004. Differential expression profiles of chemokines in resistant and susceptible catfish after infection of Edwardsiella ictaluri. Plant and Animal Genome XII, San Diego, p713. Simmons M, K. Mickett, P. Li, H. Kucuktas, and Z.J. Liu 2004. Assessing genetic diversity of wild populations of channel catfish (Ictalurus punctatus) using AFLP. Plant and Animal Genome XII, San Diego, p709. Peatman E, X. Wei, J. Feng, L. Liu, H. Kucuktas, P. Li, C. He, D. Rouse, R. Wallace R Dunham, and Z.J. Liu 2004. Development of est resources from eastern oyster (Crassostrea virginica) for identification of marine biomarkers. Plant and Animal Genome XII, San Diego, p734. Liu ZJ. 2004. Aquaculture genomics and National Animal Genome Research Program (NAGRP) NRSP-8: An introduction. WAS Conference, Hawaii. P.357. Liu ZJ, Huseyin Kucuktas, Chongbo He, Puttharat Boaprasertkul, Eric Peatman, Micah Simmons, Liqiao Chen, Jerry Serapion, Ping Li, Benjaporn Somridhivej 2004. Development of genomic resources of catfish for comparative genomic analysis. WAS Conference, Hawaii. P.357. Liu ZJ, 2003. Development of Genome Resouces in Catfish and Use of Microarrays for Analysis of Gene Expression. Abstracts of The 2nd International Symposium on Aquatic Genomics, p.26. Peatman, Eric J., Xianchao Wei, Jinian Feng, Huseyin Kucuktas, Ping Li, Chongbo He, David Rouse, Richard Wallace, Rex Dunham, Zhanjiang Liu. 2003. Development of Expressed Sequence Tags (ESTs) from Eastern Oyster (Crassostrea virginica): Lessons Learned from Previous Efforts. Abstracts of The 2nd International Symposium on Aquatic Genomics, P-18. Karsi A Waldbieser, G., Liu ZJ, and William Wolters. 2003. cDNA Cloning, Phylogenetic Analysis, and Expression of Proopiomelanocortin (POMC) gene from Channel Catfish (Ictalurus punctatus). Plant and Animal Genome XI, San Diego, P652. Attila Karsi , Geoffrey C Waldbieser , Brian Small , Zhanjiang Liu , William R Wolters. 2003. Molecular cloning, characterization, tissue expression pattern, and linkage mapping of proopiomelanocortin (POMC) gene from channel catfish (Ictalurus punctatus). Plant and Animal Genome XI, San Diego, P653. Liu ZJ. Chongbo He, Jerry Serapion, Ping Li, Geoff Waldbieser, and William Wolters 2003 Multiple alternative splicing and polyadenylation of the interleukin-8 gene from channel catfish (Ictalurus punctatus). Plant and Animal Genome XI, San Diego, P.655. 45 117. K. Mickett, M. Simmons, C. Morton, J. Feng, P. Li, R. A. Dunham, D. Cao, and Z.J. Liu 2003. Assessing genetic diversity of domestic populations of channel catfish (Ictalurus punctatus) in Alabama using AFLP markers. Plant and Animal Genome XI, San Diego, P.656. 118. Chongbo He, Liqiao Chen, Ping Li, Huseyin Kucuktas, Soonhag Kim, and Liu ZJ. 2003. Type I Single Nucleotide Polymorphism Markers of Catfish Identified by Comparative EST Analysis. Plant and Animal Genome XI, San Diego, P654. 119. Liu ZJ. & Ping Li 2003. Multiple Tc1-like transposon elements from channel catfish (Ictalurus punctatus). Plant and Animal Genome XI, San Diego, P657. 120. Liu ZJ, 2003. Catfish genomics: where are we going based on the progress of the first five years? Plant and Animal Genome XI, San Diego, W28. 121. Waters, P., R. Beam, C. Ligeon, Z.J. Liu, R. Dunham, G. Warr, A. Nichols, P. Duncan, B Argue, D. Middleton, and H. Kucuktas 2003. Comparison of direct selection, indirect selection, and transgenesis to improve bacterial disease resistance in channel catfish. Catfish Research Symposium, Catfish Farmers of America, Sandestin, Florida, p.19 122. Liu ZJ, Chongbo He, Jerry Serapion, Ping Li, Geoff Waldbieser, and William Wolters 2003. Multiple alternative splicing and polyadenylation of the interleukin-8 gene from channel catfish (Ictalurus punctatus). Catfish Research Symposium, Catfish Farmers of America, Sandestin, Florida, p. 42. 123. Simmons M, K. Mickett, C. Morton, J. Feng, P. Li, R. A. Dunham, D. Cao, and Z.J. Liu 2003. Assessing genetic diversity of domestic populations of channel catfish (Ictalurus punctatus) in Alabama using AFLP markers. Catfish Research Symposium, Catfish Farmers of America, Sandestin, Florida, p. 20. 124. He C, Chen L, Ping Li, Huseyin Kucuktas, Soonhag Kim, and Liu ZJ. 2003. Type I Single Nucleotide Polymorphism Markers of Catfish Identified by Comparative EST Analysis. Catfish Research Symposium, Catfish Farmers of America, Sandestin, Florida, p. 29. 125. Liu ZJ. & Ping Li 2003. Multiple Tc1-like transposon elements from channel catfish (Ictalurus punctatus). Catfish Research Symposium, Catfish Farmers of America, Sandestin, Florida, p. 43. 126. Karsi A, Andrea Patterson, Jinian Feng, and Liu ZJ. 2003. Translational machinery of channel catfish: a transcriptomic approach to the analysis of 32 40s ribosomal protein genes and their expression. Catfish Research Symposium, Catfish Farmers of America, Sandestin, Florida, p. 39. 127. Patterson A, Attila Karsi, Jinian Feng, and Liu ZJ. 2003. Translational machinery of channel catfish: complementary dna and expression of the complete set of 47 60s ribosomal proteins. Catfish Research Symposium, Catfish Farmers of America, Sandestin, Florida, p. 128. Liu ZJ, Attila Karsi, Ping Li, Zhenlin Ju, Arif Kocabas, Dongfeng Cao, Andrea Patterson, Huseyin Kucuktas, Rex Dunham, and Kathryn Mickett 2003. Large scale analysis of catfish genes: initial analysis of 11,000 expressed sequence tags from the channel catfish brain, head kidney, spleen, liver, skin and other tissues.Catfish Research Symposium, Catfish Farmers of America, Sandestin, Florida, p. 41. 129. Liu ZJ, Soonhag Kim, Attila Karsi 2003. Channel catfish follicle stimulating hormone and luteinizing hormone: cdna cloning and their expression during ovulation. Catfish Research Symposium, Catfish Farmers of America, Sandestin, Florida, p.41. 130. Zhenlin Ju, Rex A. Dunham, and Liu ZJ. 2003. Differentially expressed genes in the brain of channel catfish (Ictalurus punctatus) in response to cold acclimation. Catfish Research Symposium, Catfish Farmers of America, Sandestin, Florida, p.37. 46 131. Arif M. Kocabas, Huseyin Kucuktas, Rex A. Dunham, and Liu ZJ. 2003. Molecular characterization and differential expression of the myostatin gene in channel catfish (Ictalurus punctatus). Catfish Research Symposium, Catfish Farmers of America, Sandestin, Florida, p.39. 132. Liu ZJ. 2003. Catfish genomics program at auburn university. Catfish Research Symposium, Catfish Farmers of America, Sandestin, Florida, p.43. 133. Matthew J. Jenny , Gregory W. Warr , Z. J. Liu, Gerardo R. Vasta, Jose A. F. Robledo, Amy H. Ringwood, Paul S. Gross, Eric R. Lacy, Jonas Almeida, Robert W. Chapman. (2002) ECOGENOMICS: Use of Gene Expression Profiles for Evaluating the Relationship between Crassostrea virginica and the Estuarine Environment. An Abstract presented in International Conference on Shellfish Restoration, Charleston, SC. 134. Kocabas A, and Liu ZJ. 2002. Molecular Characterization of the myostatin gene in channel catfish (Ictalurus punctatus): sequence, genomic structure, and expression pattern. Poster presentation in AU Hotel and Conference Center, Graduate Council. 135. Liu, Z. J., Dunham R. (2002) Role of genetics in sustainable aquaculture. Sustainable agriculture in Alabama Symposium, Feb 24-26, 2002, Birmingham, AL. 136. Liu ZJ. (2002) Encyclopedia cDNA libraries of channel catfish and systematic analysis of genes, gene expression, and markers. Plant and Animal Genome X, San Diego, W30. 137. Karsi, A, Patterson A, Feng J, Liu, Z. (2002) Translational machinery of channel catfish: I. a transcriptomic approach to the analysis of 32 40s ribosomal protein genes and their expression. Plant and Animal Genome X, San Diego, p632. 138. Patterson A Karsi A Feng J, and Liu ZJ. (2002) Translational machinery of channel catfish: II. complementary dna and expression of the complete set of 47 60s ribosomal proteins. Plant and Animal Genome X, San Diego, p633. 139. Kocabas A, Dunham R, Liu ZJ. (2002) Comparative genomic analysis of differentially expressed genes in the brain of white catfish in response to cold acclimation. Plant and Animal Genome X, San Diego, p634. 140. Kim S, Kucuktas H, Karsi A, Liu ZJ. (2002) Multiple isoforms and an unusual cathodic isoform of creatine kinase from channel catfish (Ictalurus punctatus). Plant and Animal Genome X, San Diego, p635. 141. Ju Z, Dunham R, and Liu ZJ. (2002) Differentially expressed genes in the brain of channel catfish (Ictalurus punctatus) in response to cold acclimation. Plant and Animal Genome X, San Diego, p636. 142. Kocabas A, Kucuktas H, Dunham R, and Liu ZJ. (2002) Molecular characterization and differential expression of the myostatin gene in channel catfish (Ictalurus punctatus). Plant and Animal Genome X, San Diego, p637. 143. Karsi A, Cao D, Li P, Ju Z Kocabas A, Feng J, Waldbieser, G., Wolters W, and Liu ZJ. (2002) Transcriptome analysis of channel catfish (Ictalurus punctatus): Initial analysis of gene expression and microsatellite-containing cdnas in the skin. Plant and Animal Genome X, San Diego, p638. 144. Kocabas A, Li P, Cao D, Ju Z, Karsi A, Patterson A, Dunham R, and Liu ZJ. (2002) Identification of genes expressed in channel catfish (Ictalurus punctatus) spleen by expressed sequence tags. Plant and Animal Genome X, San Diego, p639. 145. Song, X., Feng J, Wei, X., Li P, He,C., Wallace R, Dunham R, Rouse D, and Liu ZJ. (2002) Mitochondrial genes are expressed at unusually high levels in the gill of oysters (Crassostrea virginica) as analyzed by expressed sequence tags. Plant and Animal Genome X, San Diego, p640. 47 146. Kocabas A, Li P Kim S, Karsi A and Liu Z. J. (2001) Isolation and characterization of gdf8 (myostatin) gene from channel catfish (Ictalurus punctatus). Plant and Animal Genome IX, San Diego, p621. 147. Karsi,A., Moav,B., Hackett, P., Liu , Z. J. (2001) Effects of insert size on transposition efficiency of the sleeping beauty transposon in mouse cells. Plant and Animal Genome IX, San Diego, p626. 148. Karsi A, Li P, Kocabas A, Ju Z, Mickett, K., Silvernail, T., Feng J,Cao D, Dunham R, and Liu ZJ. (2001) Production of transgenic channel catfish using the synthetic transposon system sleeping beauty. Plant and Animal Genome IX, San Diego, p618. 149. Cao D, Kocabas A, Ju Z, Karsi A, Li P, Patterson A, Feng J, and Liu ZJ. (2001) transcriptome analysis of channel catfish (Ictalurus punctatus): gene cataloguing and profiling from the head kidney. Plant and Animal Genome IX, San Diego, p620. 150. Liu ZJ. (2001) Catfish genomics: past accomplishments and challenges ahead. Plant and Animal Genome IX, San Diego, W17. 151. Kocabas A, Feng J, Patterson A, Cao D, Li P, Ju Z, Karsi A, and Liu ZJ. (2001) Comparative EST analysis and single nucleotide polymorphism identified by digital hybridization from channel catfish and blue catfish. Plant and Animal Genome IX, San Diego, p619. 152. Ju Z, Karsi A, Kocabas A, Patterson A, Li P, Cao D, Dunham R, and Liu ZJ. (2001) Transcriptome analysis of channel catfish (Ictalurus punctatus): genes and expression profile from the brain. Plant and Animal Genome IX, San Diego, p622. 153. Liu ZJ. (2000) DNA marker technologies, gene mapping, marker-assisted selection, gene cloning, genetic engineering, and integrated genetic improvement programs at Auburn University. A Compilation of Abstracts of Research on Channel Catfish, Catfish Research Forum February 19, 2000, Albuquerque, New Mexico, p.16-17. 154. Kim S, Li P, Zheng, X., Karsi A, Dunham R A., and Liu ZJ. (2000) Gene expression in the muscles of young and mature channel catfish (Ictalurus punctatus) as analyzed by expressed sequence tags and gene filters. Plant and Animal Genome VIII p426. 155. Kim S, Dunham RA., and Liu ZJ. (2000) The skeletal muscle alpha actin gene of channel catfish (Ictalurus punctatus) is associated with two classes of SINE elements. Plant and Animal Genome VIII p424. 156. Kim S, Li P, Zheng, X., Karsi A, Dunham R A., and Liu ZJ. (2000) Gene expression in the muscles of young and mature channel catfish (Ictalurus punctatus) as analyzed by expressed sequence tags and gene filters. A Compilation of Abstracts of Research on Channel Catfish, Catfish Research Forum February 19, 2000, Albuquerque, New Mexico, p.17-18. 157. Kim S, Dunham RA., and Liu ZJ. (2000) The skeletal muscle alpha actin gene of channel catfish (Ictalurus punctatus) is associated with two classes of SINE elements. A Compilation of Abstracts of Research on Channel Catfish, Catfish Research Forum February 19, 2000, Albuquerque, New Mexico, p.18. 158. Yant R, Li P, Kim S, Chatakondi, N., Li, M., Robinson, E., Dunham R, Kucuktas H, and Liu ZJ. (2000) DNA markers linked to feed conversion efficiency and growth rate for marker-assisted selection. A Compilation of Abstracts of Research on Channel Catfish, Catfish Research Forum February 19, 2000, Albuquerque, New Mexico, p.19. 159. Karsi A Kim S, Dunham RA., and Liu ZJ. (2000) Performance traits-linked DNA markers and marker-assisted selection. Plant and Animal Genome VIII p425. 48 160. Li P, Karsi A, Cao D, Ju Z, Kocabas A, Mickett, K., Kim S, Kucuktas H, Dunham R, and Liu ZJ. (2000) An AFLP-based catfish genetic linkage map and QTL detection for growth rate and disease resistance. Plant and Animal Genome VIII p423. 161. Liu ZJ. (2000) Functional genomics: performance traits-linked DNA markers and normalized cDNA libraries in catfish. Plant and Animal Genome VIII p.9 162. Karsi A, Dunham R, and Liu ZJ. (1999) Development of polymorphic EST markers suitable for genetic linkage mapping of catfish. Plant & Animal Genome VII, page 109, Scherago International, Inc., New York, NY. 163. Liu ZJ, Karsi A, Kucuktas H, and Dunham R.(1999) Transcribed dinucleotide microsatellites Development of polymorphic EST markers suitable for genetic linkage mapping of catfish. Plant & Animal Genome VII, #536, Scherago International, Inc., New York, NY. 164. Liu ZJ, Karsi A, Li P, and Dunham R.(1999)Transcriptional activities in the pituitaries of channel catfish before and after induced ovulation by injection of carp pituitary extract as revealed by expressed sequence tag analysis. Plant & Animal Genome VII, page 209, Scherago International, Inc., New York, NY. 165. Liu ZJ, and Dunham R. (1999) Catfish gene mapping, progress and perspectives. International Plant and Animal Genome Conference VII, January 17-21, 1999, San Diego. 166. Liu ZJ, and Dunham R. (1998) Progress in catfish gene mapping. International Plant and Animal Genome Conference VI, January 13-17, 1998, San Diego. 167. Liu ZJ, Li P, Argue B, and Dunham R. (1998) Inheritance of RAPD markers in channel catfish, blue catfish, and their F1, F2, and backcross hybrids. International Plant and Animal Genome Conference VI, January 13-17, 1998, San Diego. 168. Liu ZJ, Tan G, A., Karsi A, Kucuktas H, Li P, Zheng X, Kim S, and Dunham R. (1998) Conservation of microsatellite loci between channel catfish and blue catfish. International Plant and Animal Genome Conference VI, January 13-17, 1998, San Diego. 169. Liu ZJ, Nichols A, Tan G, Kucuktas H, Li P, and Dunham R. (1998) Inheritance of AFLP markers in channel catfish, blue catfish, and their hybrids. International Plant and Animal Genome Conference VI, January 13-17, 1998, San Diego. 170. Liu ZJ, Li P, Argue B, and Dunham R.A. (1997) Molecular cloning and expression of gonadotropin alpha subunit glycoproteins in channel catfish during carp pituitary-induced ovulation. 1997 Catfish Culture Research Symposium. Feb. 28, 1997. Nashville, TN. In: Research and Review, A Compilation of Abstracts of Research on Channel Catfish, Page 28. 171. Liu ZJ, Li P, and Dunham R.A. (1997) Characterization of an A/T-rich family of satellite sequences from channel catfish (Ictalurus punctatus). 1997 Catfish Culture Research Symposium. Feb. 28, 1997. Nashville, TN. In: Research and Review, A Compilation of Abstracts of Research on Channel Catfish, Page 27. 172. Yan, L., Liu ZJ, and Faras A. (1994) Effects of endogenous long terminal repeats on expression of cellular genes in line-0 chickens. Retroviruses Conference, Cold Spring Harbor, New York, May 24-29, 1994. 173. Liu ZJ, Yan, L., Wendell, S., Faras A. (1994) Molecular analysis of complexities and promoter activities of the endogenous EAV-related retroviral long terminal repeats. Retroviruses Conference, Cold Spring Harbor, New York, May 24-29, 1994. 174. Lee, H., Liu ZJ, and Faras A. J. (1993) Molecular analysis of the retroviral reverse transcriptase-related sequences in ev- chickens. Retroviruses Meeting, Cold Spring Harbor, New York, May 25-May 31, 1993. 49 175. Liu ZJ, Ostrow R, Ghai J and Faras A. (1993) The expression levels of HPV-16 E6 and E7 determines the immortalization or transformation phenotypes of BRK cells. 12th International Papillomavirus Conference, Baltimore, MD. 176. Ghai J, Liu ZJ, Ostrow R, McGlennen R, and Faras A. (1993) The E5 ORF of rhesus papillomavirus is an activator of endogenous ras in NIH 3T3 cells. 12th International Papillomavirus Conference, Baltimore, MD. 177. Bushnell W, Clark, T., Liu ZJ, Smith, A., Szabo L, and Zeyen, R. (1992) Host response gene expression in powdery mildew of barley. Extended Abstracts for the Eighth Mediterranean and European Cereal Rusts and Mildews Conference, Sept. 8-11, 1992, Freising, Germany. 178. Hackett, P., Liu ZJ, Chaldovic, L., Kapuscinski A, Faras A., and Moav B (1991) Regulation of early transgene expression in genetically engineered fish. XVII Pacific Science Congress, May 27-June 2, 1991. 179. Hackett, P., Liu ZJ, Chaldovic, L., Kapuscinski A, Faras A., and Moav B. (1991) Regulation of transgene expression in genetically engineered fish. 2nd Int. Biotechnology Conference, Baltimore, MD, October 1991. 180. Liu ZJ, Bushnell W, and Szabo L (1991) Cloning and analysis of abundant mRNA from Puccinia graminis. Phytopathology 81:1157. 181. Bushnell W, and Liu ZJ. (1991) Time of treatment with cordycepin in relation to inhibition of hypersensitive response in powdery mildew of barley. Plant Molecular Biology Congress. Tuscon. 182. Bushnell W, and Liu ZJ. (1991) Time of treatment with cycloheximide in relation to the hypersensitive response in powdery mildew of barley. In: Molecular Strategies of Pathogens and Host Plants (S. Patil, S. Ouchi, D., Mills and C. Vance, eds.), Springer Verlag, New York p. 258. 183. Guise, K., Hackett, P., Liu ZJ. (1990) Genetic engineering by gene transfer in fish. 4th University of Minnesota Research Poster Session, Minneapolis. 184. Schneider J, Hallerman E, Yoon, S.J., He L, Gross M, Liu ZJ, Faras A., Hackett, P., Kapuscinski A, and Guise, K. (1989) Transfer of the bovine growth hormone gene into northern pike, Esox lucius. J. Cell. Biochem. Suppl. 13B, 173. 185. Guise, K., Yoon, S.J., Liu ZJ, Kapuscinski A, Hackett, P., and Faras A. (1988) Transfer of neomycin resistance gene in fish. First International Symposium on Fish Endocrinology, Edmonton, Canada. 186. Yoon, S.J., Liu ZJ, Kapuscinski A, Hackett, P., Faras A., and Guise, K. (1988) Gene transfer by microinjection in fish. 3rd International Congress on Genetics in Aquaculture, Norway. 187. Yoon, S.J., Liu ZJ, Kapuscinski A, Hackett, P., Faras A., and Guise, K. (1988) Successful gene transfer in fish. UCLA Symposia on gene transfer in animals. J. Cell. Biochem., Suppl. 12B, 190. 188. Yoon, S.J., Liu ZJ, Kapuscinski A, Hackett, P., Faras A., and K. Guise (1988) Transfer of marker genes into goldfish. Genome 30, suppl. 1, 448. XVIth International Congress of Genetics, Toronto, Canada. 189. Guise, K., Schneider J, Hallerman E, Yoon, S.J., He L, Gross M, Liu ZJ, Zhu Z, Faras A., Hackett, P., and Kapuscinski A (1988) Transfer of neomycin resistance and growth hormone genes into goldfish and northern pike. 1st International Symposium on Marine Molecular Biology, Baltimore, MD. 190. Schneider J, Hallerman E, Yoon, S.J., He L, Myster, S., Gross M, Liu ZJ, Zhu Z, Hackett, P., Guise, K., Kapuscinski A, and Faras A. (1988) Microinjection and successful transfer of 50 bovine growth hormone gene into northern pike, Esox lucius. 50th Midwest Fish and Wildlife Conference, Columbus, OH. 191. Bushnell WR., Liu ZJ, and K. Mendgen (1986) Cyanine potentiometric probes for mitochondria of Erysiphe graminis. Phytopathol. 76, 1123. 192. Liu ZJ, Bushnell W R., and B. Brambl (1986) Monitoring mitochondria in plant cells with a potentiometric fluorescent probe: enhanced response with kinetin. VI International Congress of Plant Tissue and Cell Culture, page 369. Invited Lectures and Presentations 2015 2015 2015 2015 2015 2015 2015 Genomic advances and applications in aquaculture. Invited lecture, Department of Biological Sciences, Auburn University, October 2015. Genomic advances and applications in aquaculture. Invited lecture, Hunan Normal University, Changsha, China. November 2015. Catfish genomics and potential applications in aquaculture. Invited lecture, Hunan Agricultural University, Changsha, China. November 2015. Catfish genomics and potential applications in aquaculture. Invited lecture, Changsha University, Changsha, China. November 2015. Genomic advances and applications in aquaculture. Keynote lecture, ISGA XII Congress, Santiago de Compostela, June 21-26, 2015. Whole genome sequencing and annotation in catfish. Invited lecture, Central Institute of Freshwater Aquaculture (CIFA), Bhubaneswar, India, January 20, 2015. Analysis of performance traits using genomic approaches. Invited lecture, Central Institute of Freshwater Aquaculture (CIFA), Bhubaneswar, India, January 20, 2015. 2015 Trend of genomic sciences and their application in aquaculture. Invited lecture, Central Institute of Fisheries Education (CIFE), Mumbai, India, January 16, 2015. 2015 Enabling the Catfish Genome Sequences for Selection through Enhanced Scaffolding, Transcriptome Sequencing, & Analysis of Genetic Variation. PI Meeting, San Diego, January 9, 2015. 2014 Study of performance traits: Genomics made it easy, Invited Lecture, National Chengkong University, Taiwan, June 8, 2014. 2014 Genomic approaches to the understanding of disease resistance and low oxygen tolerance. Next Generation Genome View of Plants, Animals and Microbes, March 5-7, Singapore 2014 2013 Genomics approaches to the understanding of performance and production traits in catfish, Invited seminar, South China Sea Fisheries Research Institute, Guangzhou, October 25, 2013. 2013 Genomics approaches to the understanding of performance and production traits in catfish, Invited seminar, Pearl River Fisheries Research Institute, Guangzhou, October 25, 2013. 2013 Genomics approaches to the understanding of performance and production traits in catfish, Keynote for Aquatic Genomics Session, International Conference of Genomics. Qingdao, China, October 29, 2013. 2013 Genomics approaches to the understanding of disease resistance and low oxygen tolerance in catfish, Keynote for Fish Physiology Symposium, Across the Straight Symposium Series IIIV, Shanghai Ocean University, China, November 2, 2013. 51 2013 2013 2013 2013 2013 2013 2012 2012 2012 2012 2012 2012 2012 2012 2011 2011 2011 2011 2011 2011 2011 Genomics approaches to the understanding of disease resistance and innate immunity in catfish, Keynote lecture for the First International Fish and Shellfish Immunology, June 26, Vago, Spain. Genomics approaches to the understanding of disease resistance and innate immunity in catfish, Invited lecture, June 11, 2013, Dalian Ocean University, Dalian, China. Genomics approaches to the understanding of disease resistance and innate immunity in catfish, Invited lecture, June 4, 2013, Huazhong Agricultural University, Wuhan, China. Genomics approaches to the understanding of disease resistance and innate immunity in catfish, Invited lecture, June 7, 2013, Northwest A&F University, Yangling, China. Genomics approaches to the understanding of disease resistance and innate immunity in catfish, Invited lecture, June 1, 2013, Southwest University, Chongqing, China. Genomics approaches to the understanding of disease resistance and innate immunity in catfish, Invited lecture, June 1, 2013, Southwest University, Chongqing, China. Aquaculture genomics research at Auburn University, seminar at Makerere University, Kampala, Uganda. Aquaculture genomics and genome technologies for genetic improvement of aquaculture, invited lecture at Symposium for Aquaculture Genetics, Beijing, China. Challenges of genome era aquaculture genetics research, invited lecture, June 29, 2012, at the International Symposium of Aquaculture Genetics, Auburn, AL USA. Challenges of aquaculture genetics research in post-genome era. Invited lecture, October 18, at Qingdao Agricultural University, Qingdao, China. Catfish Genome Sequencing, invited lecture at Yellow Sea Fisheries, October 17, 2012, Qingdao, China. Transcriptome of channel catfish and its applications to genome research and genetic enhancement programs. The conference was to allow international scientists to communicate their research discoveries in aquaculture., Prague, Czech Republic, September 1-6, 2012. Genome infrastructure for resolving major challenges facing the catfish aquaculture industry. Invited lecture, AU-OUC Joint Symposium. Genome technologies in aquaculture, invited lecture, June 9, 2012, Shanghai Ocean University. Challenges of genome era aquaculture genetics research, Invited lecture, Hudson/Alpha Institute for Biotechnology, Huntsville, Alabama, October 25, 2011. Challenges of Aquaculture Genetics Research in the Post Genome Era. Plenary Session Keynote lecture, 2011 Forum on Fisheries Science and Technology, Beijing, China, July 15-18, 2011. Challenges of Aquaculture Genetics Research in the Post Genome Era. Plenary Session Keynote lecture, International Symposium of Genomics in Aquaculture, Heraklion, Crete, Greece. September 14-18, 2011. Genome resources for efficient genome assembly and annotation in catfish, Shanghai Ocean University, Shanghai, China, May 18, 2011. Strategies for efficient genome assembly and annotation, Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China, May 16, 2011. Liu, Z.J. How to write SCI papers: Special Considerations for Chinese scientists, College of Animal Sciences, Northwest A&F University, Yangling, China, May 13, 2011. Liu, Z.J. Strategies for the application of whole genome sequences for improving production and performance traits in catfish. Keynote lecture, Functional Genomics Session, World Aquaculture Society Conference, Natal, Brazil, June 6-10, 2011. 52 2011 2011 2011 2010 2010 2010 2010 2010 2010 2010 2009 2009 2009 2008 2008 2008 2008 2008 2007 Liu, Z.J. Strategies for efficient assembly and annotation of the catfish whole genome sequence. Finfish Genetics Session, World Aquaculture Society Conference, Natal, Brazil, June 6-10, 2011. Whole genome sequencing of catfish and potential uses of genome information for improving performance traits. World Aquaculture Society Conference, Genomics Session, February 28-March 2, 2011, New Orleans, USA. “Strategies for efficient assembly and annotation of the whole genome sequence of catfish”, Plenary lecture presented in Aquaculture Workshop, International Conference of Plant and Animal Genome Mapping XIX, January 14-19, 2010, San Diego, CA. “Trend of Aquaculture Genomics”, Hainan University, Haikou, China. “Strategies for efficient assembly and annotation of the catfish whole genome sequence”, Chinese Academy of Fisheries Science, Beijing, and televised remotely to all research institutes of the academy. “Strategies for efficient assembly and annotation of the catfish whole genome sequence”, International Workshop on Marine and Aquaculture Genomics, November 16-17, 2010, Shenzhen, China. Honors seminar to STEM students and faculty, “Catfish Genome Technologies and Applications to Aquaculture”, Alabama State University. Keynote lecture, “Genome research of catfish: from ground zero to whole genome sequencing”, Annual Graduate Symposium, Department of Animal Science, University of Maryland. “Genome research trends for aquaculture species: from ground zero to whole genome sequencing”, Shanghai Ocean University, China. “Genome research of catfish and related aquaculture species: from ground zero to whole genome sequencing”, Chinese Academy of Fisheries Science, Beijing, and televised remotely to all research institutes of the academy. “Catfish genomics: does all the hard work in the past means anything?” The Genomics in Aquaculture Symposium, Bodø University College, Bodø, Norway. “Going with trend or falling behind: Aquaculture genomics at a crossroad”, keynote lecture, International Symposium on Genetics in Aquaculture (ISGA 2009), Bangkok, Thailand. “Aquaculture functional genomics: focus, trend and challenges ahead”, keynote lecture, World Aquaculture Society Conference, Veracruz, Mexico. Invited lecture, “Monitoring phenotypes under water: Challenges and possibilities ahead. Oct 26-29, 2008. Allerton Conference IV, Confronting Phenotypes, University of Illinois, Champaign-Urbana, IL. “Motivating for grantsmanship: A faculty and an administrator’s perspective”, presentation made in the session of “Best Practices: Motivating faculty for grants, coaching, and post award management” in the South Agricultural Experiment Station Directors Meeting, Mach 31-April 2, 2008, Knoxville, TN. “Healthy aquaculture: the genomic approaches”, keynote lecture, AU-OUC Joint Symposium, Healthy Aquaculture, Qingdao, China. “Status and trends of aquaculture genomics” keynote lecture, World Aquaculture Society, Pusan, Korea. “Genomic approaches for the identification of immune responsors: you will not miss any guard”, keynote lecture, the 5th World Fisheries Congress, Yokohama, Japan, "Status and challenges of aquaculture genomics: Making reactions work without reagents". Invited lecture for the Conference of “Convergence of Genomics and the Land 53 2007 2007 2007 2007 2007 2006 2006 2006 2006 2006 2005 2005 2005 2005 2005 2005 2005 2005 2005 2004 2004 2004 Grant Mission: Emerging Trends in the Application of Genomics in Agricultural Research”, Purdue University, West Lafayette, Indiana, September 10-12, 2007. “The status of aquaculture genomics: Are the waves of the genomics revolution moving anything under the water?” Invited lecture for Intermountain Systems Biology Symposium, Logan, Utah, June 5-7, 2007. “Status and trend of aquaculture genomics: challenges ahead”, seminar given on November 26, 2007 at Kentucky State University, Frankfort, Kentucky. “How to write a scientific paper?” Shanghai Fisheries University, China. “DNA marker technologies and their application in fish breeding”, Aquaculture Research Institute of Jiangsu Province, Nanjing, China. “Trend of aquaculture genomics”, Oceanic Institute, Chinese Academy of Sciences, Qingdao, China. “status and trends in genomics and modern genetic technologies in aquaculture and fisheries”, the CAS Conference on Marine Science and Technology, Qingdao, China. “Catfish Genomics, achievements and challenges”, Human Normal University, Changsha, China. “Aquaculture genomics for genetic improvement in aquaculture”, Plenary Session keynote lecture, the International Aquaculture Genetics Conference (IAGA IX), Montpellier, France. “Aquaculture genomics: It is both aquaculture and genomics”, International Workshop on “Status and trends in aquatic genetic resources: a basis for international policy” Victoria, British Colombia, FAO of the United Nations. “A glance at catfish genomics”, invited lecture given at Simon Fraser University, Vancouver, Canada. “Pre-genome research of catfish in post-genome era”, invited lecture in the Department of Biological Sciences, University of Wisconsin, Milwaukee. “EST analysis, why such a tedious approach is optimal for fish genome studies?” invited lecture at the Great Lakes Water Institute, University of Wisconsin, Milwaukee. “Genomic studies of catfish using channel catfish x blue catfish interspecific hybrid system.” World Aquaculture Society Aquaculture America 2005. New Orleans, January 18, 2005. “Innate immunity and chemokine diversity in catfish”, the International Conference of Marine Biotechnology, New Foundland, Canada. “Functional genomics platform development”, Yellow Sea Fisheries Research Institute, Qingdao, China. “Catfish Genome Research”, Dalian Academy of Fisheries Sciences, Dalian, China. “Catfish genomics”, Helongjiang Fisheries Research Institute, Harbin, China. “Aquaculture genomics in the United States”, Shanxi Agricultural University, China. “Catfish genomics”, Institute of Hydrobiology, the Chinese Academy of Sciences, Wuhan, China. “Functional genomics: a key research area in 21st century”, Yellow Sea Fisheries Research Institute in conjunction with the establishment of the Sino-American Joint Laboratory of Fish Functional Genomics, Qingdao, China. “Marking the genome: DNA marker technologies”, keynote lecture, the Workshop of Application of Genome and Related Technologies in Aquatic Sciences, Shanghai, China. “Mapping the genome: what is best for my species?” Shanghai Fisheries University, China, for the Workshop of Application of Genome and Related Technologies in Aquatic Sciences. 54 2004 2004 2004 2004 2004 2004 2003 2003 2003 2003 2003 2003 2003 2002 2002 2002 2002 2002 2002 2002 2002 2002 “Physical mapping of genomes”, Shanghai Fisheries University, China, for the Workshop of Application of Genome and Related Technologies in Aquatic Sciences. “Are aquaculture geneticists ready for functional genomics yet?” Shanghai Fisheries University, China, for the Workshop of Application of Genome and Related Technologies in Aquatic Sciences. “Genome research in catfish”, China Oceanic University, Qingdao, China. 30,000 catfish ESTs: a case study for a set of quality standards established for the analysis of ESTs. Plant and Animal Genome XII, San Diego, W22. “Development of genomic resources of catfish”, presented in Aquaculture 2004, World Aquaculture Society Annual Conference, Honolulu, Hawaii. Mapping the catfish genome: DNA marker technologies, linkage mapping, and functional analysis. AAAS Symposium: Aquaculture: Recent Advances in Fish Culture, Breeding and the Mitigation of Environmental Impact. Seattle. “Catfish genomics: where are we going based on the progress of the first five years?” presented in Aquaculture Workshop, International Conference of Plant and Animal Genome Mapping XI, January 10-14, 2003, San Diego, CA. “Structural genomics: strategic ways of thinking for dissection of a genome”, Shanghai Fisheries University, China. "Genomics revolution: its impact in aquaculture”, Oceanic Institute, Chinese Academy of Sciences, Qingdao, China. "Functional genomics: revealing functions of genes at the genome level", Shanghai Fisheries University, China. "Genomics revolution: its impact on aquaculture”, East China Normal University, Shanghai, China. "Genomics revolution: its impact on aquaculture”, Yellow Sea Fisheries Research Institute, Qingdao, China. “Development of genome resources in catfish and use of microarray for analysis of gene expression”, the Second Aquatic Genome Symposium, Tokyo, Japan. “DNA markers and their applications in aquaculture and fisheries”, ICLARM, ABBASA Office, Abbasa, Egypt. “Genomics, a rapidly expanding branch of science”, Xian Jiaotong University, Xian, China. “QTL mapping and application of genomics in agriculture”, Northwest Sci-Tech University of Agriculture and Forestry, Yangling, China. “Functional genomics”, Sichuan University, Chengdu, China. “Catfish genomics: DNA marker technologies, linkage mapping, and functional analysis”, Aquaculture Genomics Session, World Aquaculture Society Annual Conference, Beijing, China. “Role of genetics in sustainable aquaculture” presented at the Sustainable agriculture in Alabama Symposium, Feb 24-26, 2002, Birmingham, AL. “Encyclopedia cDNA libraries of channel catfish and systematic analysis of genes, gene expression, and markers”, presented in Aquaculture Workshop, International Conference of Plant and Animal Genome Mapping X, January 11-16, 2002, San Diego, CA. “Genomic research in fish ponds: from markers to functional genomics”, invited lecture at University of Alabama-Birmingham Medical School. “Catfish Genomics: from markers to functional genomics”, seminar given in the Department of Biological Sciences, College of Science and Mathematics, Auburn University. 55 2001 2001 2001 2001 2001 2001 2001 2000 2000 2000 1999 1999 1999 1999 1999 1999 1999 1999 1999 1999 1998 1998 1998 "Catfish Genomics: achievements from the past and challenges in the future", presented in Aquaculture Workshop, International Conference of Plant and Animal Genome Mapping IX, January 13-17, 2001, San Diego, CA. "Catfish genomics, from DNA markers to functional genomics", invited lecture at Oklahoma State University. “Footsteps of catfish genomics, a short story”, invited lecture at University of New Hampshire. "Footsteps of catfish genomics", Tel Aviv University, Tel Aviv, Israel. "Functional genomics of catfish", Galilee Research Institute, Israel. "Catfish genomics, from structural genomics to functional genomics", the National Center of Mariculture, Eilat, Israel. "Genome research and its impact on aquaculture", invited lecture for INGA, Hanoi, Vietnam. “Transgenic fish: its potential and problems for the future as a food source”, Sichuan Province Fisheries Bureau, Chengdu, China. Gave invited lecture “Gene mapping, marker-assisted selection, gene cloning, and biotechnology programs at Auburn University", Albuquerque, New Mexico. “National progress in catfish gene mapping and marker-assisted selection”, presented in Aquaculture Workshop, International Conference of Plant and Animal Genome Mapping VIII, January 8-13, 2000, San Diego, CA. “Catfish genomics: from DNA markers to whole genome functionality”, USDA ARS Catfish genetics Unit. “Catfish Gene Mapping, Progress and Perspectives”, Presentation in Aquaculture Workshop, International Conference of Plant and Animal Genome Mapping VII, January 17-21, 1999, San Diego, California. “Genomics: where is it going?” Invited lecture in Sino-American Biotechnology Conference, Yangling, Shannxi, China. “Modern Concept of Agriculture and Agricultural Technologies”, the City of Yanan. “The American Political System: Perspectives and Potential Application in China”, Wuqi County. “DNA marker technologies”, Northwestern Forestry University, Shaanxi, China. “Transgenic fish research: from traditional approaches to stem cell technology”, Northwestern Agricultural University, Shaanxi, China. “Genomics: Its today and tomorrow”, China Agricultural University. “Aquaculture genomics, resource analysis, and marker-assisted selection”, Northwestern University, Xian, China. “Gene mapping, marker-assisted selection, genetic engineering, and integrated genetic improvement programs at Auburn University” The fifth International Network on Genetics in Aquaculture (INGA), University of Malaya, Kuala Lumpur, Malaysia. “Progress in Catfish Gene Mapping”, Presentation in Aquaculture Workshop, International Conference of Plant and Animal Genome Mapping VI, January 13-17, San Diego, California. “From DNA markers to functional genomics”, seminar given at the Repro Forum, Auburn University. “Hormonal changes during induced ovulation in channel catfish”, seminar given at the Repro Forum, Auburn University. 56 1997 1997 1997 1997 1997 1996 1994 1993 1991 1990 1989 1986 “New Technologies and Biotechnology Industry”, Invited lecture at Genovus Inc. Plymouth, Minnesota. “Transgenic fish technology: its past, present, and the future.” Invited lecture at the Center of Marine Biotechnology, Baltimore, MD. “Molecular cloning of an A/T-rich family of sequences from channel catfish”. Catfish Research Symposium, Nashville, TN. “Transgenic channel catfish using growth hormone gene constructs made from the "allfish" expression vectors. Catfish Research Symposium, Nashville, TN. “Catfish molecular genetics research programs”, seminar given at the Repro Forum, Auburn University. “Transgenic catfish research: where the genes are to be from?” Invited lecture at the University of Minnesota, St. Paul, MN. “Molecular analysis of promoter complexities of the endogenous retroviruses in EVchickens”. Presented at the Cold Spring Harbor Retroviruses Meeting. “Biotechnology: current status and its future”. Beijing Agricultural University, China. “Application of molecular biology in plant pathology”. Noble Foundation, Ardmore, OK. “Development of expression vectors for transgenic fish”. Seminar, Department of Genetics and Cell Biology, University of Minnesota. “Reverse cloning, a novel approach for generation of clones suitable for DNA sequencing”. Seminar, Institute of Human Genetics, University of Minnesota. “Application of molecular techniques in plant pathology”. Beijing Agricultural University, Beijing, China. 57