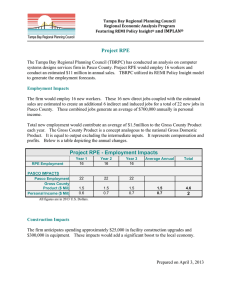
Session 517 RPE Pathology
advertisement

ARVO 2016 Annual Meeting Abstracts 517 RPE Pathology Thursday, May 05, 2016 8:00 AM–9:45 AM Exhibit/Poster Hall Poster Session Program #/Board # Range: 6027–6055/D0246–D0274 Organizing Section: Retinal Cell Biology Program Number: 6027 Poster Board Number: D0246 Presentation Time: 8:00 AM–9:45 AM Nuclear factor-erythroid 2-related factor-2 (Nrf2) and peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) deficient mice: Dissecting the antioxidant regulation pathways affects retinal autophagy Jussi J. Paterno1, 2, Adrian Smedowski2, 3, Maria Hytti2, 4, Marialaura Amandio5, Anna-Kaisa Ruotsalainen6, Henri O. Leinonen6, Heikki Tanila6, Anna-Liisa Levonen6, Pasi Tavi6, Anu Kauppinen1, 4, Kai Kaarniranta1, 2. 1Department of Ophthalmology, Kuopio University Hospital, Kuopio, Finland; 2 Department of Ophthalmology, University of Eastern Finland, Kuopio, Finland; 3Department of Physiology, Medical University of Silesia, Katowice, Poland; 4School of Pharmacy, University of Eastern Finland, Kuopio, Finland; 5Department of Drug Sciences, University of Pavia, Pavia, Italy; 6A.I. Virtanen Institute of Molecular Sciences, University of Eastern Finland, Kuopio, Finland. Purpose: Autophagic clearance participates in the regulation of proteostasis in the retina. Here, we describe a mouse model, deficient in two major transcriptional factors regulating antioxidant defense response, that mimic autophagy decline in retinal pigment epithelium (RPE). Methods: Nrf2–/– PGC-1α–/– double knock-out (DKO) mice and age-matched wild-type controls on a C57BL/6J background were housed in groups on a 12-hour light/dark cycle and given food and water ad libitum. All procedures were in compliance with the ARVO statement for the Use of Animals in Ophthalmic and Vision Research and approved by the Finnish National Animal Experiment Board. Optical coherence tomography (OCT) of retina and darkadapted electroretinogram (ERG) were recorded in young adult (3.5 to 4.5 months old) and aged (12 to 13.5 months old) DKO mice and controls. Retinal tissue sections were immunostained with primary antibodies against 4-hydroxynonenal (4-HNE), ubiquitin, human antigen R (HuR, embryonic lethal abnormal vision (ELAV)-like protein 1), sequestosome 1 (SQSTM1, nucleoporin p62), beclin-1, and microtubule-associated proteins 1A/1B light chain 3B (LC3). Results: Aged Nrf2/PGC1α–/– mice developed RPE degeneration, associated with upregulation of oxidative stress (4-HNE), protein aggregation (ubiquitin), selective autophagy of aggregate-prone proteins (HuR, p62) and overall autophagy (beclin-1, LC3). These changes were accompanied by impaired visual function as assessed by ERG. Young adult DKO (n = 6) and control (n = 6) mice did not differ in their a- and b-wave amplitudes (p > 0.05), but among the aged mice both the a- and b-wave amplitudes were smaller in DKO (n = 7) compared to control mice (n = 7, a-wave: p < 0.05; b-wave: p < 0.01). Conclusions: Our findings suggest that combined Nrf2 and PGC-1α deficiency increases oxidative stress and protein aggregation, and affects autophagy. This coincides with the retinal degeneration and decreased visual function observed in this promising animal model of age-related retinal degeneration. Commercial Relationships: Jussi J. Paterno; Adrian Smedowski, None; Maria Hytti, None; Marialaura Amandio, None; Anna-Kaisa Ruotsalainen, None; Henri O. Leinonen, None; Heikki Tanila, None; Anna-Liisa Levonen, None; Pasi Tavi, None; Anu Kauppinen, None; Kai Kaarniranta, None Support: Finnish Eye Foundation Grant Program Number: 6028 Poster Board Number: D0247 Presentation Time: 8:00 AM–9:45 AM Exogenous claudin-3 and claudin-19 rescue ARPE19 from dedifferentiation SHAO-BIN WANG1, Shaomin Peng1, 2, Deepti Singh1, Ron A. Adelman1, Bo Chen1, Lawrence J. Rizzolo1. 1 Surgery&Ophthalmology, YALE UNIVERSITY, NEW HAVEN, CT; 2 Aier School of Ophthalmology, Central South University, Changsha, China. Purpose: The ARPE19 cell line gradually loses RPE properties with cell passage. One potential cause is that ARPE19 fails to uniformly express the major claudins found in native RPE tight junctions. As tight junctions regulate cell shape, morphology and gene expression, we explored whether exogenous expression of claudin-3 or claudin-19 might restore native RPE properties. Methods: ARPE19 were transduced with adenoviral vectors that expressed claudin-3, claudin-19, or as a control, green fluorescent protein (GFP). We used the transepithelial electrical resistance (TER) to assess barrier properties; RT-PCR arrays of RPE markers or junctional proteins to assess gene expression; and immunoblotting and immunofluorescence to assess protein expression. Comparisons were made with human fetal RPE (hfRPE). A scratch assay and inhibitors of EGF-stimulated pathways were used to evaluate wound-healing. Results: PCR arrays revealed substantial quantitative differences between hfRPE and ARPE19 with respect to RPE signature/ maturation genes. Exogenous claudin-3 and claudin-19 increased the TER and the expression of ~50% of the genes that were underexpressed in ARPE19. The claudins had little effect on the expression of other tight, adherens, and gap junction proteins. Expression of the EGF receptor was decreased, which prompted us to examine wound-healing. In ARPE19, EGF stimulated cell proliferation and wound-healing using the AKT arm of the EGF-signaling pathway. In contrast, wound-healing was retarded in claudin-expressing ARPE19 and was insensitive to EGF. Instead, AKT was constitutively activated. Wound-healing was more epithelial-like in that wounds were slowly closed by a combination of cell spreading and cell proliferation. Conclusions: Directly or indirectly, claudin-3 and -19 effected changes in gene expression that partially restored a native RPE phenotype. The results suggest claudins and AKT signaling might be therapeutic targets for treating proliferative vitreoretinopathy. Commercial Relationships: SHAO-BIN WANG, None; Shaomin Peng, None; Deepti Singh, None; Ron A. Adelman, None; Bo Chen, None; Lawrence J. Rizzolo, None Support: Dept. of Defense MR130036; CT Regen Med Rsch Fund 14-SCB-Yale-18; Leir Family Fund; Alonzo Family Fund Program Number: 6029 Poster Board Number: D0248 Presentation Time: 8:00 AM–9:45 AM The role of IP-10 in RPE degeneration during AMD progression Jacob Roney, Kabhilan Mohan, Kyung Jung, Dingyuan Lou, Subhash C. Prajapati, Jennifer Brown, Mark E. Kleinman. Ophthalmology & Visual Sciences, University of Kentucky, Lexington, KY. Purpose: Interferon gamma-induced protein 10 (IP-10/CXCL10) is a pro-inflammatory cytokine that is upregulated in the retinal pigment epithelium (RPE) during progression of age-related macular degeneration (AMD). Previous data suggest significantly enhanced expression of IP-10 after RPE treatment with doublestranded RNA (dsRNA) which is also increased in eyes with AMD. In this study, we set out to determine whether this cytokine is critical to dsRNA mediated RPE degeneration in a mouse model These abstracts are licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International License. Go to http://iovs.arvojournals.org/ to access the versions of record. ARVO 2016 Annual Meeting Abstracts of Ip10 deficiency as well as phenotypic features and cytokine expression in the aged mouse. Methods: Ip10 deficient mice (Jackson Labs, C57BL/6J background, young (4-8 weeks) and aged (12-18 months) were compared to age-matched Wild type C57BL/6J mice (N=6-8). Intravitreous injections (PBS, poly I:C or Alu RNA; both 1μg) were performed in Wild-type and Ip10-/- mice (N=4-6, 4-6 weeks) and analyzed at day 7. Fundus photography and autofluorescence were obtained using Topcon TRC-50IX; electroretinograms (ERG; Espion) were obtained under scotopic conditions. RPE/choroid flatmounts were prepared and immuno-stained for the tight junction protein, zona occludens-1 (ZO-1). RPE levels of inflammatory cytokines in the aging model were analyzed via multiplex bead arrays (Luminex, N=3). The Ip10-/mouse strain was sequenced for spontaneous rd8 mutations (N=3). Results: Ip10-/- mice are not protected from dsRNA induced RPE degeneration. Funduscopy, autofluorescent imaging, and posterior segment OCT revealed widespread moderate retinal degeneration in the aged Ip10-/- mice compared to age-matched controls. Protein studies revealed significantly decreased levels of multiple proinflammatory cytokines in the RPE of aged Ip10-/- mice. Sequencing for rd8 mutations was positive. Conclusions: A mouse model of Ip10 deficiency was not protected from dsRNA induced RPE degeneration suggesting that this cytokine is not critical during AMD progression. Aged Ip10-/mice developed spontaneous retinal degeneration compared to age matched Wild-type BLC57/6J mice. We discovered that this strain harbors a previously unreported rd8 mutation which may account for this abnormal retinal phenotype. Program Number: 6030 Poster Board Number: D0249 Presentation Time: 8:00 AM–9:45 AM Retinal pigment epithelium atrophy in the Cln3Δex7/8 knock-in mouse model of juvenile neuronal ceroid lipofuscinosis Kabhilan Mohan1, Yu Zhong2, Jinpeng Liu3, Ahmad Al-Attar4, Huijuan Liu2, Kyung Jung1, Morgan Dow4, Jinze Liu3, Teresa Fan4, Qing Jun Wang2, Mark E. Kleinman1. 1Ophthalmol & Visual Sciences, University of Kentucky, Lexington, KY; 2Molecular and Cellular Biochemistry, University of Kentucky, Lexington, KY; 3Computer Science, University of Kentucky, Lexington, KY; 4 University of Kentucky, Lexington, KY. Purpose: Juvenile neuronal ceroid lipofuscinosis (JNCL or Batten’s disease) is a blinding pigmented retinal dystrophy with homozygous deletion of the exons 7 and 8 of the CLN3 gene in a large population of JNCL patients. Although JNCL is primarily a lysosomal storage disease, the retinal pigment epithelium (RPE) atrophy and the role of autophagy in mediating such atrophy remains unclear. In this study, we aim to characterize RPE pathology and the status of various autophagic markers in the Cln3Δex7/8 knock-in mouse model of JNCL. Methods: Cln3Δex7/8 mutant mice (Jackson Labs) were analyzed for spontaneous rd1/rd8 mutation; 8-month (n=5) and 18-monthold (n=5) mutant mice were compared to age-matched Wild type C57BL/6J mice (8 month, n=4; 18 month; n=3). Fundus photography and autofluorescence were obtained using Topcon TRC-50IX; electroretinograms (ERG; Espion) were obtained under scotopic conditions. RPE/choroid flatmounts were prepared and immunostained for the tight junction protein, zona occludens-1 (ZO-1). RNA interference of the CLN3 gene was performed in the RPE-1 cell line (ATCC). Autophagic flux was studied with Western blot for the known markers p62 and LC3II. Autophagy and lysosome gene expression profiles were captured by RNA-Seq and qPCR. Results: No rd1/rd8 mutations were found in the Cln3Δex7/8 strain. Funduscopic analyses revealed a mottled appearance with extensive hypopigmentation in aged Cln3Δex7/8 mutant mice. Hyper-autofluorescent aggregates were observed in Cln3Δex7/8 mice at both ages. Significant losses in both a- and b-waves of aged mutant mice were observed compared to age-matched controls. ZO-1 immunofluorescence revealed widespread disruption of RPE tight junctions exclusively in the aged mutant mice. CLN3 siRNA treatment of RPE-1 cells led to greatly enhanced autophagic flux as well as autophagy and lysosome gene expression alterations that are consistent with increased autophagic and lysosomal degradation. Conclusions: We have observed extensive RPE atrophy and gradual loss of retinal function loss in the Cln3Δex7/8 mouse model; interestingly, autofluorescent aggregates occur even in young mutant mice. Cell culture studies show enhanced autophagic activity resulting from CLN3 deficiency. Our data suggest a role for Cln3 function in autophagy which may contribute to RPE degeneration in JNCL and serve as a novel therapeutic in this blinding disease. Commercial Relationships: Jacob Roney, None; Kabhilan Mohan; Kyung Jung, None; Dingyuan Lou, None; Subhash C. Prajapati, None; Jennifer Brown, None; Mark E. Kleinman, None These abstracts are licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International License. Go to http://iovs.arvojournals.org/ to access the versions of record. ARVO 2016 Annual Meeting Abstracts Human umbilical tissue-derived cell (hUTC) is an allogeneic cellbased medicinal product for the treatment of geographic atrophy, an advanced form of dry AMD. We previously demonstrated that subretinal injection of hUTC preserves visual function and retinal architecture in the RCS rat with reduced apoptosis in the retina, suggesting that hUTC may have a protective effect on retina. The retina is particularly susceptible to oxidative stress and this plays an important role in the pathogenesis of AMD and contributes to RPE damage. Here, we aimed to determine whether hUTC could improve the health of RPE cells exposed to oxidative stress. Methods: The ARPE-19 cell line was obtained from American Type Culture Collection. Primary human RPE (hRPE) cultures were prepared from postmortem human eyes obtained from donors and the purity was confirmed by cytokeratin staining. The MTT and crystal violet assays were performed to evaluate the cell viability of ARPE-19 and hRPE exposed to a lethal dose (1500 µM) of hydrogen peroxide (H2O2) after co-culturing with hUTC for 72 hours in transwell inserts. Western blots were preformed to examine the level of protein carbonylation, a key marker of oxidative stress induced damage, in ARPE-19 cells co-cultured with hUTC and then subject to sublethal doses of H2O2 (200 and 400 µM) for 3 and 6 hours, respectively. Results: ARPE-19 co-cultured with hUTC for 72 hours showed significantly improved viability following treatment with 1500 µM H2O2 compared to untreated control cells, as assessed by both MTT and crystal violet assays. Similar results were obtained in hRPE. Moreover, a significantly lower level of protein carbonylation was observed in hUTC pretreated ARPE-19 cells exposed to 400 µM H2O2 for 6 hours as determined by Western blot analysis. Conclusions: These findings suggest that hUTC may secrete factors that can potentially protect RPE from oxidative damage. Investigation is currently underway to examine the mechanisms of action. Commercial Relationships: Sayak K. Mitter, Janssen R&D (F); Michael E. Boulton, Janssen R&D (F); Ian Harris; Jing Cao, Janssen R&D Support: The work described was performed under a sponsored research agreement between Indiana University and Janssen R&D and a Research to Prevent Blindness Unrestricted Grant. Commercial Relationships: Kabhilan Mohan, None; Yu Zhong, None; Jinpeng Liu, None; Ahmad Al-Attar, None; Huijuan Liu, None; Kyung Jung, None; Morgan Dow, None; Jinze Liu, None; Teresa Fan, None; Qing Jun Wang, None; Mark E. Kleinman, None Support: Mark Kleinman - K08EY021757; Foundation Fighting Blindness; Research to Prevent Blindness. Qingjun Wang - Ellison Medical Foundation Program Number: 6031 Poster Board Number: D0250 Presentation Time: 8:00 AM–9:45 AM Human umbilical tissue-derived cells protect retinal pigment epithelial cells from oxidative damage Sayak K. Mitter1, Michael E. Boulton1, Ian Harris2, Jing Cao2. 1 Ophthalmology, Indiana University School of Medicine, Indianapolis, IN; 2Janssen R&D, Spring House, PA. Purpose: Retinal pigment epithelium (RPE) cells perform many functions crucial for retinal homeostasis and vision. RPE cell dysfunction results in various retinal degenerative diseases including age-related macular degeneration (AMD) for which there are currently no effective treatment. Cell therapies targeting RPE cells are being developed in the clinic for the treatment of retinal degeneration. Program Number: 6032 Poster Board Number: D0251 Presentation Time: 8:00 AM–9:45 AM Neuroprotectin D1 (NPD1) modulates amyloid precursor protein (APP) processing in human retinal pigment epithelial cells (RPE) Khanh Do, Jorgelina M. Calandria, Nicolas G. Bazan. Neuroscience Center of Excellence, Louisiana State University Health, NEW ORLEANS, LA. Purpose: Amyloid beta is a major component of drusen in agerelated macular degeneration (AMD) (Ratnayaka et al; Eye, 2015). In neuronal cells in culture from human brain expressing APP mutations that cause familial forms of Alzheimer’s disease, NPD1 shifts APP processing from the tandem beta and gamma secretase to alpha secretase (Zhao et al; PLOS One, 2011). We hypothesize that NPD1 deters formation of amyloid-β towards the prosurvival sAPPa, as it occurs in human brain cells. Here we have studied the bioactivity of NPD1 on APP processing in RPE cells. Methods: Plasmid construct containing Swedish double mutant APP protein (APPsw), which dominantly produce sAPPβ (a precursor of amyloid beta), was transfected into human ARPE-19 cells. After 24h, NPD1 was added at 50, 100 and 500 nM concentrations in the oxidative stress-inducing condition (10 ng/mL of tumor necrosis factor alpha (TNFα) plus 600 nM H2O2). The synthesized nonamyloidogenic sAPPα and amyloidogenic sAPPβ were analyzed by western blot after 48h of NPD1 treatment. Non-transfected and These abstracts are licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International License. Go to http://iovs.arvojournals.org/ to access the versions of record. ARVO 2016 Annual Meeting Abstracts non-oxidative stress conditions were used as negative controls. Furthermore, we also investigated the anti-oxidant activity of 2 nM sAPPα protein in preventing oxidative stress-induced apoptosis in ARPE-19 cells. The cell survival ratio was analyzed by Hoechst staining. Results: At 48h, higher sAPPα production was observed in the increase of NPD1 concentration. The sAPPα expression peaked 1.5 times higher than the non-treated control at 500 nM of NPD1. In contrast, reverse production of sAPPβ was observed, where the highest amount of sAPPβ was detected in the absence of NPD1. An equal amount of holo-APP, regarded as total of all sAPP forms, was detected in all studied condition. Also, we saw that NPD1 decreased the level of amyloid-β secretion to the control level. For anti-oxidant action, our results showed that the presence of 2 nM sAPPα could prevent oxidative stress-induced apoptosis in RPE cells. Conclusions: We found an NPD1 concentration-dependent increase of non-amyloidogenic sAPPα and decrease of amyloidogenic sAPPβ in RPE cells. These results suggest that NPD1 shifts production of amyloidogenic to the non-amyloidogenic pathway in human RPE cells. Additionally, the protective activity of sAPPα modulated by NPD1 represents a new RPE cell survival route. Commercial Relationships: Khanh Do, None; Jorgelina M. Calandria, None; Nicolas G. Bazan, None Support: NIGMS grant P30 GM103340, NEI grant R01 EY005121 (NGB) and Research to Prevent Blindness Program Number: 6033 Poster Board Number: D0252 Presentation Time: 8:00 AM–9:45 AM Bruch’s Membrane is Leaky and RPE lacks Apoptosis, Autophagy and Lysosomes in AMD Mones S. Abu-Asab, Christopher P. Ardeljan, Maria M. Campos. Histology Section, National Eye Institute, Bethesda, MD. Purpose: This study aims to interpret the ultrastructural aberrations in RPE and Bruch’s membrane of AMD in order to identify contributing factors to its etiology. Very few published ultrastructural reports on AMD have identified pathological abnormalities in RPE and Bruch’s membrane. We have observed that some aberrations could cause the accumulation of drusen and cellular debris, as well as other abnormalities. We are also investigating whether changes within the macula differ from those in the peripheral fundus. Methods: Ultrastructural examination of Bruch’s membrane and RPE, from the macula and peripheral fundus, was performed on the eyes of 7 AMD donors, all of whom were over 80 years old. Eyes were fixed in formalin and samples from the macula and temporal fundus were dissected out. Preparation for TEM was done as follows: tissues were embedded in epoxy resin, sectioned, double-stained with uranyl acetate and lead citrate, and viewed with a JOEL JM-1010 TEM. Results: Ultrastructural aberrations were seen in all of the examined specimens. Normal and necrotic RPE cells appeared without lysosomes or autophagosomes, and none of the specimens had apoptotic RPE cells. In some specimens, the RPE layer had doubled by producing a new healthy layer of cells on top of the older degenerate one. Drusen and debris of necrotic cells were present below the RPE layer. Bruch’s membrane was leaky, allowing drusen contents and cellular debris of necrotic RPE cells to pass from the RPE side onto the choroidal side; these encompassed lipid droplets, lipofuscin, chromatin vesicles, degenerate melanosomes, and aqueous vesicles. Additionally, Bruch’s membrane showed collagen degeneration and loss of elastin. Conclusions: Our survey of AMD specimens revealed a number of ultrastructural aberrations, some of which have not been previously described in AMD. These RPE aberrations indicate dysfunctional cellular clearance mechanisms such as apoptosis, lysosomes, and autophagy. Lack of apoptosis seems to be forcing RPE cells into necrosis leading to an accumulation of cellular debris on top of and within Bruch’s membrane. Debris is also being passed onto the choroidal side of Bruch’s membrane. Necrosis and not apoptosis appears to be the prominent type of cell death in AMD. Both the macular and peripheral regions shared the same abnormalities and there was no significant differences between them. Commercial Relationships: Mones S. Abu-Asab; Christopher P. Ardeljan, None; Maria M. Campos, None Program Number: 6034 Poster Board Number: D0253 Presentation Time: 8:00 AM–9:45 AM Altered Transepithelial Resistance of Induced Pluripotent Stem Cell-derived Retinal Pigment Epithelium Obtained from Age-related Macular Degeneration (AMD) Patients Ernesto F. Moreira, Jie Gong, Sibylle Rosendahl, Amanda Barrett, Mark A. Fields, Zsolt Ablonczy, Baerbel Rohrer, Lucian V. Del Priore. Ophthalmology, Medical University of South Carolina, Charleston, SC. Purpose: We have previously reported the generation of induced pluripotent stem cells (iPSC) derived from fibroblasts of patients with advanced macular degeneration, as well as the differentiation of iPSCs into retinal pigment epithelium (RPE). Here, we report the transepithelial resistance (TER) analysis of iPSC-derived RPE cells from AMD patients and unaffected controls. Methods: Fibroblasts were grown and expanded from punch biopsies obtained from patients with advanced (dry or wet) AMD and unaffected controls. Sendai virus technology (ThermoFisher Scientific) was used to deliver Yamanaka factors to reprogram fibroblasts into iPSCs. Subsequently, iPSCs were differentiated into RPE using our recently published protocol (Gong et al, 2015). Pure iPSC-RPE cells, grown in transwell plates for 1-2 months, were characterized morphologically (honeycomb monolayer appearance and pigmentation) and by immunocytochemistry for RPE specific molecules (i.e., OTX2, MITF, Bestrophin). RPE trans-epithelial resistance (TER) was assessed with a Voltohmmeter averaging three measurements in each (duplicate) transwell. Results: iPSC-RPE cell lines derived from AMD patients (N=4) and controls (N=2) formed characteristic monolayers showing typical honeycomb organization and pigmentation. These cells also expressed specific RPE cell markers important for their differentiation and function, including MITF, RPE65. Baseline TER analysis showed that dry AMD samples had a significantly reduced TER (29 +/-10 (#1) and 37 +/-16 (#2) and 117 +/-21 (#3)) as compared to the wet AMD and controls (411 +/-16 (wet AMD); 629 +/-26 (control #1); 330 +/- 48 (control #2)). Conclusions: These studies suggest that intrinsic differences in RPE cells between AMD patients and unaffected controls may exist in the ability to make tight junctions necessary for RPE barrier function. In addition, these results further indicate the potential use of these cells to investigate features of AMD based on the genotype and/ or phenotype of the patients. Ongoing experiments are aimed at analyzing complement activation and VEGF levels in the supernatant of iPSC-RPE before and after challenging them with specific AMDrelevant stressors, in addition to determining expression of tightjunction markers for these cells. Commercial Relationships: Ernesto F. Moreira, None; Jie Gong, None; Sibylle Rosendahl, None; Amanda Barrett, None; Mark A. Fields, None; Zsolt Ablonczy, None; Baerbel Rohrer; Lucian V. Del Priore, Mesoblast (C), Angioblast (C), Advanced Cell Technology (C), Ocata (C) These abstracts are licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International License. Go to http://iovs.arvojournals.org/ to access the versions of record. ARVO 2016 Annual Meeting Abstracts Support: This work is supported in part by a Brightfocus Foundation grant and a faculty funding program from the Dean’s office of the Medical University of South Carolina (EFM); NIH grants EY019320 (BR), EY019065 (ZA), and an unrestricted grant to the Ophthalmology Department, MUSC, from the Research to Prevent Blindness. BR is also sponsored in part by a Department of Veterans Affairs merit award RX000444, and the Feldberg Endowment. LVDP is supported in part by a grant from the Foundation Fighting Blindness. Program Number: 6035 Poster Board Number: D0254 Presentation Time: 8:00 AM–9:45 AM Compromised phagosome maturation underlies defective RPE clearance in an in vitro model of Smith-Lemli-Opitz Syndrome Sriganesh Ramachandra Rao1, 3, Nestor Mas Gomez2, Bruce A. Pfeffer1, 3, Aryn Rowsam1, 3, Claire H. Mitchell2, Steven J. Fliesler1, 3. 1Ophthalmology and Biochemistry, SUNY-Buffalo Sch Med & Biomed Sci and SUNY Eye Institute, Buffalo, NY; 2Anatomy & Cell Biology, University of Pennsylvania Sch Dental Med, Philadelphia, PA; 3Research Service, VAWNYHSBuffalo VAMC, Buffalo, NY. Purpose: The retinal pigmented epithelium (RPE) in the AY9944 rat model of Smith-Lemli-Opitz syndrome (SLOS), a genetic disorder of cholesterol biosynthesis, exhibits accumulation of phagosomes and other inclusions, compared to controls [Fliesler et al., 2004, Arch. Ophthalmol.]. We examined SLOS patient iPSC-derived (SLOS RPE), vs. normal human embryonic stem cell-derived (nhRPE), cells in vitro to determine the underlying mechanism of this defect. Methods: SLOS RPE (harboring both T93M and IVS8 G-C DHCR7 mutations) and nhRPE were treated with bovine rod outer segments (ROS) for 48 h; rhodopsin levels were quantified by Western blotting/ densitometry (WB/D; probed with 1D4 MAb) to assess phagosome clearance. Lysosomal protease (mature Cathepsin-D), and markers of autophagic flux (p62 and LC3-I/II) were assessed by WB/D, normalized to GAPDH levels, with corresponding antibodies. SLOS RPE and nhRPE lysosomal pH was measured using LysoSensor Yellow/Blue DND-160, and lysosomal Cathepsin-D activity was estimated using a BODIPY-Pepstatin assay kit (Molecular Probes). Statistical significance of mean/S.E. values was determined by paired Student’s t-test (criterion, p ≤ 0.05). Results: Autophagic marker levels in SLOS RPE (expressed as % change, vs. nhRPE) were as follows (p<0.05): Beclin-1, +62%; p62, +18%; LC3-II, -50%. Mature Cathepsin-D levels were unaltered. Lysosomal pH in SLOS hRPE was statistically more acidic (4.50 ± 0.02, p<0.05) compared to nhRPE (4.56 ± 0.01), but Cathepsin-D activity was not significantly altered. However, heterophagic clearance of exogenous ROS was defective in SLOS RPE, as evidenced by persistence of 1D4+ material (rod opsin C-terminus), compared to nhRPE cells. Conclusions: Decreased LC3-II levels, elevated p62 levels, and persistence of 1D4+ material indicate compromised phagosome maturation in SLOS RPE compared to nhRPE. Elevated Beclin1 levels obviate defective initiation of autophagy/heterophagy; also, compromised lysosomal physiology was not involved. However, increased 7-dehydrocholesterol (7DHC) levels (a SLOS hallmark) and/or oxysterols derived therefrom may underlie the SLOSassociated RPE defect. Commercial Relationships: Sriganesh Ramachandra Rao, None; Nestor Mas Gomez, None; Bruce A. Pfeffer, None; Aryn Rowsam, None; Claire H. Mitchell, None; Steven J. Fliesler, None Support: NIH RO1 EY007361 (SJF); RPB Unrestricted Grant (SJF); VAWNYHS facilities and resources (SRR, BAP, AMR, SJF). NIH R01 EY013434 (CHM) Program Number: 6036 Poster Board Number: D0255 Presentation Time: 8:00 AM–9:45 AM Semi-automated analysis of lipofuscine and melanolipofuscine granules in the RPE using structured illumination microscopy Nil Celik1, Gerrit Best2, Florian Schock2, Alena Bakulina3, Gerd U. Auffarth1, Christoph Cremer2, Juergen Hesser3, Stefan Dithmar4. 1Department of Ophthalmology, University Hospital Heidelberg, Heidleberg, Germany; 2Institute of Molecular Biology, Mainz, Germany; 3University Medical Center Mannheim, Experimental Radiation Oncology, Mannheim, Germany; 4 Department of Ophthalmology, Dr. Horst Schmidt Kliniken, Wiesbaden, Germany. Purpose: We already introduced a new analyzing tool for lipofuscine granules (LG) in retinal pigment epithelium (RPE) cells in highly resolved 3D-Structured illumination microscopy (SIM) images automatically. In a further analysis we additionally investigated melanolipofuscine granules (MLG). Methods: 3D-SIM images were examined for LG and MLG using three different excitation wavelengths (488, 568 and 647 nm). Our automated software identifies and quantifies (number, size and position) single granules in RPE flatmounts. The LG and MLG of an 88-year-old donor were examined for the macular region. Results: From 541 granules, 411 (76.0%) were identified as LG and 130 (24.0%) as MLG. LG and MLG were distributed towards the cell edge. Conclusions: Highly resolved 3D-images of LG as well as MLG are obtainable with SIM. LG and MLG can be differentiated and characterized regarding their number and position with a semiautomated analysis of the established SIM data. Quantitative characteristics of autofluorescent granules help to understand cellular processes within the RPE. Commercial Relationships: Nil Celik; Gerrit Best, None; Florian Schock, None; Alena Bakulina, None; Gerd U. Auffarth, Abbott Medical Optics, Alcon Laboratories, Inc., Alimera Sciences, Inc., Bausch & Lomb Surgical, Carl Zeiss Meditec, Contamac, Glaukos Corporation, Heidelberg Engineering, HumanOptics, Novartis Pharmaceuticals Corporation, Oculentis, Physiol, Power Vision, Rayner Intraocular Lenses Ltd, Technolas (F), Abbott Medical Optics, Alcon Laboratories, Inc., Allergan, Bausch & Lomb Surgical, Bayer Healthcare Pharmaceuticals, Carl Zeiss Meditec, Hoya, HumanOptics, Kowa, Oculentis, OPHTEC, Physiol, Rayner Intraocular Lenses Ltd, Technolas (R), Abbott Medical Optics, Alcon Laboratories, Inc., Allergan, Bausch & Lomb Surgical, Novartis Pharmaceuticals Corporation, Rayner Intraocular Lenses Ltd, Technolas (C); Christoph Cremer, None; Juergen Hesser, None; Stefan Dithmar, None Support: Supported by Gertrud Kusen Foundation Program Number: 6037 Poster Board Number: D0256 Presentation Time: 8:00 AM–9:45 AM Mutations in the Mitf gene affect autophagy in mouse primary RPE cells Andrea García-Llorca1, Margret Helga Ogmunsdottir2, Eirikur Steingrimsson2, Thor Eysteinsson1. 1Physiology, University of Iceland, Reykjavík, Iceland; 2Biochemistry and Molecular Biology, University of Iceland, Reykjavík, Iceland. Purpose: Mutations at the mouse microphthalmia locus (Mitf) affect the development of different cell types including melanocytes, retinal pigment epithelium and osteoclasts. The MITF protein is a member of the MYC supergene family of basic-helix-loop-helix-leucinezipper (bHLHZip) transcription factors and is known to regulate the expression of cell-specific target genes by binding DNA as homodimer or as heterodimer with related proteins. The purpose of These abstracts are licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International License. Go to http://iovs.arvojournals.org/ to access the versions of record. ARVO 2016 Annual Meeting Abstracts this study was to examine the role of Mitf in regulating autophagy in mouse primary RPE cells by studying mouse Mitf mutations. Methods: Primary RPE cells from wild type and MITF mutant mice (Mitfmi-enu122(398), MitfMi-Wh/+ and MitfMi-Wh/Mitfmi-mi) were isolated by enzymatic dissociation. The characteristic morphology and the expression of RPE65 of these RPE cells were observed by antibody staining and microscopy. Expression of LC3 and MITF was analyzed with western blot analysis and confocal microscopy in primary RPE cells from C57BL/6J mice, untreated or treated with the mTOR inhibitor Torin1 for 3 hours or after incubation in starvation media for 1 hour. Torin1 and starvation are known activators of autophagy. The levels of LC3 and MITF were measured and compared by western blot in RPE cells from wild type and the Mitf mutant mice. Results: Wild type RPE cells express MITF and also basal levels of LC3. The treatment with starvation media and Torin1 treatment increased the levels of LC3 in RPE cells. Furthermore, both starvation and Torin1treatment resulted in reduced MITF protein levels. Wild type treated RPE cells with starvation media showed phagosomes around the nuclei. Only the LC3II protein was detected in RPE cells from MITF mutant whereas wild type RPE cells showed both LC3I and II, suggesting that the degradation pathway of LC3 is stalled in the RPE from Mitf mutant mice. Conclusions: This study suggests that autophagy is affected in Mitf mutant mice. This is consistent with in vitro data showing that MITF regulates expression of genes involved in autophagy. Commercial Relationships: Andrea García-Llorca, None; Margret Helga Ogmunsdottir, None; Eirikur Steingrimsson, None; Thor Eysteinsson, None Support: Icelandic Research Council Grants, Helga Jonsdottir and Sigurlidi Kristjansson Memorial Fund Program Number: 6038 Poster Board Number: D0257 Presentation Time: 8:00 AM–9:45 AM Age-related degeneration of the retinal pigment epithelium (RPE) of DJ-1 mice Vera L. Bonilha1, 2, Brent A. Bell1, Mary E. Rayborn1, Joe G. Hollyfield1, 2, Ivy S. Samuels1, 3, Chengsong Xie4, Huaibin Cai4. 1 Ophthalmic Research, Cleveland Clinic, Cleveland, OH; 2 Ophthalmology, Cole Eye Institute/CCLCM, Cleveland, OH; 3 Research Service, Louis Stokes Cleveland Veterans Affairs Medical Center, Cleveland, OH; 4Lab. of Neurogenetics, National Institute of Aging/ NIH, Bethesda, MD. Purpose: DJ-1 is ubiquitously expressed in many tissues including the brain where it functions as an antioxidant, redox-sensitive molecular chaperone and transcription regulator robustly protecting cells from oxidative stress. We previously reported mild structural and physiological changes including RPE thinning, central decrease in red/green cone opsin staining, decreased labeling of ezrin, broader distribution of ribeye labeling, decreased tyrosine hydroxylase in dopaminergic neurons, and increased DNA oxidation in the retinas of 3 and 6 month old DJ-1 knockout (KO) in comparison to control mice. The severity of these changes increased with age; therefore, in the present study we extended our analysis to older (18 month-old) mice and compared the results to young (3 month-old) DJ-1 KO and control mice. Methods: The effects of DJ-1 deletion were examined in KO mice through non-invasive, fundus imaging in vivo using SLO and OCT. A profile analysis of the outer retina was performed from the four retinal quadrants to assess OCT signal reflectivity and lamina morphology. Photoreceptor and RPE function were assessed by ERG. Histological and immunohistological evaluation of the retinas of DJ-1 KO and control mice were also performed in cryosections as well as whole-mounted retina and RPE/choroid. Retina/RPE lysates were assayed for oxidation using antibodies. Results: SLO showed no differences between the fundus of DJ-1 KO and control mice. OCT imaging demonstrated all retinal layers in both young and older DJ-1 KO retinas. The profile analysis revealed significant hyper-reflectivity of photoreceptor outer segments and outer retina thinning in DJ-1 KO in comparison to control mice for all retinal quadrants examined. ERG studies of older DJ-1 KO mice noted significant decreased values under dark- and light-adapted conditions. Histologically, changes in the RPE of the DJ-1 KO mice progressed from increased presence of vacuoles at 3 months of age to the presence of vacuoles and lesions at 18 months of age. The RPE lesions were also visible by OCT and in whole mounted RPE/choroid samples labeled with phalloidin. Photoreceptors were degenerated when opposing the RPE lesions. Increased protein carbonyl derivatives were detected in retina/RPE lysates from older DJ-1 KO mice. Conclusions: DJ-1 KO mice display progressive signs of retinal/ RPE degeneration in association with increased levels of oxidative stress markers. Commercial Relationships: Vera L. Bonilha, None; Brent A. Bell, None; Mary E. Rayborn, None; Joe G. Hollyfield, None; Ivy S. Samuels, None; Chengsong Xie; Huaibin Cai, None Support: Research to Prevent Blindness, Wolf Family Foundation,National Eye Institute (EY014240-08) and a VA Merit Award i01-BX002754 from Biomedical Laboratory Research and Development (ISS). Program Number: 6039 Poster Board Number: D0258 Presentation Time: 8:00 AM–9:45 AM TFEB-mediated clearance of the lipofuscin fluorophore A2E Ivana Trapani1, Elisabetta Toriello1, Renato Minopoli1, Alberto Auricchio1, 2. 1Telethon Institute of Genetics and Medicine (TIGEM), Pozzuoli, Italy; 2Medical Genetics, Department of Translational Medicine, Federico II University, Naples, Italy. Purpose: Abnormal accumulation of various by-products of the visual cycle, including the diretinoid-pyridinium-ethanolamine (A2E), in retinal pigment epithelium (RPE) is an hallmark of both Stargardt disease (STGD) and age-related macular degeneration (AMD). This is responsible for RPE and, consequently, photoreceptor (PR) cell death. A2E storage in RPE lysosomes has been shown to reduce both the capacity of RPE to degrade phagocytized PR outer segments and autophagosome biogenesis, trafficking and autophagic flux. The transcription factor EB (TFEB) is a master regulator of cellular clearance. Here we tested if TFEB overexpression induces A2E clearance from the RPE cells. Methods: We have generated a plasmid encoding for a double-serine mutant (S142A, S211A) form of TFEB, which is constitutively active. We have transfected this plasmid in the human RPE-derived ARPE19 cells to evaluate TFEB activation of its transcriptional targets and ability to clear A2E after loading. Results: We found that TFEB overexpression in ARPE19 cells results in the induction of TFEB transcriptional targets involved in cellular clearance. Importantly, TFEB overexpression in ARPE19 cells was associated with reduction of intracellular A2E accumulation. Conclusions: Taken together these results suggest that TFEB overexpression is an effective strategy to promote A2E clearance in vitro. Further studies will clarify if this holds true in vivo. These results may have implications for the therapy of both STGD and the more common AMD. Commercial Relationships: Ivana Trapani, None; Elisabetta Toriello, None; Renato Minopoli, None; Alberto Auricchio, None Support: Italian Telethon Foundation grant TGM11MT1 These abstracts are licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International License. Go to http://iovs.arvojournals.org/ to access the versions of record. ARVO 2016 Annual Meeting Abstracts Program Number: 6040 Poster Board Number: D0259 Presentation Time: 8:00 AM–9:45 AM Chloroquine toxicity of the retinal pigment epithelium and retina in vitro and a mouse model of retinal toxicity Dhanesh Amarnani1, 2, Leo A. Kim2, Patricia A. D’Amore2, Lindsay Wong2. 1Ophthalmology, Harvard Medical School, Boston, MA; 2Retina, Schepens Eye Research Institute/ Massachusetts Eye and Ear Infirmary, Boston, MA. Purpose: To evaluate the cell death mechanisms of chloroquineinduced toxicity of human cultured epithelial cells(RPE). To develop and characterize a murine model of chloroquine-induced retinal toxicity Methods: ARPE-19 cells were cultured until confluence, treated with 500uM chloroquine for 24 hours and cell death was measured by lactate dehydrogenase(LDH) release. Cytoskeletal structure and lysosomal permeability were assessed using phalloidin and lysosomal-associated membrane protein(LAMP-1) staining, respectively. The roles of lysosomal enzymes cathepsin B and L were examined by blocking their activity using specific cell permeable inhibitors. The contribution of different cell death mediators was evaluated by specific inhibition of caspase-1, 3, 8, 9 and receptorinteracting protein(RIP) kinases. To develop an animal model of retina chloroquine toxicity, subretinal chloroquine (0.2 mg/kg) injections were performed. Phenotypic changes were evaluated using optical coherence tomography(OCT) and functional assessment was performed by electroretinography(ERG). Histological changes to the RPE were assessed. Results: 500uM chloroquine induced 30-40% of cell death at 24 hours. Lysosomal permeabilization and cytoskeletal structure disruption were observed at 3 and 6 hours. Blockage of cathepsin B/L activity resulted in a significant decrease in cell death, indicating that lysosomal destabilization and resulting cathepsin release are upstream of these cell death pathways. Partial rescue of cell death was observed with caspases and RIP kinase inhibitors. Preliminary results from OCT demonstrate loss of the RPE layer and thinning of the outer retina within two weeks of injection. ERG revealed a decreased scotopic response in both the A and B-waves of the ERG waveform. H&E staining of retinal cross sections in chloroquine-treated mice revealed loss of RPE and disruption of outer segments. ZO-1 staining of RPE flat mounts demonstrated RPE cells death, enlarged cells and loss of cell junctions. Conclusions: Chloroquine-induced cell death occurs through multiple cell death mechanisms. Inhibition of upstream activators such as cathepisn B/L may be a feasible approach to block multiple cell death pathways. Some of the phenotypic and functional changes observed in our mouse model are similar to those observed in human subjects with chloroquine toxicity, and may be model of RPE and retinal degeneration. Commercial Relationships: Dhanesh Amarnani, None; Leo A. Kim, None; Patricia A. D'Amore, None; Lindsay Wong Program Number: 6041 Poster Board Number: D0260 Presentation Time: 8:00 AM–9:45 AM Prominin 1 Plays a Central Role in Regulating Stress-induced Autophagy in the Retinal Pigment Epithelium Edward Chaum, Christina S. Winborn, Sujoy Bhattacharya, Jinggang Yin. Ophthalmology, Univ of Tennessee Health Sci Ctr, Memphis, TN. Purpose: Prominin1 (Prom1) is a transmembrane glycoprotein that localizes to plasma membrane protrusions and is a known cancer stem cell marker. It also plays a role in photoreceptor disk morphogenesis; mutations in the Prom1 gene cause Type 4 Stargardt disease. Our previous studies have shown that Prom1 is also a novel regulator of autophagy in the RPE. The purpose of this study was to examine the cellular mechanisms of Prom1-mediated autophagy. We show that loss of Prom1 expression impairs stress-induced autophagy in the human RPE. Methods: We used the CRISPR/CAS9 nuclease system to knockout (KO) Prom1 in ARPE19 cells. FlowSight imaging cytometry and confocal immunofluorescence assays were used to quantify LC3 puncta accumulation in the presence and absence of chemical modulators of autophagy in control APRE19 cells and in Prom1 KO cells. We also monitored autophagosome formation in response to autophagic regulators using a tandem RFP-GFP-LC3 construct containing a GFP marker that is quenched when the autophagosome fuses with the lysosome. Results: We show that knocking out Prom1 expression significantly reduces starvation-induced LC3-puncta accumulation in the human RPE. Flow cytometry and confocal microscopy demonstrated that Prom1 KO; 1) decreases LC3 puncta accumulation in response to starvation or rapamycin, 2) reduces autophagy flux and increases chloroquine-induced LC3-puncta accumulation, and 3) delays autophagosome maturation. Conclusions: Prom1 expression plays in important role in the activation of stress-induced autophagy in the human RPE. Commercial Relationships: Edward Chaum, None; Christina S. Winborn, None; Sujoy Bhattacharya, None; Jinggang Yin, None Support: The Shulsky Foundation, New York, NY, an unrestricted UTHSC departmental grant from Research to Prevent Blindness, New York, NY, The Lions of Arkansas Foundation Inc., and the Plough Foundation, Memphis, TN. UTHSC Hamilton Eye Institute Core Grant for Vision Research, supported by the National Eye Institute (P30 EY013080). Program Number: 6042 Poster Board Number: D0261 Presentation Time: 8:00 AM–9:45 AM Molecular Mechanisms Regulating Prom1-dependent Autophagy in Human Retinal Pigment Epithelial Cells Sujoy Bhattacharya, Qiuhua Zhang, Jinggang Yin, Christina S. Winborn, Edward Chaum. Ophthalmology, University of Tennessee Health Science Center Hamilton Eye Institute, Memphis, TN. Purpose: Beyond its role in photoreceptor disk morphogenesis, our previous studies have demonstrated that Prom1 (CD133) is a novel regulator of autophagy in the RPE. However, the cellular and molecular mechanisms governing Prom1-mediated RPE autophagy have not previously been determined. We show that Prom1 modulates autophagy in the RPE through inhibition of Akt/mTOR activities and through its association with the selective autophagy receptor, sequestosome (SQSTM1)/p62. Methods: The CRISPR/Cas 9 nuclease system was used to knockout Prom1 in the RPE and Prom1 lentiviral constructs were also used to overexpress Prom1 in ARPE19 cells. Western blotting and flow cytometry assays were used to quantify LC3-II processing and punctae formation in the presence and absence of lysosomal inhibitors as cellular readouts for autophagic flux. Immunoprecipitation was used to detect Prom1 interacting proteins and Western blotting was performed to analyze the mTOR and Akt pathway-associated proteins: p-ULK1 (Ser555), p-AMPKα (Thr172), p-mTOR (Ser2448), pAkt (Ser473), p4EBP1(Thr37/46), and LC3-II/ LC3-I after stimulation of autophagy. Results: Overexpression of Prom1, significantly reduced phosphorylation of S6 ribosomal protein at Ser235/236 and Akt at Ser473, surrogate markers for mTORC1 and mTORC2 activities, respectively, and decreased p62 levels. Prom1 overexpression increased Atg5, and Atg7 expression, LC3I/LC3-II turnover, EBSS- These abstracts are licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International License. Go to http://iovs.arvojournals.org/ to access the versions of record. ARVO 2016 Annual Meeting Abstracts induced autophagy flux, and decreased Bafilomycin-induced p62 accumulation. Immunoprecipitation experiments revealed that Prom1 interacted with p62 and HDAC6, both critical mediators of autophagy flux and autophagosome maturation. Conversely, the Prom1 KO increased mTOR activity, delayed autophagosome trafficking to the lysosome, increased p62 accumulation, and impaired stress-induced autophagy. Conclusions: We demonstrate a novel regulation of Prom1dependent autophagy via upstream inhibition of the mTOR/Akt pathway, and through the protein acting as a molecular scaffold for p62/HDAC6 association in the autophagosomal-lysosomal fusion pathway. Commercial Relationships: Sujoy Bhattacharya, None; Qiuhua Zhang, None; Jinggang Yin, None; Christina S. Winborn, None; Edward Chaum, None Support: The Shulsky Foundation, New York, NY; an unrestrcited UTHSC departmental grant from Research to Prevent Blindness, New York, NY; The Lions of Arkansas Foundation Inc., and the Plough Foundation, Memphis, TN; UTHSC Hamilton Eye Institute Core Grant for Vision Research; supported by the National Eye Institute (P30 EY013080) Program Number: 6043 Poster Board Number: D0262 Presentation Time: 8:00 AM–9:45 AM Loss of MREG dependent LC3 associated phagocytosis (LAP) by the RPE leads to altered intracellular lipid processing Desiree Alexander1, Anuradha Dhingra1, William C. Gordon2, Bokkyoo Jun2, Nicolas G. Bazan2, Kathleen Boesze-Battaglia1, Alvina Bragin1. 1Biochemistry, University of Pennsylvania, Philadelphia, PA; 2Neuroscience Center of Excellence School of Medicine, Louisiana State University Health, New Orleans, LA. Purpose: RPE cells utilize a hybrid process that incorporates aspects of phagocytosis and autophagy to efficiently degrade ingested photoreceptor outer segments. Our previous studies have established that this LC3 dependent process is mediated by a cargo sorting protein, melanoregulin (MREG). The purpose of this study was to determine if defective phagosome maturation as observed in the absence of MREG and LC3B contributes to alterations in lipid deposition in the RPE. Methods: In all studies we utilized RPE isolated from Mreg -/-, LC3B -/- and C57Bl6/J mice. Accumulation of cholesterol and cholesterol esters was assessed by filipin staining followed by multi-color confocal microscopy. Levels of cholesterol and 7-ketocholesterol were determined using LC/MS and docosahexaenoic acid (DHA) levels by LC MS/MS. iPLA2-VIA activity was determined using a BODIPY® FL-based fluorometric assay. Results: Cholesterol accumulated in the Mreg -/- RPE as indicated by the prevalence of filipin positive structures. Biochemical analyses also showed elevated cholesterol levels in Mreg -/- RPE, however the most significant increases were in 7-ketocholesterol, in which case loss of MREG was associated with 5- fold increase in this immuno-modulatory lipid. Moreover, an increase in cholesterol accumulation (or filipin positive structures) was also seen in LC3B/- RPE. OS lipids provide a massive daily supply of DHA esterified to phospholipids for cleavage by the iPLA2-VIA phospholipase A2 isoform. Defective phagosome maturation in Mreg -/- RPE also contributed to a decrease in free DHA, with a majority of the DHA found to be esterified to phospholipids. This decrease in phospholipid hydrolysis was not due to alterations in iPLA2-VIA activity but likely due to defective phagocyte processing. Conclusions: Our results suggest that defective LC3 associated phagocytosis, specifically regulated by MREG, leads to accumulation of cholesterol over the long term. Furthermore, loss of MREG leads to a decrease in the intracellular DHA pool, likely compromising the generation of protective docosanoids. Commercial Relationships: Desiree Alexander, None; Anuradha Dhingra, None; William C. Gordon, None; Bokkyoo Jun, None; Nicolas G. Bazan, None; Kathleen Boesze-Battaglia, None; Alvina Bragin, None Support: NEI grant(s) EY-10420 (KBB) and EY-005121 (NGB). Program Number: 6044 Poster Board Number: D0263 Presentation Time: 8:00 AM–9:45 AM Aging and antioxidants differentially modulate the effect of phagocytized lipofuscin granules on expression of antioxidant enzymes in ARPE-19 cells Anna K. Pilat1, Michal Sarna1, Dawid Wnuk2, Magdalena Olchawa1, Tadeusz J. Sarna1. 1Biophysics, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, Krakow, Poland; 2Cell Biology, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, Krakow, Poland. Purpose: Aged RPE cells are at elevated risk of oxidative stress due accumulation of lipofuscin (LF). It was postulated that chronic oxidative stress, mediated in the human RPE by LF, could contribute to the development of age-related macular degeneration (AMD). We have previously shown that LF under sub-lethal photic stress conditions photooxidizes cellular proteins in ARPE-19 cells. Here we ask whether LF-mediated oxidative stress modulates expression of antioxidant enzyme proteins such as heme oxygenase-1 (HO-1) and catalase in ARPE-19 cells. Methods: Lipofuscin granules (LF), isolated from human RPEs from two age groups, were enriched with a combination of zeaxathin and alpha tocopherol (An). Control or antioxidant enriched lipofuscin granules (An-LF) were introduced to APRE-19 cells by phagocytosis. Control cells, An-treated cells or cells with LF and An-LF, irradiated with blue light for selected time intervals, were analyzed by Western blot for selected antioxidant enzymes. To monitor sub-lethal oxidative stress in ARPE-19 cells mediated by phagocytized human RPE lipofuscin atomic force microscopy (AFM) and laser–scanning confocal fluorescence microscopy were employed. Results: Irradiation of LF containing cells with blue light induced HO-1 protein with the effect being light dose dependent. The extent of HO-1 expression by lipofuscin was higher for lipofuscin isolated from older donors (age: 50-59) compared to younger donors (age: 18-29). Enrichment of lipofuscin granules isolated from both age groups with zeaxathin and alpha tocopherol reduced HO-1protein level. AFM analysis revealed that after blue light-treatment, cells with lipofuscin granules, were significantly softer. Fluorescence analysis showed that lipofuscin-mediated photic stress to RPE cells induced significant reduction in the formation of actin stress fibers. Conclusions: HO-1 upregulation may help to protect RPE cells form LF-mediated oxidative stress. Phototoxicity of lipofuscin granules in RPE can be modulated by combination of antioxidants. AFM and fluorescence image analysis could be employed as sensitive methods for detecting early sub-lethal photic changes in RPE cells mediated by LF. Commercial Relationships: Anna K. Pilat, None; Michal Sarna, None; Dawid Wnuk, None; Magdalena Olchawa, None; Tadeusz J. Sarna, None Support: Poland National Science Center (grant Maestro 2013/08/A/ NZ1/00194). Faculty of Biochemistry, Biophysics and Biotechnology of Jagiellonian University is a partner of the Leading National Research Center (KNOW) supported by the Ministry of Science and Higher Education. These abstracts are licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International License. Go to http://iovs.arvojournals.org/ to access the versions of record. ARVO 2016 Annual Meeting Abstracts Program Number: 6045 Poster Board Number: D0264 Presentation Time: 8:00 AM–9:45 AM Proteomic changes in ARPE-19 cells stimulated with complement serum and UV-irradiated photoreceptor outer segments (UV-POS) Martin Busch1, Susanne Wasmuth1, Albrecht Lommatzsch2, Solon Thanos3, Daniel Pauleikhoff2. 1Ophtha-Lab, Department of Ophthalmology at St. Franziskus Hospital, Muenster, Germany; 2 Department of Ophthalmology at St. Franziskus Hospital, Muenster, Germany; 3Institute of Experimental Ophthalmology, University of Muenster, Muenster, Germany. Purpose: ARPE-19 cells as a model for retinal pigment epithelial (RPE) cells were shown to undergo proinflammatory and proangiogenic changes upon complement stimulation and UV-POS treatment suggesting a role in AMD pathology. The aim of our study was to reveal potential intracellular signaling pathways mediating these functional changes of RPE cells using proteomics and stimulation with complement serum and UV-POS. Methods: For complement stimulation, ARPE-19 cells were incubated in DMEM/F12 medium supplemented with 5 % human complement serum (HCS) for 24 hours. Medium alone or supplemented with either 5 % heat-inactivated HCS or C7-deficient serum were used as controls. Further groups of cells were pre-treated every other day with 10 µg/ml UV-POS for one week to a total of three pre-treatments. Afterwards the cells were washed and incubated in medium containing 5 % HCS or medium alone. Proteins were separated by 2D-gel electrophoresis and differentially occurring protein spots were peptide mapped with MALDI-TOF mass spectrometry. Results: Some protein spots were differentially expressed in the various treatment groups and differed between two independent experimental sets. From the two experimental sets, a total of 27 protein spots were selected and processed for MALDI-TOF analysis that identified a total of 56 different proteins, which could be grouped into 7 functional entities including metabolic (12), structural (16), globular (3), cell-cell and cell-matrix interaction-associated proteins (2), gene expression (4), proteins related to inflammatory processes and angiogenesis (7), and proteins involved in signal transduction (10). 60 percent of the identified proteins participating in signal transduction are associated with cellular and oxidative stress response. Out of two cases, expression of these proteins was observed in response to HCS, UV-POS, or their combination. Conclusions: The results show that complement stimulation and UV-POS uptake regulate the cell proteome. Further experiments are required to clarify how far the identified differentially expressed proteins associated with signal transduction are involved in the mediation of the functional changes of RPE cells in response to complement stimulation and UV-POS. Commercial Relationships: Martin Busch, None; Susanne Wasmuth, None; Albrecht Lommatzsch, None; Solon Thanos, None; Daniel Pauleikhoff, None Support: Dr. Werner Jackstädt Foundation and Voltmann Foundation Program Number: 6046 Poster Board Number: D0265 Presentation Time: 8:00 AM–9:45 AM Mutations in CFH perturb RPE homeostasis in vitro Roni Hazim, David S. Williams. Jules Stein Eye Institute, UCLA, Los Angeles, CA. Purpose: Single nucleotide polymorphisms (SNPs) in the complement factor H gene (CFH) are closely linked with increased risk of age-related macular degeneration (AMD). In particular, SNPs in exons 2 and 9, responsible for I62V and Y402H amino acid substitutions, respectively, constitute a genetic haplotype that increases the risk of developing AMD in late adulthood by 7-fold. Individually, both of these SNPs have been shown to compromise the function of CFH. Although most abundant in the circulating plasma, CFH is locally expressed and secreted by the retinal pigment epithelium (RPE), a primary site of insult in AMD pathogenesis. Numerous models have been made to elucidate the role of CFH, yet the full extent of CFH function and the underlying cellular mechanisms responsible for perturbing RPE homeostasis remain elusive. The purpose of this study is to investigate the role of CFH, in relation to RPE biology and pathology, using an in vitro model of human RPE cells. Methods: We used the genome-editing technology of the CRISPR/ Cas9 system to specifically target and modify the CFH locus in an immortalized human RPE cell line (ARPE-19). Two stable cell lines were generated, one of which exhibited a knockdown of CFH while the other expressed the mutated form of the protein (VV62, HH402). Brightfield microscopy was used to assess the morphology of the stable cell lines, and immunocytochemistry and western blotting were used to analyze cytoskeletal organization and to test for signs of inflammation and cellular stress. Results: Sanger sequencing verified the genetic modifications to the CFH locus, and western blotting confirmed the reduction in CFH protein levels in the CFH-knockdown line. Brightfield microscopy revealed a striking morphological phenotype in the CFH-knockdown line, which exhibited an elongated, fusiform shape. Immunolabeling of α-tubulin confirmed an abnormal arrangement of the microtubule cytoskeleton in these cells. Additionally, both CFH-mutant cell lines were found to possess an aberrant expression of ZO-1, a tight junction protein known to associate with actin filaments. Characteristics of AMD, including inflammation and cellular stress, were also more prominent in the CFH-knockdown line, as indicated by increased expression of C5b-9 and 3-NT, respectively. Conclusions: Our results indicate that compromised CFH function has a significant effect on RPE morphology, as well as increasing inflammation and cellular stress. Commercial Relationships: Roni Hazim, None; David S. Williams, None Support: NIH grant EY07042 Program Number: 6047 Poster Board Number: D0266 Presentation Time: 8:00 AM–9:45 AM Growth differentiation factor 6 (GDF6) is a novel inducer of the epithelial-to-mesenchymal transition in retinal pigmented epithelial cells Amanda M. Hurley1, Monte J. Radeke2, Pete J. Coffey2. 1Molecular, Cellular, and Developmental Biology, University of California, Santa Barbara, Santa Barbara, CA; 2Neuroscience Research Institute, University of California, Santa Barbara, Santa Barbara, CA. Purpose: As retinal pigmented epithelium (RPE) cells are passaged, they undergo an irreversible epithelial-to-mesenchymal transition (EMT). We have shown previously, that GDF6, a member of the transforming growth factor-beta (TGFβ) family, is highly upregulated in RPE cells that have undergone EMT. We hypothesize that GDF6 plays an integral role in the irreversible transition of an RPE cell from an epithelial state to a mesenchymal state. Methods: Passage 0 fetal RPE cells were transduced with a lentiviral construct encoding GDF6 or an empty vector control. The GDF6 positive cells were collected, re-plated at a high density, and allowed to differentiate for 32 days. At this point images were obtained for pigmentation analysis and cells were harvested for RNA. Pigmentation was analyzed using Matlab software. The images were binarized and the number of back pixels was divided by the total number of pixels. qRT-PCR was performed using a variety of EMT These abstracts are licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International License. Go to http://iovs.arvojournals.org/ to access the versions of record. ARVO 2016 Annual Meeting Abstracts and RPE markers. Student’s T tests were used to determine statistical significance Results: RPE transduced with GDF6 produce a significantly lower level of pigmentation (21%) than cells infected with an empty vector control (68%, p=0.0001). This reduction in pigmentation is accompanied with a change in cell morphology; where the control cells maintain a cuboidal morphology and the GDF6 secreting cells have a spindle-like morphology. qRT-PCR analysis reveals that RPE transduced with GDF6 significantly upregulate known EMT markers like ACTA2, CTGF, and COL1A1 and downregulate classical RPE markers such as LRAT, APOE, and PMEL compared to control cells. Conclusions: GDF6 is a novel inducer of EMT in RPE. Cells that overexpress GDF6 will prematurely undergo EMT, simultaneously downregulating traditional RPE markers while upregulating EMT and wound response markers. Inhibition of GDF6 may be integral in prolonging the integrity of RPE cultures. Commercial Relationships: Amanda M. Hurley, None; Monte J. Radeke; Pete J. Coffey, None Support: CIRM LA1-02086, Garland Initiative for Vision Research, Arnold & Mable Beckman Initiative for Macular Research Program Number: 6048 Poster Board Number: D0267 Presentation Time: 8:00 AM–9:45 AM Cellular effects of lysosomal dysfunction in retinal pigment epithelial cells Mallika Valapala2, Abdulla Alamri2, Rhonda Grebe1, J S. Zigler1, James T. Handa1, Debasish Sinha1. 1UVEA, Johns Hopkins Univ Sch of Med, Baltimore, MD; 2Optometry, Indiana University, Bloomington, IN. Purpose: The retinal pigment epithelial (RPE) is not only one of the most active phagocytic cell types in the body but also has a high rate of autophagy. Lysosomes are involved in the terminal stages of phagocytosis and autophagy, hence dysfunctional lysosomes lead to impaired cellular clearance in the RPE. The objective of this study was to analyze if the transcriptional program regulating lysosomal biogenesis and function can be induced in the RPE following lysosomal disruption by treatment with ammonium chloride. We also examined the cellular effects of lysosomal dysfunction in the RPE using Cryba1 (gene encoding βA3/A1-crystallin) knockout mice. Methods: Confluent monolayers of ARPE-19 cells were treated with 1 mM ammonium chloride (to block endolysosomal acidification) for 18 hrs. Ammonium chloride was later removed and the cells were subjected to a 6 hr recovery period. Quantitative real time PCR (qPCR) analysis was performed to study the expression of the following genes involved in lysosomal biogenesis and function: Lysosome associated membrane protein 1(LAMP-1), Cathepsin D (CTSD), Cathepsin A (CTSA), Cathepsin F (CTSF), hexosaminidase A, and ATPase, H+ transporting, lysosomal V0 subunit a1 (ATP6V0A1) encoding for a subunit of V-ATPase. Immunofluorescence and immunoblotting was used to determine the expression of the lysosomal marker, LAMP-1. Cellular ultrastructure of the Cryba1 KO RPE was studied by transmission electron microscopy (TEM). Results: qPCR analysis of ARPE-19 cells treated with ammonium chloride and subjected to a 6 hr recovery period revealed significant induction of genes regulating both lysosomal biogenesis (LAMP-1,HexA) and function (CTSD, CTSA,CTSF and ATP6V0A1) compared to untreated cells. Immunoblotting and immunofluorescence studies showed increased expression of LAMP1 in cells recovered from ammonium chloride treatment compared to untreated cells. TEM of 25 month old Cryba1 KO mice revealed many vacuole-like structures containing undigested cellular material and photoreceptor outer segments. Conclusions: Our studies suggest that perturbation of lysosomal function by pre-treatment with ammonium chloride followed by a recovery period results in a rapid induction of the transcriptional program that induces lysosomal biogenesis and function in ARPE-19 cells. Impaired lysosomal function seen in the Cryba1 KO mice leads to RPE cellular abnormalities and eventual degeneration Commercial Relationships: Mallika Valapala, None; Abdulla Alamri, None; Rhonda Grebe, None; J S. Zigler, None; James T. Handa, None; Debasish Sinha, None Program Number: 6049 Poster Board Number: D0268 Presentation Time: 8:00 AM–9:45 AM Nrf2 and Hif1a have opposite responses to oxidative stress in ARPE-19 cells Hong Wei, James T. Handa. OPHTHALMOLOGY, JOHNS HOPKINS UNIVERSITY, BALTIMORE, MD. Purpose: Smoking causes oxidative stress and damage to the retina, reduces blood flow in eye tissue, and promotes ischemia, hypoxia, and micro-infarctions. It also induces cell death to retinal pigment epithelial (RPE) cells. This study explores how the RPE cell responds to stress induced by cigarette smoking and hypoxia, and how the transcription factor NF-E2-related factor 2 (Nrf2) and Hypoxia Inducible Factor 1 alpha (Hif1a), which has been reported to be induced by oxidative stress in addition to hypoxia, can influence the anti-oxidant response through JNK and p38 signaling. Methods: ARPE-19 cells were grown to confluence, serum starved for 24h, and then treated with cigarette smoke extract (CSE) and 1% O2 for 24h. The transcription factors Nrf2 and Hif1a proteins and MAPK pathway signaling were analyzed by Western blotting. Results: CSE induced a significant dose dependent increase in NRF2 protein (p<0.01) and a remarkable reduction in Hif1a (p<0.001) in ARPE-19 cells, while treatment with 1% O2 resulted in a significant increase in Hif1a (p<0.01) with no change in Nrf2. Since oxidative stress and hypoxia can either induce or impair JNK or p38 signaling, we next looked at phosphorylated JNK and p38 after CSE treatment. CSE decreased phospho-JNK (p<0.01) and increased phosphor-p38 (p<0.01) compared to control treated cells while 1% O2 increased phospho-JNK (p<0.01) and decreased phospho-p38 (p<0.05). Conclusions: NRF2 and Hif1a respond differently to oxidative stress in RPE cells, with differential signaling though Phospho-JNK and Phospho-p38, which can mediate inflammation, neovascularization, and apoptosis. Commercial Relationships: Hong Wei; James T. Handa, None Support: : EY019044, EY14005, RPB Physician Scientist Award, Unrestricted grant from RPB. Dr. Handa is the Robert Bond Welch Professor. Program Number: 6050 Poster Board Number: D0269 Presentation Time: 8:00 AM–9:45 AM Impaired lysosomal calcium signaling in RPE cells with an in vitro model of chloroquine retinopathy Nestor Mas Gomez2, Wennan Lu2, Jason Lim2, Alan Laties1, Kirill Kiselyov3, Claire H. Mitchell2, 4. 1Ophthalmology, University of Pennsylvania, Philadelphia, PA; 2Department of anatomy and cell biology, School of dental medicine, University of Pennsylvania, Philadelphia, PA; 3Department of Biological Sciences, University of Pittsburgh, Pittsburgh, PA; 4Physiology, University of Pennsylvania, Philadelphia, PA. Purpose: The incidence of chloroquine retinopathy is increasing, with patients experiencing a loss of central vision and photoreceptors. While the initial step in the pathology is widely acknowledged to involve lysosomal alkalinization by chloroquine, the links between alkalinization and photoreceptor death remain unknown. These abstracts are licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International License. Go to http://iovs.arvojournals.org/ to access the versions of record. ARVO 2016 Annual Meeting Abstracts The lysosome is a major source of intracellular Ca2+, and release of lysosomal Ca2+ through the TRPML1 channel is necessary for signaling and lysosomal exocytosis. In this study we examined the effect of an in vitro model of chloroquine retinopathy on the release of Ca2+ through TRPML1 in RPE cells and asked whether lysosomal exocytosis contains protective components Methods: For Ca2+ measurements, RPE cells from C57Bl6J mice and ARPE19 cells were grown on coverslips, loaded with fura-2 and the 340/380 nm signal was recorded using microscope or fluorimeter based readings. Exocytosis was recorded from mouse cells grown on transwell filters. IL-6 was measured with an Elisa kit and acid phosphatase (AP) with a colorimetric assay. TRPML1 mRNA levels were determined with qPCR. The in vitro model of chloroquine retinopathy consisted of ARPE-19 cells treated with 10 µM CHQ for 7 days and 1 µM U18666A for 1 day Results: The TRPML1 channel agonist ML-SA1 led to reversible and repeatable elevations in cytoplasmic Ca2+ in mouse RPE and ARPE19 cells. The response to ML-SA1 was significantly reduced in the chloroquine model. Treatment increased TRPML1 mRNA levels, while lysosomal Ca2+ levels released by glycyl-L-phenylalanine-betanaphthylamide were not altered; together this suggests the reduced Ca2+ efflux in the chloroquine model reflected a reduction in channel function. In polarized mouse RPE cells, ML-SA1 triggered a release of lysosomal enzyme AP across the apical membrane. Importantly, the cytokine IL-6 was also released in the same samples, suggesting lysosomal release of IL-6 following activation of TRPML1 Conclusions: The ability of ML-SA1 to trigger lysosomal Ca2+ release and concurrent exocytosis of acid phosphatase and IL-6 implies that lysosomal Ca2+ contributes to exocytosis in RPE cells. The reduced response to ML-SA1 in the in vitro chloroquine retinopathy model predicts that IL-6 release will also be reduced. As IL-6 is reported to protect photoreceptors, this reduced signal may be detrimental, although this awaits direct confirmation Commercial Relationships: Nestor Mas Gomez, None; Wennan Lu, None; Jason Lim, None; Alan Laties, None; Kirill Kiselyov, None; Claire H. Mitchell, None Support: EY-013434 Program Number: 6051 Poster Board Number: D0270 Presentation Time: 8:00 AM–9:45 AM The tyrosine phosphatase receptor PTPRZ1 inhibits β-catenindependent gene expression in RPE cells Jorgelina M. Calandria, Khanh Do, Swornim Shrestha, Nicolas G. Bazan. Neuroscience Center of Excellence, LSU Health Sciences Center, New Orleans, LA. Purpose: PTPRZ1 is a tyrosine phosphatase receptor that stabilizes the cadherin/catenin complex at cell-cell junctions. The retinal pigment epithelium (RPE) relies on these cell-cell interactions to maintain the integrity and polarization of the monolayer in the retina. β-catenin may be released from the complex and translocated into the nucleus. Neuroprotectin D1 (NPD1), a bioactive derivative of docosahexaenoic acid (DHA), modulates gene expression upon stress conditions (Calandria et al, 2015. Cell Death Differ). We propose that the inhibition of PTPRZ1 by HMGB1 promotes the release of β-catenin from the cell-cell junction complex and, along with NPD1, favors its nuclear translocation to modulate gene expression and promote survival of RPE. Methods: Primary RPE cells were exposed to 600 µM H2O2 and 10ng/ml TNFα to induce oxidative stress. β-catenin release and translocation was assessed by immunocytochemistry. Gene expression was measured by real-time PCR. Silencing and overexpression of PTPRZ1 and HMGB1 was carried out by transfecting RPE cells with siRNA and vectors carrying the open reading frame. TCF/LEF-β-catenin activity was measured using a TOP Flash system driving the expression of luciferase. Results: The addition of HMGB1 in the medium induced an increase in the mobilization of β-catenin from the membrane to the cytoplasm. The silencing of PTPRZ1 induced the same effect, suggesting that HMGB1 acts through the inhibition of the tyrosine phosphatase receptor. NPD1 and HMGB1 separately increased the nuclear translocation of β-catenin. Intriguingly, HMGB1, and not NPD1, induced a rise in TCF/LEF activity, suggesting that both effects proceed through different pathways. The wnt/β-catenin canonical genes CCND1, CLDN1 and MMP2 were modulated by HMGB1 but not affected by NPD1. On the other hand, FOXO3a-driven genes BIM, CDKN1A and CDKN1B expression was influenced by HMGB1 and NPD1 in an additive manner. Conclusions: Altogether, these results suggest that NPD1 and HMGB1 act cooperatively, but through different pathways, to modulate β-catenin activity. Suppression of PTPRZ1 plus addition of NPD1 lead to an increased mobilization and nuclear translocation of β-catenin, affecting its activity in a differently. The modulation of the β-catenin activity may be used as a therapeutic instrument to promote retinal cell remodeling and survival. Commercial Relationships: Jorgelina M. Calandria, None; Khanh Do, None; Swornim Shrestha, None; Nicolas G. Bazan, None Support: Supported by NIGMS grant P30 GM103340 and NEI grant R01 EY005121 (NGB) and Research to Prevent Blindness Program Number: 6052 Poster Board Number: D0271 Presentation Time: 8:00 AM–9:45 AM Effects of Anticancer Drug Cisplatin on Cybrids Containing A, B, D or L Haplogroup mtDNA Gregory Yung, Kunal Thaker, Marilyn Chwa, Sina Abedi, Tej Patel, Theresa Thai, Cristina M. Kenney. Gavin Herbert Eye Institute-UCI, Irvine, CA. Purpose: Mitochondria (mt) DNA can be categorized into haplogroups according to the accumulation of single nucleic polymorphisms (SNPs). While the influence of mitochondria in metabolism has been established, the extent to which mtDNA haplogroups influence intercellular metabolic activities is not well understood. It is recognized that different racial/ethnic populations have different susceptibilities to diseases and responses to medication. The present study characterizes the response of transmitochondrial cybrids containing either A (non-white Hispanic, B (non-white Hispanic), D (East Asian) and L (African origin) haplogroups to Cisplatin, a commonly used cancer drug. Methods: Cybrids were created by fusing platelets from donors with different ethnic/racial backgrounds with Rho0 ARPE-19 cells (lacking mtDNA). Cybrids containing either A (n=2), B (n=2), D (n=3), or L(n=5) haplogroups were treated with 40 µM Cisplatin for 48 hours and then cell viabilities were measured with the MTT assay. Data were calculated based upon 6-12 wells per cell line. The untreated cybrid values were normalized to 100% to serve as the baseline. An unpaired t-test was used to analyze the data (GraphPad Prism 5). Results: The L cybrids showed significantly reduced viability between Untreated cybrids (100% ± 5.42) and Cisplatin-treated groups (69.75 ± 5.81%, p = 0.0003). D cybrids had decreased cell viability after Cisplatin treatment (77.39 ± 2.13%, p < 0.0001) compared to Untreated D cybrids (100% ± 1.58). The A cybrids also showed lower cell viability (80.11 ± 3.75%) compared to Untreated A cybrids (100% ± 3.83, p = 0.0007). In contrast, Cisplatin-treated B cybrids had similar levels of cell viability compared to Untreated B cybrids (95.27 ± 4.96% versus 100% ± 8.09, p = 0.62). These abstracts are licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International License. Go to http://iovs.arvojournals.org/ to access the versions of record. ARVO 2016 Annual Meeting Abstracts Conclusions: Our results show the degree of cytotoxic effects of Cisplatin can vary depending upon the mtDNA haplogroup contained within the cells; L (African origin) cybrids > D (Asian origin) cybrids > A (non-white Hispanic) cybrids > B (non-white Hispanic) cybrids. These data suggest an individual’s mtDNA background may play a greater role than previously thought in responses and side effects to Cisplatin and may redefine the appropriate anticancer treatment and chemotherapy for different racial groups. Commercial Relationships: Gregory Yung, None; Kunal Thaker, None; Marilyn Chwa, None; Sina Abedi, None; Tej Patel, None; Theresa Thai, None; Cristina M. Kenney, None Support: Discovery Eye Foundation, Guenther Foundation, Beckman Initiative for Macular Research, Polly and Michael Smith Foundation, Max Factor Family Foundation, Iris and B. Gerald Cantor Foundation Program Number: 6053 Poster Board Number: D0272 Presentation Time: 8:00 AM–9:45 AM Varying Mitochondrial Haplogroups Show Differential Expression Levels of Histone Proteins Jon L. Norman, Shari R. Atilano, Marilyn Chwa, Kunal Thaker, Cristina M. Kenney. Gavin Herbert Eye Institute - UCI, Irvine, CA. Purpose: Gene expression can be regulated by how tightly DNA associates with the nuclear proteins called histones, which are responsible for nucleosome structure integrity and gene regulation in eukaryotes. Recent studies show that cybrids (cytoplasmic hybrids), with identical nuclei but different mitochondrial (mt) DNA, differentially express methylation and acetylation genes. Our hypothesis is that different mtDNA haplogroups, representing populations from different geographic origins, have altered expression levels of histone proteins genes. The present study was conducted to determine if cybrids with mtDNA haplogroups H (European origin), L (African origin), and K (European origin – most common in Ashkenazi Jews) show differences in expression levels for histone genes. Methods: Cybrids were created by fusing Rho0 ARPE-19 cells (devoid of mtDNA) with platelets isolated from Haplogroup H (n=8), L (n=6), or K (n=7) individuals. Haplogroups were determined by DNA analyses with PCR/restriction enzyme digestion and sequencing. Gene expression was analyzed by RT-qPCR for four histone genes using ALASv1 as a reference gene. Results: L cybrids had lower expression levels for the histone genes HIST1H3F (2-fold, p=0.0003), HIST1H3H (0.44-fold, p=0.008), and HIST1H4H (0.63-fold, p=0.003) when compared to haplogroup H cybrids. K cybrids had lower expression levels for the histone genes HIST1H3A (0.3-fold, p<0.0001), HIST1H3H (0.28-fold, p<0.0001), and HIST1H4H (0.5-fold, p<0.0001) when compared to haplogroup H cybrids. The H cybrids values were assigned a value of 1. Conclusions: Cybrids, which have identical nuclei but either H, L, or K haplogroup mtDNA, have differing RNA expression levels for histone genes. Previous studies have demonstrated that different mtDNA haplogroups mediate different expression levels of genes related to complement, inflammation, and apoptosis, but the mechanism(s) of retrograde signaling are not understood. Our results show that H, L and K cybrids also have unique expression levels of HIST1H3A, HIST1H3F, HIST1H3H and HIST1H4H, which would lead to variability in the nucleosome core and may influence acetylation/methylation status of cells and their retrograde signaling abilities. These factors may play a key role in gene regulation and in determining why cells from unique ethnic/racial origins are more susceptible to certain eye diseases. Commercial Relationships: Jon L. Norman, None; Shari R. Atilano, None; Marilyn Chwa, None; Kunal Thaker, None; Cristina M. Kenney, None Support: Discovery Eye Foundation, Guenther Foundation, Beckman Initiative for Macular Research, Polly and Michael Smith Foundation, Max Factor Family Foundation, Iris and B. Gerald Cantor Foundation. Program Number: 6054 Poster Board Number: D0273 Presentation Time: 8:00 AM–9:45 AM Comparison of Lipid and Cholesterol-Biosynthesis Gene Products in H- and K-mt-DNA haplogroup cybrids Christine Garabetian1, Kunal Thaker1, Javier Cáceres-del-Carpio1, Shari R. Atilano1, Marilyn Chwa1, Anthony B. Nesburn1, 2, Cristina M. Kenney1, 3. 1Gavin Herbert Eye Institute, University of California Irvine, Irvine, CA; 2Cedars-Sinai Medical Center, Los Angeles, CA; 3Department of Pathology and Laboratory Medicine, University of California Irvine, Irvine, CA. Purpose: Cholesterol has been associated in the pathogenesis of neurodegenerative and systemic pathologies, and certain populations have higher incidence of cholesterol-linked familial diseases. The purpose of this experiment was to determine if mitochondrial DNA (mt-DNA) haplogroup H and K transmitochondrial cybrids regulate the cholesterol and lipid biosynthesis pathways differently. Using transmitochondrial cybrids, cell lines with identical nuclei but either H or K mtDNA, we looked at the relationship between the gene expression levels of cholesterol-related genes and neutral lipid deposits in H and K cybrids. Methods: Transmitochondrial cybrids were created by fusing mitochondria (mt) isolated from human platelets and cells lacking mtDNA; thus these cybrids have identical nuclei but either H or K mtDNA. RNA from H and K cybrids were analyzed using GeneChip arrays (n=3). Specific cholesterol-related genes FDFT1, SQLE, LSS, CYP51A1, EBP, and DHCR24 were analyzed using Q-PCR (H, n=7; K, n=9). Corresponding reference genes were used as housekeepers for each of the six genes studied. H and K cybrids (n=10) were seeded at 104/well and stained with HCS LipidTox Green Neutral Lipid Stain. Statistical analyses were performed using student’s t-test. Results: Q-PCR showed insignificant differences of the cholesterol biosynthesis genes in H vs. K; however, one K cybrid (CY#13-57) showed elevated expression of 4 out of 6 cholesterol biosynthesis genes compared to H cybrids; (17.4-fold increase in FDFT1 expression, a 31-fold increase in SQLE expression, a 10.8-fold increase in CYP51A1 expression, and a 5.4-fold increase in EBP). LipidTox staining showed cytoplasmic distribution of the lipids with significantly elevated intensity in the CY#13-57 compared to H cybrids or other K cybrids (p<0.001). Conclusions: Due to all cybrids differing only in their mtDNA content, our findings suggest that H- and K-haplogroup mt-DNA do not necessarily affects genes involved in cholesterol biosynthesis. However, the increased expression of cholesterol-related genes and larger and more vibrant lipid deposits seen in CY#13-57 suggest the mt-DNA in that specific K cybrid is different than other K cybrids and is mediating cholesterol production and lipid transport through some mechanism. Commercial Relationships: Christine Garabetian, None; Kunal Thaker, None; Javier Cáceres-del-Carpio, None; Shari R. Atilano, None; Marilyn Chwa, None; Anthony B. Nesburn, None; Cristina M. Kenney, None These abstracts are licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International License. Go to http://iovs.arvojournals.org/ to access the versions of record. ARVO 2016 Annual Meeting Abstracts Support: Discovery Eye Foundation, Guenther Foundation, Beckman Initiative for Macular Research, Polly and Michael Smith Foundation, Max Factor Family Foundation, Iris and B. Gerald Cantor Foundation, J.C.d.C. was the David & Julianna Pyott PanAmerican-Retina Research Fellowship Program Number: 6055 Poster Board Number: D0274 Presentation Time: 8:00 AM–9:45 AM Pax6 role in the regulation of retinal pigmented epithelium maturation Yamit Cohen1, Pablo Blinder2, Maria Idelson3, Benjamin Reubinoff3, Shalev Itzkovitz4, Ruth Ashery-Padan1. 1Department of Molecular Genetics and Biochemistry, Tel-Aviv University, Tel-Aviv, Israel; 2 Department of Neurobiology, Tel Aviv University, Tel Aviv, Israel; 3 Department of Gynecology, Hadassah-Hebrew University Medical Center, Jerusalem, Israel; 4Department of Molecular Cell Biology, Weizmann Institute of Science, Rehovot, Israel. Purpose: Haploinsufficiency of Pax6 was long shown to cause aniridia and was reported to involve neovascularization of the cornea. However, Pax6 involvement in vascularization remained elusive and lack of imaging techniques for the choroid vasculature limited further inquiry regarding the effect on this tissue. This research aims to determine the molecular mechanism in which Pax6 regulates retinal pigmented epithelium (RPE) maturation and choroid development using somatic mutagenesis in mouse models and RPE cells derived from human embryonic stem cells (hES-RPE). Methods: To this purpose, conditional mutations are induced in mice and gain of function analysis is conducted using sub-retinal injections followed by electroporation. Quantitative expression levels are measured using single molecule FISH and the choroidal phenotype is analyzed using a novel perfusion technique. Further investigation of the molecular mechanism involved in this regulation is performed using hES-RPE cells and includes chip-seq, RNA-seq and knockdown using viral infections. Results: Pax6 deletion in the specified RPE resulted in a phenotype of aniridia and among the changes in gene expression we observed an up-regulation in Sox9 expression level, a key transcription factor in organogenesis which is related to late stages of RPE differentiation. Pax6 and Sox9 expression patterns were determined in course of RPE differentiation in wild type mice and mice with Pax6 specific ablation from the RPE. Quantitative expression analysis illustrated opposite correlation and gain of function analysis confirmed this observation since miss-expression of Pax6 resulted in inhibition of Sox9 expression. Conclusions: This study is the first to document Pax6 role in timing RPE differentiation through its regulatory relations with Sox9. Future efforts are aimed to elucidate the molecular mechanism of this regulation and examine a possible effect on the choroid vasculature. Commercial Relationships: Yamit Cohen, None; Pablo Blinder, None; Maria Idelson, None; Benjamin reubinoff, None; shalev Itzkovitz, None; Ruth Ashery-Padan, None Support: ISVER travel grant award for best presentation These abstracts are licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International License. Go to http://iovs.arvojournals.org/ to access the versions of record.