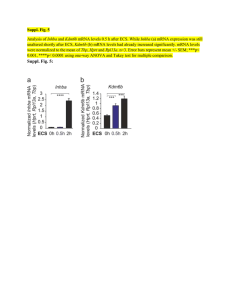
Inferring Genetic Architecture of Complex Biological Processes
advertisement

Inferring Genetic Architecture
of Complex Biological Processes
Brian S. Yandell12, Christina Kendziorski13, Hong Lan4,
Jessica Byers45, Elias Chaibub1, Alan D. Attie4
CIBM Training Program Retreat 2004
1
Department of Statistics
2 Department of Horticulture
3 Department of Biostatistics & Medical Informatics
4 Department of Biochemistry
5 Department of Nutritrional Sciences
University of Wisconsin-Madison
http://www.stat.wisc.edu/~yandell/statgen
13 October 2004
Statistics: Yandell © 2004
1
Gene mapping infers the relationship between genotype and
phenotype in a segregating population. We map thousands of mRNA
expression phenotypes, or expression QTL, using dimension
reduction methods to uncover correlated genetic architecture,
including number and location of genomic regions as well as gene
action and epistasis. We show a novel blending of principal
components and discriminant analysis with functional information
to detect multiple expression QTL that together may affect the
expression of many correlated mRNA. These common patterns of
gene action are largely overlooked by simple interval mapping when
conducted separately for each mRNA. In our current study with 60
F2 mice from a B6-BTBR ob/ob model of diabetes and over 40,000
mRNA measured with Affymetrix chips, we find three pairs of
genomic regions of particular interest associated with signal
transduction, apoptosis, and lipid metabolism. We propose to join
genetic architecture with graphical models of biochemical activity.
Our approach is directly applicable to gene mapping for other
“omic” measurements on the horizon.
13 October 2004
Statistics: Yandell © 2004
2
glucose
Statistics: Yandell © 2004
(courtesy AD Attie)
13 October 2004
insulin
3
studying diabetes in an F2
• mouse model: segregating panel from inbred lines
– B6.ob x BTBR.ob F1 F2
– selected mice with ob/ob alleles at leptin gene (Chr 6)
– sacrificed at 14 weeks, tissues preserved
• physiological study (Stoehr et al. 2000 Diabetes)
– mapped body weight, insulin, glucose at various ages
• gene expression studies
– RT-PCR for a few mRNA on 108 F2 mice liver tissues
• (Lan et al. 2003 Diabetes; Lan et al. 2003 Genetics)
– Affymetrix microarrays on 60 F2 mice liver tissues
• U47 A & B chips, RMA normalization
• design: selective phenotyping (Jin et al. 2004 Genetics)
13 October 2004
Statistics: Yandell © 2004
4
The intercross (from K Broman)
13 October 2004
Statistics: Yandell © 2004
5
interval mapping basics
•
observed measurements
– Y = phenotypic trait
– X = markers & linkage map
observed
• i = individual index 1,…,n
•
missing
• alleles QQ, Qq, or qq at locus
Q
unknown quantities
– M = genetic architecture
– = QT locus (or loci)
– = phenotype model parameters
•
Y
missing data
– missing marker data
– Q = QT genotypes
•
X
unknown
pr(Q=q|X,) genotype model
after
Sen & Churchill
(2001 Genetics)
– grounded by linkage map, experimental cross
– recombination yields multinomial for Q given X
•
f(Y|q) phenotype model
– distribution shape (assumed normal here)
– unknown parameters (could be non-parametric)
13 October 2004
Statistics: Yandell © 2004
M
6
genetic architecture: heterogeneity
•
heterogeneity: many genes can affect phenotype
–
–
–
•
different allelic combinations can yield similar phenotypes
multiple genes can affecting phenotype in subtle ways
multiple genes can interact (epistasis)
genetic architecture: model for explained genetic variation
–
–
loci (genomic regions) that affect trait
genotypic effects of loci, including possible epistasis
M = {1, 2, 3, (1, 2)}
= 3 loci with epistasis between two
q = 0 + 1q + 2q + 3q + (1,2)q
= linear model for genotypic mean
= (1, 2, 3)
= loci in model M
= possible genotype at loci
= genotype for each individual at loci
q = (q1, q2, q3)
Q = (Q1, Q2, Q3)
13 October 2004
Statistics: Yandell © 2004
7
hetereogeneity: many genes affect each trait
3
(modifiers)
minor
QTL
polygenes
1
2
major
QTL
0
3
additive effect
major QTL on
linkage map
2
1
13 October 2004
0
4
5
5
10
15
20
25
30
rank order of QTL
Statistics: Yandell © 2004
8
SCD mRNA expression phenotype
2-D scan for QTL (R/qtl)
epistasis
LOD
peaks
13 October 2004
joint
LOD
peaks
Statistics: Yandell © 2004
9
2M observations
30,000 traits
60 mice
13 October 2004
Statistics: Yandell © 2004
10
modern high throughput biology
• measuring the molecular dogma of biology
– DNA RNA protein metabolites
– measured one at a time only a few years ago
• massive array of measurements on whole systems (“omics”)
– thousands measured per individual (experimental unit)
– all (or most) components of system measured simultaneously
•
•
•
•
whole genome of DNA: genes, promoters, etc.
all expressed RNA in a tissue or cell
all proteins
all metabolites
• systems biology: focus on network interconnections
– chains of behavior in ecological community
– underlying biochemical pathways
• genetics as one experimental tool
– perturb system by creating new experimental cross
– each individual is a unique mosaic
13 October 2004
Statistics: Yandell © 2004
11
finding heritable traits
(from Christina Kendziorski)
•
reduce 30,000 traits to 300-3,000 heritable traits
•
probability a trait is heritable
pr(H|Y,Q) = pr(Y|Q,H) pr(H|Q) / pr(Y|Q)
Bayes rule
pr(Y|Q) = pr(Y|Q,H) pr(H|Q) + pr(Y|Q, not H) pr(not H|Q)
•
phenotype given genotype
pr(Y|Q, not H) = f0(Y) = sum f(Y| ) pr()
pr(Y|Q, H) = f1(Y|Q) = productq f0(Yq )
if not H
if heritable
Yq = {Yi | Qi =q} = trait values with genotype Q=q
13 October 2004
Statistics: Yandell © 2004
12
hierarchical model for expression phenotypes
(EB arrays: Christina Kendziorski)
YQQ QQ ~ f QQ
YQq Qq ~ f Qq
mRNA phenotype models
given genotypic mean q
Yqq qq ~ f qq
QQ
qq
Qq
common prior on q across all mRNA
(use empirical Bayes to estimate prior)
q ~ pr
QQ
13 October 2004
Qq
qq
Statistics: Yandell © 2004
13
expression meta-traits: pleiotropy
• reduce 3,000 heritable traits to 3 meta-traits(!)
• what are expression meta-traits?
– pleiotropy: a few genes can affect many traits
• transcription factors, regulators
– weighted averages: Z = YW
• principle components, discriminant analysis
• infer genetic architecture of meta-traits
– model selection issues are subtle
• missing data, non-linear search
• what is the best criterion for model selection?
– time consuming process
• heavy computation load for many traits
• subjective judgement on what is best
13 October 2004
Statistics: Yandell © 2004
14
7.6
-0.2
7.8
-0.1
8.0
ettf1
8.2
PC2 (7%)
0.0
0.1
8.4
0.2
8.6
PC for two correlated mRNA
8.2
8.4
8.6
13 October 2004
8.8
9.0
etif3s6
9.2
9.4
Statistics: Yandell © 2004
-0.5
0.0
PC1 (93%)
0.5
15
PC across microarray functional groups
Affy chips on 60 mice
~40,000 mRNA
2500+ mRNA show DE
(via EB arrays with
marker regression)
1500+ organized in
85 functional groups
2-35 mRNA / group
which are interesting?
examine PC1, PC2
circle size = # unique mRNA
13 October 2004
Statistics: Yandell © 2004
16
84 PC meta-traits by functional group
focus on 2 interesting groups
13 October 2004
Statistics: Yandell © 2004
17
red lines: peak
for PC meta-trait
black/blue: peaks
for mRNA traits
arrows: cis-action?
13 October 2004
Statistics: Yandell © 2004
18
DA meta-traits: separate pleiotropy
from environmental correlation
pleiotropy only
13 October 2004
environmental
correlation only
Statistics: Yandell © 2004
both
Korol et al. (2001)
19
interaction plots for DA meta-traits
DA for all pairs of markers:
separate 9 genotypes based on markers
(a) same locus pair found with PC meta-traits
(b) Chr 2 region interesting from biochemistry (Jessica Byers)
(c) Chr 5 & Chr 9 identified as important for insulin, SCD
13 October 2004
Statistics: Yandell © 2004
20
B.H
PC ignores genotype
2
A.B
A.B
H.H
H.B
H.H
H.HA.H
A.H A.H
H.B
B.H
H.H
H.A
A.A H.HB.B
H.H
H.B
H.A
H.H
A.H
-10
-10
0
5
10
1
B.H
A.B
A.B
H.HB.H
A.A
B.AH.A
A.A
A.A H.H
H.B
H.H
H.A H.H H.H
H.B A.B H.A A.B
H.A
H.H
H.BH.H
B.H
B.H
H.H
H.A
B.H
B.H
B.H B.H
H.B
H.H
-10
2
3
4
H.B
B.A
2
B.H
H.A
A.H
1
H.H
B.H
H.H
H.B
A.HH.B
H.H H.B
H.H
H.A
H.H
H.H
B.B
A.A
B.A
H.A
B.H
H.HH.A
H.H
B.A
B.H
B.H
A.H
B.H
A.H
A.A
H.H
B.H
A.B
A.A
H.H
A.B
A.B
A.B
B.H
1
H.A
-2
H.H
A.B B.H
H.B
A.B
H.HH.A
H.B
0
H.B
0
DA1 (37%)
H.B
A.H
A.HA.HA.H
A.H
H.H
A.A
A.H
H.A
A.H B.A
-1
A.H
note better
spread of circles
B.B
-2
A.H
A.H
H.A
H.A
B.H
A.A B.H
A.H
A.B
B.H
H.H
A.H
A.HH.H
B.H
A.B
A.H
B.B
-3
1
0
B.A
A.BB.H
-3
A.H
A.H
A.H
A.H
A.B
-1
A.H
correlation of
PC and DA meta-traits
B.B
15
3
PC1 (25%)
B.B
-2
H.H
H.A
H.H
B.H
B.B
A.B
A.A
H.H
A.A
H.H A.A
A.B
DA2 (18%)
2
-5
A.H
H.B
A.A
H.H
H.A
B.A
H.H
H.H
H.H
B.H
H.H
A.H
-1
DA1 (37%)
3
4
-15
-3
B.H
B.H
B.A
H.H
H.A
-3
H.B
H.A
-15
13 October 2004
H.H
H.A
A.H
H.B
H.A
A.B
H.B
DA creates best
separation by
genotype
0
H.A
B.A
A.A A.A
H.B
H.B
B.H H.H
B.H
B.H
-1
B.H
H.H
B.A
B.H H.A
H.H
B.H
B.H
A.H
A.H B.A
DA2 (18%)
H.A
B.H
A.H
H.H
B.H
B.H
-2
10
5
0
B.B A.B
A.B
B.H
B.H
-5
PC2 (12%)
A.H
B.H
B.A
B.H
H.A
H.A
A.HA.A
B.H
A.B
DA uses genotype
H.B
H.A
H.B
A.H
PC captures
spread without
genotype
H.B
A.B
H.H
H.H
genotypes from
Chr 4/Chr 15
locus pair
(circle=centroid)
3
comparison of PC and DA meta-traits on 1500+ mRNA traits
B.H
A.B
-5
0
PC1 (25%)
5
10
15
Statistics: Yandell © 2004
-10
-5
0
PC2 (12%)
5
10
21
SCD trait
log2 expression
DA meta-trait
standard units
relating meta-traits to mRNA traits
13 October 2004
Statistics: Yandell © 2004
22
graphical models
(with Elias Chaibub)
QTL
DNA
RNA
QTL
D1
R1
D2
13 October 2004
R2
Statistics: Yandell © 2004
unobservable
protein
meta-trait
P1
observable
cis-action?
P2
observable
trans-action
23
building graphical models
• infer genetic architecture of meta-trait
• find mRNA traits correlated with meta-trait
• apply meta-trait genetic architecture to mRNA
– expect subset of QTL to affect each mRNA
• build graphical models QTL RNA1 RNA2
– class of possible models
– find best model as putative biochemical pathway
• parallel biochemical investigation
– candidate genes in QTL regions
– laboratory experiments on pathway components
13 October 2004
Statistics: Yandell © 2004
24