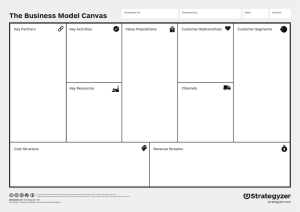
>> Lee Dirks: So I'd like to introduce Peter... the exciting and very interesting work that they're doing with...
advertisement

>> Lee Dirks: So I'd like to introduce Peter Binfield from PLoS. And to talk about the exciting and very interesting work that they're doing with article-level metrics at PLoS ONE. >> Peter Binfield: Thanks. So is the mic here -- is this working, microphone? Okay. So Pete Binfield of PLoS. I actually run PLoS ONE. I've been working at PLoS for a couple of years on PLoS ONE. And you'll see I've put the slides up on that URL down there. So if anyone wants to follow along at home, that's where the slides are. Okay. So I'll give you a quick introduction to the Public Library of Science and then talk a little bit about how we're effectively what we think re-thinking the way the academic journal works and should work. And probably the best example of us doing that at the moment is our article-level metrics program. So all of us today I think are here for, you know, a couple of good reasons. We want to accelerate new proved science, this is the science commons event, for the benefit of everyone, society, all of mankind. And I'm here basically to discuss this in the context of academic scholarly publishing. A lot of what we've heard this morning have been real scientists talking about the real science. So I'm at the other end of the chain, taking this stuff and making it public. So Public Library of Science is six years old now. We're an open access accomplisher. We're web native, which means we were created in the web era. All of our content is born digital, so it -- there's no paper component where we have an online only delivery. We have an net friendly business model, which is the Open Access business model, which is quite scalable in web environment. We're right now the largest not-for-profit Open Access publisher. Published seven OA journals. And you know, we're -- although in the grand scheme of things in scholarly publishing, we're actually a small publisher. We're making a lot of -- a lot of noisy think in the environment. And I'll talk about some of that. We're based in San Francisco and Cambridge UK. So these two, those are the first two journals ever published. I think there's some debate as to which one was really the first real journal, but a lot of people claim the philosophical transactions. Wikipedia claims it's the Le Journal des Scavans. But basically these are back in 1660, 1665ish. The journal was invented and really hasn't changed and awful lot since then. There's really typically people recognize there's four functions of a journal. Registration, so registering whether you're the first person to publish that work or come up with something. Certification. Some sort of seal of authority. Dissemination. You have to get it out there. People have to read it. And archiving. People have to find it in the future. And historically these have been the 4 main functions the journal does and has done since 1665 and continues to do now. But also I think journals now do a couple of other things. So perhaps you know, they're not necessarily doing the right things, but I think they're filtering for quality as well, so when you read a journal like Nature they have filtered for quality. They've preselected some papers that they think are the best in their field. And they journal for topic. So if you read the journal of obscure subject X you know everything in that journal is in that type of skill topic. And sometimes that's referred to as scope. Scope of the journal as well. So let's drill down into a few of these because I think, you know, they need some debate. Registration. That's just -- that's pretty trivial in today's environment of course. I think -- and I think there's actually very few debates now where people would argue that they have the first discovery of a thing by going to the journal literature. I think they probably go to a blog or a Twitter feed or something like that. But registering whether or not you are the first person to do something is quite easy in the current web environment. Dissemination, web dissemination is obviously trivial these days. You could publish something in the work and read it seconds later. Archiving I would also saying is pretty trivial these days. Although you know the archiving problem is by no means solved. It's a difficult problem. I think the fact that there are multiple copies of web pages or content around the world, you know, means that you don't need to pay -- publish a paper version of a journal and physically send it to a thousand libraries around the world in order to save archived. There are electronic archiving solutions. I think as well the filtering for topic. I think that again is something that's perhaps trivial now almost. With a search engine, do you need to actually go to a journal on a specific topic to find everything on that topic or can you just type it into the search and get everything? Or can you go to a fielded search and drill down by the topic hierarchy, for instance? You don't need to go anymore to a journal to find everything on that topic. So if we take out all those things that I think are easy to do now or easy, you know, you've got these couple of things left. Certification, filtering for quality. Certification. That's basically peer review in this setup. So peer review is pre-publication evaluation work. It's the opinion of a small number of people, usually a couple of people. It's a very confidential, very secretive process for some good reasons. It's often very subjective and it's often based on quite ill-defined criteria. So peer review, is my peer review paper differently depending on which individuals they are or which journal they're working for or what they've been told to look for? It's at the risk of bias, I think, based on decisions which have nothing to do with the science. So the peer reviews may not necessarily be commenting on whether the science is good or bad, they may be saying it's not within the scope of the journal or it's not of a high enough quality. And it's really supposed to be about the science. But often it's not. So peer review is an issue, I think, in this sort of list of things the journal does. And they also filter for quality. So this is what happens to a typical paper or an -a not untypical paper. The following process happens: It gets submitted, usually to Nature. It gets reviewed possibly and rejected straight away. It goes to another one down. They revise the paper, the submit it to the next journal down in their imaginary hierarchy of journals. It gets reviewed, submitted, rejected, reviewed, submitted, rejected, so on, so on. Repeat until successful and finally journal X will publish your paper. And it's depressing how often this happens. So, you know, that paper found a home. Great. How long did that take? You know, how many months were spent going through that chain of events? How many people had to look at that paper, waste time peer reviewing it only for it to be rejected out of scope or not good enough quality. How much opportunity cost was wasted? You know, the authors wanted to move on to something else. They didn't want to spend their entire life trying to get this paper published. In addition, that paper was filtered. So a journal publishes. So you could say a filtering happened there. Was that actually a good way to do the filtering? Or is this filter failure? You know, it's not -- it's the typical quote it's not information overload, it's filter failure. And I think this is filter failure. These things can take months or years to happen. And you know, I'm not making this stuff up. So this is a paper. I put a call out on FriendFeed this week for a couple of examples of this happening. This paper was submitted to Nature in 2003, rejected as out of scope. It was then submitted to five more journals. Going down that chain it was repeatedly rejected. Finally the authors were told to split the paper in two before somebody would publish them. The most recent journal that actually rejected them as being out of scope went on to publish the competing paper a few weeks later. The two halves were finally published at the end of 2006 in two different journals. And one of those halves, one of those papers actually made the cover of the journal. It can't have been that bad. But it took them four years to get that paper published in an unsatisfying way. This is another example. Cameron might recognize this paper. It was reject by Nature, Science, Nature Biotech, Nature Chemical Biology. Went through multiple rewrites; got cut in half. One-half finally got published 18 months later in NAR. This is Cameron's paper. It now has over 80 citations which is, you know, a very high number, even for a Nature publication. Cameron claims that, you know, if it had been published earlier it would have advanced the field. Why did it have to wait 18 months going through this chain of events? The other half went to 17 journals before it finally appeared in a journal which nobody reads because it's not even online. This was a horrible experience, Cameron. Okay. So how did this process actually accelerate and improve science for the benefit of humanity? It didn't, did it? And who benefited? The authors didn't benefit here. The society didn't benefit. That knowledge was locked up for years. Science didn't benefit. You could argue that the papers were approved by these multiple rewrites. But to the extent that this amount of wastage happened, these are not unusual stories. So what is the answer? Well, it is PLoS ONE. Of course. We satisfy many of those criteria of the traditional definitions of a journal, and we do it, we think, in a superior way. So we're Open Access. We have the widest possible dissemination of our content. We're online only. There's no size limitations to our papers. We've published papers that are 200 pages long. A paper journal cannot do that because they have a limitation of a number of pages to publish. We have no topic or scope limitations. We set ourselves a scope of the whole of science. Although in reality we're mostly in the biomedical areas. And we have a scalable business model. So the business model is that there's a publication fee which is charged after acceptance and upon publication basically. So that's scalable. Each individual publication pays its own costs in that model. However, I think the two really interesting things that we do, we have a different type of peer review question that is asked. All we ask our peer reviewers is is it scientific? Is the science sound? Is it publishable? Would this paper be published somewhere after going all the way down that train to journal X? And that's all we ask them. We don't ask and how could it be improved or is it a major advance in the field or anything like that. They can choose to answer those questions but that's not part of the acceptance criteria. And in addition, apart from these basic acceptance criteria, we do have seven criteria and it has to be scientific. The data has to follow the method. You know, it has to be in English. There's some pretty basic criteria. But other than that, there's no filtering for quality. So we're really not asking our peer reviewers is this a major advance because we only want to publish the very best stuff. What we want to publish is everything that is publishable. So basically everything that passes our peer review and is therefore publishable is published. And we think that this way we're getting good science in front of the right people as fast as possible. And I think those are the two elements of PLoS ONE that have made it so stand out. And I think have made it the success that it is right now. So is it a success? Well, it is. This journal is absolutely unparalleled in the history of the industry. We launched in December 2006, so we're now four years old. And this year we're the largest journal in the world. 2009 we were the third largest journal in the world. So this is our statistics here. We're publishing last year 4404 articles. There's only two journals that did more than that that year. And the final column is interesting. We last year published a half a percent of everything that was published in PubMed. Can anyone think what that number might be for PubMed Central, PMC? We were almost eight percent of PMC last year in one journal. We have amazing community acceptance. 50,000 authors have published with us now. We have 1,000 academic editors. Several are in this room. And we believe we're promoting a real paradigm shift here with what we're doing. We believe we're allowing people to move from thinking about the journal to the article. In the past from 60 to 65 until four years ago, it's all about the journal, the journal being a container or a package for the content. We're moving past that now. And we really think that we're accelerating the scientific process by doing this. We're doing people a great benefit we think. Okay. So how are we doing that? Well, one of the really interesting things that we're doing I think is article-level metrics, which is what this is now moving on to. We're attempting to instead of evaluating a journal via an impact factor, we are evaluating articles via something more meaningful than the impact effect of the journal that they happened to have made it into via that random route of making it down to the right journal. So does anyone know what this is? This is the past. This was the first journal published. This is an era when dinosaurs stalked the Earth, stomping small mammals under their feet. And people didn't even have cell phones back then. It was a very dark and dismal era. But we don't live in the past. We live in the future. And this is product being put out from some company in California. We live in the future and we shouldn't have to accept the way that the industry or the business scientific publishing has been set up. We have better tools now and faster tools. And that's what we're trying to promote here. So if we start to think just about the article, how could we measure the impact or the quality or the degree of advance or whatever you want to call it of an article? Degree of relevance to myself. All of those kind of things at the moment are just packaged up into the journal. But the -- for academic publishing, the unit of publication is the article, not the journal. And perhaps in the future it's something else. But right now we worry about articles. So we could track citations, web usage, expert ratings, social bookmarking, community rating, media coverage, blog coverage, commenting activity and more potentially. And there's been papers published with a big long list of what you could do here. And the fact is that it's only now in the web environment that this is possible. There's an entire ecosystem now of third parties that are basically doing a lot of this stuff. They're starting to track a lot of this data, specifically for academic papers. So the obvious one, the one that I think most people still across is probably the gold standard is citations. Citations are obviously tracked by, you know, some big people. Scopus, Web of Science, PubMed Central, Cross-Ref, and so on. So we track citations to all of our articles from Scopus, PubMed Central and CrossRef. We generate the web usage statistics for every one of our articles. And we provide that in three formats, HTML, PDF, and XML usage to certain COUNTER standards which people in the library world would have heard of here. Although, those standards were not developed for article-level metrics, they were developed for journal-level. We don't have expert ratings yet. But there's people who do that factor of a thousand, for instance. We track social bookmarking activity on a couple of big social bookmarking cites. In the academic world CiteULike and Connotea are the equivalent of Delicious, for instance. We allow people to leave star ratings on our articles in three different categories. We track media and blog coverage from four major blog aggregators in the scientific field. Postgenomic there is actually the largest aggregator that generates this data for us. And we allow commenting activity on all of our articles so people can leave notes, comments a discussion forum on every article. And I'm about to show you some of this. All of this data is openly available except the web usage, which is generated from our own web blogs. So there's no reason that any other publisher couldn't do exactly what we're doing. It's all via open APIs. We've published the list of the APIs we use. We've told everyone how we've done this. Anyone can do this. It's not rocket science. Maybe generating your web usage data is. So the important thing here is these things are not just about citations and usage. This is a whole basket of metrics, which the assumption is that in some way this vast metrics provides you with some insight into the article. And I always put the word impact here. It's not just impact we're talking about, it's degree of advance, relevance to myself, that kind of thing. It's at the article level, not the journal level. It's for every assemble article we've ever published, going back through our corpus. And it's not just about that evaluation of whatever quality relevance, it's also a way to filter and discover. So in the future, and this is coming down the line in the next few months, we'll be adding for instance the ability to search new results and sort them based on this article or metrics data. So perhaps you want to search that says just show me all the articles with more than 10 social bookmarks, for instance, and rank it by usage. We're going to be doing that very soon. And we're the first people to really do this properly we think. And wear really hoping that everyone else does it as well. Because we think this is a big deal. We think it makes a difference. And, you know, hopefully standards will evolve and people will be able to, you know, compare these metrics against different publishers. Okay. So what does it look like? So I'm going to attempt to use the web here. They always say never work with children, dogs, or the live web. Here we go. So this is an interesting article we published. Very relevant to today's topics. Apparently sharing detailed research data increases your citation rate, which is nice. Okay. So when you're in the article, you have the plain HTML page here. But you'll see at the top there's some tabs. Metrics -- I'm sorry. Metrics related content and comment. And you'll also see there's a bit of a summary of some of the metrics here. But everything interesting here is happening under the metrics tab. So if we click here, you'll see that graph dynamically built. So this is a graph of all the usage over time. This article has had 13,206 downloads broken down into these different view types. If you hover over each point, it gives you the monthly breakdown and the total breakdown. >>: [inaudible]. >> Peter Binfield: A what? >>: 13,000, is that a lot. >> Peter Binfield: That's a good question. It is a lot. And we provide some information because nobody knows whether 13,000 is a big or a small number in this field, because nobody's done it before. So we've actually provided some summary table showing what is the average number of downloads per year per title per subject area. And actually I don't really have time to show those screen shots. But normally it's part of my presentation. It's been cited 11 times. CrossRef has found 11 citations, PubMed Central five and Scopus 12. If you click on each of these, you go to a landing page at that third party which then gives you the information. Note here Scopus is a subscription product, however, they know the deal. They're sending you to a preview. So without the subscription to Scopus which costs a lot of money, they show you the first 20 citations which is great. And we feel that, you know, we're not -- we're not completely, you know, stupid. That that's as good as we can expect out of Scopus. Web of Science don't have the equivalent landing page, as far as we know, so we're not linking to the Web of Science data. But CrossRef is obviously that the just a page of all the CrossRef data. Again, you see it dynamically generated out of our database. And because it's CrossRef and we're a CrossRef member, all of this stuff sits actually on our side. But usually what we're doing is we're sending our people out to a landing page on the third party site. Scroll down. These are the user ratings. These particular articles only have one user rating, but if it did have more, you can click into here and you could have got a list of all the ratings with a detailed breakout of what the ratings were, any comments they had, who actually left them and so on. Okay. Then we have comments. So this paper has the ability to leave comments on the entire article and make a note on the specific part of the article. And here are the comments and the notes. And you can see there's been debates and discussion about this. Here's somebody called JC Bradley. And he's having a conversation about this article and somebody called Cameron Neylon also got involved in that article. So this now is a permanent record of the discussion that happened about the article. Anyone coming to read the article can now read this discussion, decide whether, you know, that gives them some extra information about how that article is relevant to them. And then if we care on down. CiteULike. These are the social bookmarking cites. 17 users of CiteULike have bookmarked this article. And you click here and you go to CiteULike landing page. And if it loads -- here we go. So these are all the users that bookmarked that article in CiteULike. Here's their user name. And here's what they tagged it under. And somewhere down here there's that Cameron Neylon's appearing again. Here we go. Camera Neylon bookmarked this article. And the beauty of this kind of system actually is you can click-through and see everything else that Cameron's bookmarked, what he's interested in, and you can surf through it. So again it gives you some rich contextual information about this article and about what the people who are interested in this article are also interested in. And, you know, all of this stuff could have been found by a Google search as well. But we're putting it on the article. Postgenomic is a backlog aggregator. These people went out and found four blog posts that were written about this article and for their own purposes they have a landing page for that. And so we just link to it. And we to it all by the DOI. So all of this, the unique identifier for us is the DOI. And it's all done via open APIs. And then in addition we have the ability to leave trail backs and so on. So that's what article-level metrics looks like from our side. And going back to the presentation, if I just get through -- this is in case the Internet wasn't working. Okay. So how have they been received? So we launched this -- we launched it really first in March last year, but then we added the usage date of September. So September we considered it to be a sort of 1.0 release and got a lot of coverage for it. As an author, I would love to see this kind of service, a substantial value add. This is a PLoS ONE author. This person send me this quote unsolicited, although it reads like I wrote it for him. Your innovation of the article-level metrics is an extremely promising development in the evaluation of scientific publications. We are hopeful this will transform the way impact is assessed. This was an interesting one. So now people submit to us and quote their article-level metrics back to us to prove what a high-quality author they are and why we should publish them. So this is a quote directly from somebody's cover letter, pointing out how many downloads they have had and bookmarks and so on. And this was a fascinating one by Duncan Hull, who is a blogger in the UK: As paying customers of commercial publishers, should scientists and their funders be demanding this could of information in the future? I reckon they should. And that's our opinion as well. You know, this stuff was not hard to generate. It shows the granular article level, what was interesting about that content, and why shouldn't people provide that? People are now taking this and evaluating the data. So the data is open. We allow people to download the individual data for an article via download XML data link for the article. And we also provide a 23 megabyte spreadsheet of all the data for the entire corpus very granular. And some people have taken this and they started evaluating it. So this is somebody who has put this -- that data up on get help, which is an open source software development environment. And he's basically an incredibly detailed advance search, [inaudible] but effective. He took the same data and made some visualizations. He put this into many eyes. So here's some of the visualizations he created just out of that data, and this is what they look like. So PLoS article citations per day covered by publication year and broken out by journal. Article downloads per day. And the center one is the most number of downloads per day. That was actually our [inaudible] article. The big article we published last year. So it got a huge amount of downloads. And this data that he was working off was as of July. So massive number of downloads in just a couple of months of data. So that's why it's appearing there. But that's somebody called Mike Chalen is doing that, and he's doing a great job. Other people have taken the commenting data and attempted to evaluate that as well. The commenting data is just free text, so it's quite hard to actually evaluate it. But some people, and this is [inaudible] at Nature, took our data and crowd sourced the interpretation of the data, so they just put up the title of the article and the text of the comment and then gave crowd source options to random users to say what type of article or what type of comment that was and got the entire corpus categorized I think in a couple of weeks and then did an analysis of that. So 40 percent of the comments were from authors. 11 percent were requests for clarification. Direct criticism was 13 percent. So they were able to do some pretty -- pretty nice semantic I guess analysis of what the commenting data is telling us. Like I say, we're not the only people doing this, but we think we're doing it the most comparatively. But there are other people doing it, elements of this. The Frontier series of journals which is also open access provide much more sophisticated usage analytics at their paper. So they show you time spent on sites and a number of repeat visitors. Institutional repositories are also working on this. So David Palmer, he's in Hong Kong. What's important for an IR apparently is not so much the usage of an article but the usage of the authors because you can aggregate authors up to show the impact of your research institution. So they're working to get data in at the author level. And this is the association for computing machinery who are doing exactly that. So you'll see here they've used their massive database of ATM content to find everything that Stuart Feldman has published, and then they show you how many downloads Stuart Feldman has from their corpus in the last six weeks, 2800. And his list of papers that scrolls down with the individual data, which is great. He may want to change his photo. But this is auto generated data for them. And they allow -- they do allow actually the individuals to go in and verify that the list is correct and put some information about themselves. And so I think that's the way this is going. This is article level metrics, but there's no reason they can't also be people level metrics and institution level metrics once you start aggregating it out. And so what's missing? Well, all of these metrics really you have to wait for the article to be out for a while and people to start bookmarking it or citing it or using it even. So we don't really have any predictive metrics. And I think appearance in the journal is at least some sort of predictive metric. You know that if an article appears in nature somebody looked at it before it was published and said it's going to be quite good this article. So you've -- you sort of as an individual you know anything in nature's probably quite good. You don't have to wait a few weeks to find out if it was any good. So we missed that. But we could, for instance, have our editorial board do exactly that. When they're reducing a paper they could say I think this is going to be in the top 10 percent of all papers. And we could put that up as an article level metric. We're missing expert ratings. And what I just described is basically that but also a factor of a thousand ratings we'd like to get in there. Media coverage is really hard. So blog coverage we can do because there's people aggregating blog coverage, and they're interested in scientific bloggers. But nobody really aggregates all the New York Times coverage of PLoS. And all of the coverage by, you know, the guardian in the UK or something. And the reason is I think often they don't reference the DOI, they don't even mention the title of the article or the author name, so it's actually very hard to sort of computationally get at that. We'd like to have more sophisticated usage metrics. We'd like to track conversations outside the publisher, so we have this commenting and note making functionality on the site. It's not very well used. But we know that people are out there Twittering about our content or discussing it FriendFeeds, so we'd like to take some of those discussions and bring them back to the article as well. You know, we're not -- we're not sort of arrogant as to think everyone should come to our site to do the commenting. We'll let it happen wherever people are comfortable letting it happen. And reputation metrics. So none of that's built into our system at the moment. But if you are a particularly good commenter, one of the things you get is a reputation in the sort of real world, and we'd like to have that as well so that that would encourage more commenting and you would be able to see whether you trust the comments of an individual. And the stuff that still needs to be done is we need to add these filtering and navigation tools that we're doing the next few months. We need to put an API on top of our data which we're doing in the next few months. We need to add more data sources. Sort of things like the factor of a thousand. As we find them, we add them. We need to track new -- entirely new metrics. Perhaps we need to track how many times people are using this in the mental environment and using it to write their next paper, for example. That's a pretty strong correlation that there will be a citation to this in a future publication. Do we need to deduplicate this data? I don't know. All of those citation sources are basically overlapping data sets. Probably need deduplicating. We'd like more people to do some expert analysis. More of that sort of many eyes visualization, more looking for correlations in the data. We're not going to do that ourselves. We're just making the data available and hoping that the world figures it out for us. We'd like standards to evolve. At the moment, again, we're just sort of making up as we go along, but if at some point you want to consider whether, you know, five social bookmarks in PLoS is better than four social bookmarks in another journal, different publisher, you need to know they've found that data using the same methodology. And NISO is a body that might be able to help us there. And we need people to actually understand this stuff. So the one thing with the impact factor is that everyone thinks they understand it and everyone uses it, so it's widely adopted by academia, which is a great shame because it's a pretty distorting measure within academia. But perhaps instead of giving people a basket of, you know, 20 different metrics and say figure it out for yourself, somebody can come up with some clever, you know, what is one number that combines all of these into some, you know, impact factor for your article or something that actually means something. It needs to be valued by tenure committees. They need to actually -- instead of asking you how many impact factor publications do you have, how many publications do you have with more than 10,000 downloads? That's what they need to ask. People need to quote it in their resumes. And we need publishers to start using and adopting these kind of standards. So you already think via combination of this sort of great success of PLoS ONE and this movement away from thinking about a journal to thinking about an article, this could be a real break through in academic publishing. I think we could be seeing a paradigm shift, you know, if this were to take off towards the right things. And I think to misquote from Jerry Maguire, people are going to be saying: Show me the metrics, hopefully. Authors are going to be going to [inaudible] and saying, you know, PLoS can do it. I know I can get 13,000 downloads if I publish there. How many will I get in your journal? And, you know, they may be embarrassed because their journal only has 25 subscribers, for instance. And this is an opportunity now to push that agenda. And that is what we're doing and where we think it's going. So thank you. [applause]. >> Lee Dirks: Yes, sir for Peter? >>: I had a comment in terms of things that are possible in this environment that aren't possible elsewhere. Since you have registered users for comments and so on, could you actually follow up with a retraction [inaudible] a particular article and then if so how the data changed or made, you know, so that it would be important that we had that information? >> Peter Binfield: Yeah. So I guess the question was we have a knowledge of who comments on a paper, can we actually then make changes to the paper as a result of something that comes out in the discussion. Is that right? >>: My sort of thinking is that the author themselves realize that there's and error. >> Peter Binfield: Okay. >>: Then can you follow up with people who have read that article? >> Peter Binfield: You can, yes. And that -- that actually happens a lot. So on our articles, anyone can actually read the article. And they've got the option to leave a comment, what you do basically is you highlight a bit of text, click here to add a note, and see am I logged in? I don't know. But you actually get the opportunity to say what kind of a note this is. So here we go. Let's try that again. So I'm going to comment on this bit of article. Continue. So this is a note or a correction. So as a reader I could say I spotted an error. It's a correction. And then the authors are alerted, they get an e-mail alert there's been a comment. And you know if it's a correction that they basically agree with, they can escalate it up to us and say, you know, you know, I don't -- I agree that this is a error that needs to be corrected and we have the e-mail address of the commenter and if necessary we can put them in touch. Although that e-mail address is not made public. So we do have that ability to have that sort of feedback loop. >>: I guess what I'm asking is one further step from that. We had the issue presented earlier where there was some data was in error. And in science it would be ideal within the scientific community that there was this paper that they had sort of all read at one point in fact was in error, especially if it came up later, right? >> Peter Binfield: Okay. So ->>: Would there be a way that the community of readers who read that article could get ->> Peter Binfield: Not at the moment, because obviously anyone can just anonymously use the sites. We don't know who's read it. But you're talking about formal corrections and even retractions in extreme situations. So that information -- yeah. That information is made very public on the page. So if there's a formal correction when you come to read it the next time you read it, there's a big red bar that says there's a formal correction. This is what it is. And that data is also propagated out through PubMed Central, Med L ine, so on. So all those people get that information. But, yeah, there's no sort of auto notification of past readers that something has changed that you need to be aware of, unfortunately. >>: So looking at your data, incredible growth but only .5 percent market share, so to speak. Seems like there's quite a bit of room to grow. >> Peter Binfield: There is. There's 25,000 journals out there. And we're just one. But I think we see ourselves almost doubling in size every year right now which is fantastic. At some point, the rest of the industry will push back. We're not a balloon that the going to expand forever. But ->>: [inaudible] first. >> Peter Binfield: It's a scalable business model. Every article pays for itself. I think the thing that limits it actually is community adoption and things like the ed board, URL ed board. We have a thousand people. If we doubled the amount of output we would have to double our Ed board potentially. At some point it's just unscalable from a sort of human point of view. I think people do like their communities. They like to publish in their society journal of X, you know, because it's not just about publication, it's about supporting your society and things like that. So I think there will be things to limit us, but we see that exponential growth we've been seeing in many other situations today, almost carrying on for the next few years hopefully. >> Lee Dirks: Maybe one last question. >> Peter Binfield: Yes. And you had a question. >>: One of your metrics show that some of the comments were spam. Do you foresee that -- I mean that becoming a greater problem as you look towards PLoS as a place to go and people venting. >> Peter Binfield: Yeah. And maybe. We actually we have guidelines for good commenting and we reserve the right to pull down anything we can moderate, although we don't moderate pre-posting, so everything that's post goes live immediately and then people can flag that comment as offensive and we'll moderate it and pull it down. But, yes, spam is becoming an issue with some of the big services as well. So 2collab for instance and people like that are having their own spam issues. And these are being run by people at [inaudible]. So anything I think where you allow sort of open -- commenting open login, you know, is going to have issues. And unfortunately I think we'll see them as well. But at the moment, the spam is so interested in highly specialized scientific content. Okay. >> Lee Dirks: Thank you very much, Peter. [applause] >> Lee Dirks: I'm going to hand it over to Lisa to say a few words. >> Lisa Green: Well, I have just a little announcement before John starts. You can hear me without the microphone. So I'm not sure how many people in the room are aware, but we did a T-shirt project. We went with a new T-shirt for science causes and the prize was that the winning design, the person would come here, we would fly them here and give them [inaudible]. Unfortunately the person who designed the winning T-shirt was not able to attend, had a schedule conflict. But we do have the design to show you. So this is the first people have seen of it. Like I said, I'm not sure how many of you knew about the contest, but those who were [inaudible] contest were quite heated over what won. And there we go. So what we have is a scalable code that will take you to science comment -- pardon me, creative problems at work. Then we have a robot whose claws whose [inaudible]. So if you would like one of these shirts, let me know, and I can [inaudible]. >>: The back of it is a [inaudible] to the science comments ->> Lisa Green: Science comments or creative comments. [inaudible]. >> John Wilbanks: So, yeah, thank you to everybody that contributed. It's turned off, so it should be okay. Okay. So before I get into my talk, I just wanted to thank Microsoft Research for hosting us today. And I wanted to thank Lisa for putting this together and also Hope for the work that she's done in promoting this year and to all the speakers. I travel way too much. Some of you know that. And coming here today is sort of like coming home. We spent so much time together in so many weird places. I don't think we've ever actually all been on the same program before. So it's really neat, you know, to be this far away and have it feel like home. So I don't usually have to speak with so many people who use slides that we have all written together. So like the registration, certification, dissemination, I've used those slides. I've used slides that Cameron has used. We take from each other just as many as we take from anybody else and as much as we create in this community. And so I had a unique challenge today, which is I have to say stuff you haven't heard yet. And I don't usually have that problem. Because usually I'm sort of off by myself. And it's also been the five year anniversary of the Science Commons project in the last year, and so we've been doing a lot of reflecting. And so I hope you'll give me a chance to be a little more expansive and a little less detailed than I usually am. Because I'm trying to think about why do we exist at Creative Commons, what is it that we do at Creative Commons and Science Commons that makes us different? And what are we going to do for the next five years? And so it's good to start with where you came from. So Creative Commons is and organization came into existence in many ways because of the reality that on the Internet consumers do more than consume. Consumers make thing now. And it didn't used to be like that. And copyright wasn't set up that way, so -- I know this is Microsoft, but trust me, I'm going to come back and make an Apple comparison later that you like. But Apple in the late '90s came out with the iMac and with iTunes and they made this argument that you should rip, mix, and burn. Afterall it's your music is what the add said. And if you read the fine print, it's not true. And what happened that we created an economy in which the vast majority of daily creation was illegal under copyright law. That's what Creative Commons came into existence to deal with was how do we create an alternative world in which the daily act of creation is not criminal? So we had this goal as an organization to decriminalize creativity through the creation of a commons. And our actual original ethos was we were going to make a database of open copyrighted stuff and then people would contribute to it and they would get tax credits. And we write these licenses as a side effect of that operation. The database didn't work out. And so we made the decision to release the licenses and say, you know what, the web's the database. This was called a cop-out by some people that were in the room at the time. That's just -- you know, you're just flailing. And you know, the damndest thing happened. It worked. And so we've seen a lot of exponential graphs. This is my favorite exponential graph because it shows the adoption of the licenses. And the last year there is a half year. We can actually no longer effectively count the licenses. We assume it's over a billion at this point. And we're having to look for new ways to actually count and assign metrics to what we do. >>: [inaudible]. >> John Wilbanks: Millions. So we're somewhere in the 800 million to a billion licensed objects on the web range at this point based on what we can tell. But we can't really stitch the numbers together effectively anymore. Because people are beginning to embed the metadata directly into objects like PDFs and photographs instead of just listing it on their web page which we can then dereference through Google and through other search engines like Bing perhaps. So this shows that, you know, in the search for decriminalization, something else happened. Which is that people really wanted this for lots of reasons. And it's gone international. This was not planned, right? This is an example of what I would call catastrophic success. Right. Sometimes I show the clip from adjust when they say we're going to need a bigger boat, right? Because that's what's happened with the Digital Commons, is it's turned out that the way that we wrote these licenses was powerful and adaptable enough that it's gone far beyond anything we ever expected or designed it to do. And Joey Ito, who is our CEO, likes to analogize what's happening with the Creative Commons licenses to the different stacks of the network so that we started with Ethernet that connected physical networks. And before then you had to call consultants to come and wire together your computer networks. And then on top of that or almost simultaneous to it we got TCP/IP which actually connected the bits that moved across those networks. And then sort of again we get on top of that HTML and HTTP and many of the other standards that allow us to connect documents. And if we're going to make the jump from documents to knowledge, we need to have another layer, which is the Digital Commons. It's the next piece of the network. And so Creative Commons in many ways is a set of network engineers. We're sort of analogous to the IETF, except we're doing it to lawyers instead of to IBM. And five years ago when I got into this, it was a real dodgy thing. People thought I was a little bit crazy or a lot crazy to jump from a fairly nice career in technology consulting and startups to do this. But I bought into this at the beginning. And what we've seen is that just as IBM in 1992 said no one will ever build a corporate Internet on TCP/IP because it's open, we've seen the sort of criticisms of lawyers that were applied to us five years ago fade. Because of that scale we've been achieving and because the problems that we solve are problems that are being felt by companies, by nonprofits, by publishers, and by the community at large. And so I saw this in Cameron's slides, but this was the one slide I'm going to throw back at you because I like it. So why is there a Science Commons? Why is there a Science Commons projects at Creative Commons? And it's because unlike in culture where we had a criminalization problem, in science this is actually how it's always been, that we create property by giving it away to people. And the commons is uniquely structured as a flexible way to actually deal with this transformation in which you create private property by claiming credit for it when you give it to someone else. And that's why open access is becoming the new normal. That's why open data, if we can ever get past the technical infrastructure and legal issues will become the new normal, because that's how science has always worked. It's just been a really inefficient technological society that's based on paper. And so that's what we do at Creative Commons in the science project. But it's funny because I was looking back in the founding documents, and this is the best description I can have of the -- of what I was given when Science Commons started, which is we want that for science. And it wasn't a lot more detailed than that. And so the first thing we did was ask what that was. And the most common answer we got back was Wikipedia, which is that we want Wikipedia for science because Wikipedia has taken this thing that used to be a scarce resource and made it current, valuable, free of charge and created by the world. And but when we dug into it, we found out that people wanted a lot of other things. They wanted things like the PC in the '80s. Right? A generic platform where you could write applications. They wanted things like libraries used to be but on the Internet, places where you could go get information. Right? They wanted things like eBay and Amazon for science. Right? What that was was actually the entire Internet that we take for granted in our daily lives but for science. And so it wasn't about open science or really decriminalized science like it was in culture, it was about creating this innovation ecosystem that we take for granted every day as cultural consumers and as business consumers on the web. Right? That's what that was. That's what people wanted out of the commons and the sciences. And so what we're trying to do is to spark generative science. And generative is the word I'm going to stick with today because open and free are terms that come loaded from software, from culture, from other places. And open and free are tools that help us achieve generative systems. But they aren't the only tools that we use to achieve generative systems. So science audience, let's get definitions. Generativity is a concept that comes from a guy name Jonathan Zittrain. He's a professor at Harvard Law School. He hired me in 1998 and introduced me to this whole community. And the whole idea is you want to actually measure whether or not a system can produce unanticipated change through unfiltered contributions. And it's about people you don't know doing things you don't expect. Now, this is really weird for scientists to think about when you put it up because they say, well, if they're not from the guild, if I don't know who they are, I don't want their contribution. And the whole point is that if you have enough people and a large enough system, even if 999 out of 1,000 things fail, the 1,000th is Wikipedia. You know, we've not got that in science. So failure's very expensive in science. It's incredibly expensive. If you fail to get a paper out after three years of research, you may never get another grant. And so science has inherently resisted this sort of generativity. All right? So Zittrain has a great set of rules of thumb for technology. So this telescope is more generative because it's easy for you and me to use without any training and because we can use it as a bat or a door handle if we need to, than this telescope. This is more powerful but less generative because it's not accessible, it's only useable for what it's useable for. And it's very difficult to master. Now, the Internet is sort of the classic example of a generative system. So this is where the Internet comes from, this paper is where it begins. And it was to connect computers that looked like this together. I would say that this computer is the equivalent of your lab, right? You had to be at the university to have a PDP-10. You had to have funding to work on the PDP-10. You had to have permission. You had to write papers that came out of it. You had to justify every use you made because it was so dear. And that paper and these communities turned into this tiny little network, right? Almost always a generative system starts off small, specific, and very, very, very nerdy. Right? And that's a good thing. Because it's about the solving of the problem of hooking together those PDP-10s in a way that could be opened up later. And so the first key principle of generativity is leverage, which is does it do the thing you want it to do, does it do it well, and can it be leveraged for other things besides the thing you created it for? And because TCP/IP and Ethernet were open enough, they could be leveraged for things beyond connecting PDPs together inside Darpa's offices. They could be used to do things like e-mail. So this is the map in 1977. Now, at this point we've got e-mail, right? The first e-mail message has been sent, although it's been lost at this point. And what the happening in the background is hacker conventions are taking place and people like Steve Jobs and Bill Gates are beginning to wire together circuit boards that over time in the '80s turn into microcomputers. So the second key principle of generativity, adaptability. Because the network does not assume you have a PDP-10, it is adaptable to the microcomputer when it comes out. It is adaptable to the World Wide Web when the web comes out. The web embodies the same principles of leverage and adaptability which means then when Mosaic comes out, we can run browser on it. So at every point we've got a system that's highly powerful and changeable. It can add e-mail, it can add Gofer, it can add the web. And then the web itself can add visual browser, which are a hell of a lot better than the line browser that I used, yes, to get Grateful Dead set lists from the FTP archive at Cal Berkeley, which was my introduction to the Internet. So I do have a tie here with Heather. Right? Whatever it is that brings us to technology, right, is the passion. But the ability to actually use that and adapt it to our own uses is key to whether or not it's generative. Now, there's three other key factors which I'm not going to belabor quite as much, which are these ideas that is it accessible, is it easy to master, and can you transfer that mastery to someone else? And so when you think about it in terms of technology, right, it's the change between that kind of climbing and that kind of climbing. It's not about making an escalator. It doesn't have to be dead simple. You might actually have to learn how to do a little bit of metadata markup or edit and HTML page in the beginning. But if you can make the transition from extreme rock climbing gear to at least stairs carved into the face of the rock, that's the primary transition you need to make to make a system really generative. And that's what allowed this sketch of the Internet to become the point where two guys can make Twitter in two weeks and launch it at south by southwest. The cost of failure in technology is so low that you can start a company in two weeks with the right people and the right idea. And what we're trying to do at Science Commons is to bring that to science, to lower the cost of failure and the cost of collaboration to the point where you can actually have this sort of generative system. And the classic drawing of this is the hour glass. And so what you see is no matter what you have at the bottom, whether it's copper or radio or wireless or tin cans and string, you can connect that up through the hour glass at the most simple, stupid layer, which is the Internet protocol. And this only works when you make a simple, scalable layer at the core. And this is why it's important to think about the commons not just as something that the abstract concept we believe in but something that requires technical levels of diligence and scalability. Because on top of this you want eBay, Amazon, the web, e-mail, everything. And the smarter the network is at its core, the less likely it is to achieve scale. This is why smart grids and smart networks failed in comparison to dumb, open networks where the intelligence was at the ends. Right? This gets recap it late in the computer architecture. So you can put the PC and the operating system in the middle. And it didn't matter whether you connect a monitor or a scanner or a keyboard at the bottom or whether you run an application like Firefox or Quicken or Word other anything at the top. This is where the combination of Windows plus Office was a very powerful generative system, even though it wasn't open source. Because anyone could write any application they wanted to the PC without asking for permission, it created a platform for innovation and unanticipated increases in capacity that, for example, the Apple Computers of the '80s failed to achieve. And I would argue that's a big part of why Apple's market share in OS suffered over time. So these are the five elements of what makes something generative. And if we want them in science, it requires active intervention because science has a lot of institutional, cultural, financial and purely scientific barriers to the adoption of these sort of five key elements of a system. And so that's what we do at Science Commons. That's why we exist is to actively intervene to promote those five concepts of generativity. So the first thing is if we want this. So I'm assuming you agree with me that this is a good thing. If not, that's fine. But we want it. And so the first thing you have to do to do that is to deal with property rights. So you can't ignore the law over time. Right? It's a really bad assumption that if you ignore the law everything is just going to work out, right? You can talk to Lisa's friend Jordan, who is the technical architect of Napster. He's a Science Commons' fellow. But ignoring the law at Napster didn't scale over time. That's why there is a supreme court case with the word Napster in it. Right? And it didn't go well for the little guy. So the law interacts with science in at least three core classes of works. So you make data in science, you make tools in science, and you make narratives in science. Doesn't matter whether the narrative's a blog or a journal or a lab notebook or an e-mail or a Twitter, that's narrative from the law of perspective. Copyright governs it. Tools are typically covered by contracts and patents, all right? So tools would be things -- anything from a stem cell line to a man to a piece of software in many cases. Now, data, it's typically secrecy. There's also what are called sui generis rights. These are national rights created by funky laws across the world. And unlike narratives where copyright rules, the laws for tools and data are very radically different country to country, jurisdiction to jurisdiction. And one of the ironies of open and free, which is one of the reasons we're not going to use those words a lot here, is that open and free work in copyright because copyright is a very powerful international regime. It means it works relatively the same everywhere. It means public licenses work relatively the same way every where. And we have this temptation to try to recapitulate that in data and in tools because it worked so well in copyright. But the irony is in the absence of that powerful right that makes things criminal, the public license that makes things decriminal doesn't work very well. And in fact, it can actually have unintended consequences of breaking the commons in those spaces. So we started in open access. So the CC licenses are a natural way to implement for free -- this was the philosophies in open access. Heather mentioned the Budapest declaration. I've recapitulated it here. I won't read it to you. There's a couple thing I would point out though. So one is in the middle of the first paragraph, pass the literature as data to software. This was visionary in 2001, when this was written. All right? And it's become probably the most important argument for access to literature which is that if we can't index it, hyperlink it, tag it, structure it, then it's not useful. It's not machine readable if we do that. And the second is that the constraint and the role of copyright is the role of attribution, acknowledgement and citation. And that's basically word for word what the Creative Commons attribution license does is it says you are free to copy, distribute, transmit, adapt, but you've got to give credit where credit is due. So the CC license sort of happened into this role as the free legal implementation of the philosophy behind open access. The first thing that happened is we got pulled towards data. Now copyright in relation to data, a picture says a thousand words. Trying to put data into copyright licenses breaks. And trying to license data in an international context the way we license copyrights in an international context breaks. Because if we take the sorts of database rights that exist in the EU or in Australia -- actually I shouldn't say Australia because Australia last week set data are not copyrightable. It was a wonderful court decision. So we say EU and UK. Let's use them. Not to beat up on the UK. But if we license those rights even in the context of freedom, we propagate them to places where they don't exist. So if I take a data set that Cameron puts out under a data license in the UK and I put it in the United States, I've brought, I've imported a control on data in the name of freedom. If I put a contract on it, I've exported a control in the name of freedom. So we don't have this powerful sense of stuff that needs to be decriminalized. And so we don't need sort of powerful tools to make decriminalization happen. And it sort of gets worse. Things like copyleft, Share Alike, the GNU GPL, the Creative Commons Share Like licenses, these things work really nicely in copyrighted works because copyrights allow you to enable someone to do stuff and then you can control them through that enablement. I enable you to make a copy, but I control you by saying if you make a change to it, I want it brought back. But copyrighted software doesn't have to deal with things like national laws on data privacy or consumer rights about their own health information. And so if I have a copyleft license on a database of health information or actually even a different base, a database of ethnographic information and I want to combine it with health information, I can be in essentially a catch 22. Because I'm under an obligation from database one, which is ethnographic, to share any derivative data work that I do. But I'm bound by a lot of release, no data tied to health privacy. So it becomes illegal to put those two databases together because of the conflict between Share Alike and privacy. I've given you the most simple example. We're working on this with the folks from Sage for their governance project and we've begun a series of interviews with national data experts. And I can tell you that national policy on general data sharing and privacy makes a sort of health information privacy rules in the United States trivial. I can also tell you that Share Alike obligations that connect to the patriot law in the United States are not very well regarded in the UK and in the EU. These are laws that were never intended to work together that things like Share Alike activate as an accident. And so we spent, you know, years trying to figure this out. And we finally came to the conclusion that it was sort of like oil and water. If you shake it really hard, you can make an emulsion that looks like it's integrated. But if you leave it alone for five minutes, it's going to settle back. These things aren't meant to go together, property rights and data. If you try to deal with them, you're actually going to likely break the ability to do the sort of technical integration that we're talking about. And so although copyleft is essential to decriminalizing in strong copyright context, it can actually be negative in a different context. And that applies to patents just as well as it applies to data actually. So we had started all this because we wanted to use our licenses for data. It would be awesome to be able to recommend that it would be so easy to use Creative Commons licenses for data, right, they were already so well adopted elsewhere. So we said, you know, at a minimum we can do attribution, right? That can't be problematic. And then the Wikipedia guys reminded us that this is what one page of automated attribution to Wikipedia looks like when you print it. And there are 27 pages. Wikipedia in 69 years will still be under the same copyright it is today. You can imagine how long the attribution pages will be. And you can imagine in a world like the one Steven talks about in which the world is driven by citation into networks and models where machines take the models that exist, the 50 models that are built on 50 data sets and in five years we have 500,000 models all generated by machines and in 10 years five million, five billion. Right? Making it illegal to fail to attribute, giving people the right to bring the entire system down through an injunction is what happens when you use the law as opposed to using the basic norms and ethos of science, which is citation is different than attribution. Attribution happens when you make a copy and you've got to say where you got the copy. Citation says I give you credit because your ideas inspired me. So citation can scale on a context here where attribution can't. The other big tease in data licensing is if you've got really big hairy data you're probably going to cache it someplace like Microsoft Research that's got supercomputer servers and massive pipes. Well, if nobody's making a copy of it, you're not triggering any copyright or database rights. Because those rights only accrue to the copying of things. So in a world where you're caching massively large datasets, reliance on licenses fails. So we came up with what we called a protocol on how to deal with this, which is essentially if you cannot use public licenses to make things work, the only solution is to make the law go away. So first you want to waive the rights necessary for extraction and reuse. Ideally this means waiving your copyrights and your database rights, putting it into the public domain, or making it interoperable with the public domain. Second is you conditional impose any obligations on downstream reuse. So one of these things would be something like a Share Alike, another would be a contract, and I'll give you a good example of that later, that would relimit the downstream use. Because not only do I need to be able to give it to Peter, Peter needs to be able to give it to the web without any obligations. We don't want to create Achille's heels down the line that can be exploited by people who don't like the open world. And that can only be published through unambiguous one to many grants of rights. And last is to the behavior request. We've gotten addicted to requesting behavior through licenses. And the idea is we want to request those through norms, which are very powerful, at least in the sciences, and not through the law. So we've made a tool that does this. It's called CC0. The way that this works is it didn't actually put something in the public domain. It makes it interoperable with things that are in the public domain. What you agree to do is not to assert the rights that you have. Doesn't make them magically go away, but you guys could say I'm not going to sue you, right, I promise that I've waived that right to sue you. So to the extent I have a copyright, I've waived it, to the extent I have a database right, I've waived it. If I'm in a jurisdiction where I'm not allowed to do this, I agree not to sue you. So it's an international single tool. It's like the middle of the hour glass for the law when it comes to data. Because we want any kind of copyrighted or data product to go in, and we want millions of applications on top. It's a very simple, clean, and dumb in a good way standard. Because it means the only thing you have to worry about is the technical and the scientific part. And that's complicated enough, as we have heard. So we didn't know what reaction we would get. People really wanted data licenses. People really, really want easy answers to data. And even though this is an easy answer, it's not an easy answer. Because you're losing the security blanket. But we've seen really impressive uptake from the life sciences community in particular. So the tropical disease initiative has put an enormous amount of information about potential compounds that attack tropical diseases under CC0. Personal Genome Project, which I'll come back to. They have approval from the institutional review boards at Harvard an other schools to sequence the full genomes of 100,000 individuals and release them on the web and into the public domain under CC0. So the tool made it through IRB approval at Harvard Med, which is more complicated and painful than you might know. They've also got the complete health histories of those individuals in the public domain. Because even though those health histories are potentially narratives with copyrights, they need to be treated as data later. The Europeans, we were actually pretty surprised to see the EMBL adopt this for their database of drug side effects because the EU has a strong database, right, associated with it, and they don't typically like to waive it. So we were very gratified to see that happen. And we even saw this emerge in Nature where Nature in an editorial explicitly recommends using the CC0 domain approach for the life sciences and data. And it's because it actually -- even though it's a hard choice to make to let go of all of your rights on something like data, it works. And that's really the real test in the end is not if people want it, it's whether or not it works and whether or not it scales. Now, I know that the patent principles have been mentioned today, so I'm not going to belabor it. This is important in many ways because of two things to me. One is that scientists were involved in its drafting unlike almost everything else that affects science and policy. So Cameron and Peter were involved in its drafting, and Jenny, and others. The other is that Creative Commons and the Open Knowledge Foundation found agreement. So the Open Knowledge Foundation actually makes data licenses. You can guess that we're not big fans of them because of the research that we've done. But they're a good group, and they've put a really hard amount of work into this. What we did was come to the agreement that those sorts of tools, whether ours or theirs are inappropriate in the sciences. Other than the public domain tools. And so even though we have disagreements and inside the open movement we can have disagreements that make those, we have what the close world seemed tame, it's incredible how hard you argue over a tiny point with someone you basically agree with, that we could actually come to agreement on the things that really matter. And so the patent principles are a nice example, both of the science community and the policy community coming together but I hope they're also an example of how when we argue inside the commons we should remember the things that we have in common more than the things that we have in disagreement. So that's the property right piece of this. And that's where we started. That's where Creative Commons was. But the funny thing is when you deal with sciences that if you actually want to affect science in the real world, you very quickly get dragged out of the digital. So if you really want to make a change happen, it's not enough to have the literature and the data be open, you've got to actually deal with tools and inventions. And this is much more complicated because these are rights that are held by institutions, not individuals, by technology transfer offices, by governments, by funders, by businesses. And there's a lot of money at stake, not just the academic reputations and credit. And although the libraries think 25,000 a year for nuclear physics, B, is expensive, and it, to a library it's nothing compared to the cost of licensing the BRCA patent if you want to do breast cancer diagnostics. Or how difficult it is in terms of time and effort to access a line of stem cells as being competitively withheld at a university in the middle of the country that starts with a W, rhymes with miss con son or something. So if you actually want to achieve this, you've got to go after the tools. So what we did was say, all right, we're going to build some tools that chief the same things that Creative Commons licenses achieve but for biological materials. So we had to integrate existing agreement like the Universal Uniform Biological Materials Transfer Agreement, the NIH's simple letter agreement. These are the sorts of things that govern biological materials movement. We had to come up with modular concepts like no clinical use or no commercial use or if you have something that's a DNA product you can't make more of it and then redistribute it. Then we had to come up with icons for these things. Took us three or four years to actually get from the simplicity of doing a legal piece of drafting to get to an actual released product. And it has legal code. We also have human readable machine, readable code and all that good stuff. It's just like a Creative Commons copyright licenses but there's no IP. This is for the vast majority of tools and interventions that never get patented and don't have copyrights, which is basically everything our tax dollars pay for in laboratories. Things like plasmids, right? Commons have to deal with physical property that's not intellectual, just as much as they have to deal with copyrights and they have to deal with narratives. Now, the law is the easy part. Integrating this into systems that will likely have on the web is the complicated part. So this is what's called the iBridge network of technology transfer offices. There's about 50 universities in the US that have signed up to basically list on a catalog like Amazon affiliates the sorts of laboratory [inaudible] that we're talking about under these one-click contracts. Now, this is just the beginning of this. It took us about two years just to get the integration. And the idea was that you should actually be able to simply buy a plasmid or a vector the way that you would buy a book on Amazon. Of course you would have to be registered. We don't want to send these out just to you know -- they don't want to send them to my house. But if you are an academic at a regular university, the only rule you have to get is the ability to register that you're a part of a credited research institution. So we have removed the competitive barrier. We've removed the legal barrier. What's left is what we would call the fulfillment barrier, which is that I, as a scientist, don't get funded to send you copies of my stem cells or to spend my time making them for you. When I talk about every time you solve a problem in the commons you basically find the next one. So after three years of working on this, we got this integration, we got foundations to implement it and everything stopped because the scientists said we don't get paid to make things for other people, we get paid to make discoveries and write papers. So we had to reboot the entire project and start working with the biological resource centers that actually store, copy, manufacture, and forward biological materials like the Coriell Cell Culture Repository. . And this is what it looks like. So these are actually real examples. You can click through these if you want. So the Huntington's community has probably always been the most progressive community we've worked with in the disease space. There's almost a hundred million a year now going into HD out of 1 foundation, the Cure Huntington's Disease Initiative. That's a stung amount of money. But it's not even nearly enough to get a drug. The Gates Foundation puts 500 million a year into malaria, and there still isn't a cure. Right? The richest people in the world can't buy cures to diseases. What they can do is begin to be interoperable with other people that are looking at neurodegeneration. So they can expose their tools for anyone else who wants to do research on neurodegenerative diseases. And they can now open this resource up because the sum cost of letting other people put stuff in it is very low at this point. They've already spent the money. But if you put it in, it's got to be available to their researchers, too. So they're beginning to create a commons for neurodegenerative research and Huntington's research that begins to that I can that 100 million dollars and invest it, as opposed to simply spend it. And you can click your way through. It's just like a catalog. And all you have to do is click on the MTA and do some online ordering. All right? It's incredible how hard it is to make things this simple. And this isn't the stuff that gets talked about when we talk about the commons for the most part, this is just as boring as doing deep network hacking. No one at the edges of the hour glass cares how hard it is to make these sorts of agreements happen. And the people who are in charge of the system for the most part benefit from it. They don't have a big reason to change it. So you've got to work from the bottom up at almost every level in these systems to achieve this sort of change. And a lot of the work that we do involves taking the profile we get from our digital work and reapplying it in this space. Whatever social capital we earn by being cool and having a billion digital objects under our licenses, we spend that and more and run deficits to try to achieve change in the tools and intervention space. Probably the biggest win was the PGP. So I mentioned their data earlier. There's going to be 100,000 people in this project. So it's not just their genomes and their health interviews, you'll be able to buy their stem cells under Science Commons Materials Transfers Agreements. And not only that, the most liberal Science Commons Materials Transfers Agreements. So the commercial price to buy the stem cells is $85. The non-commercial price is $85. If you want to sell them, you're allowed to. If you want to use them in the clinic, you're allowed to. There will be 100,000 lines of stem cells that are this free. Tied to the data, the full sequence genome of the individual, and tied to their health interview. So if I want to do a profile on drugs and I need to find 30 sets of stem cells for Caucasian males in their late 30s who travel too much, we'll be able to order those for $85 bucks a pop and test on them. All right? That's the sort of thing that begins to get us out of the trap we're in. Because opening up access to the data in the literature and not opening up access to the tools required to do follow-on research just moved the problem. And it moves it to a place where the scientists can hide behind the institution. All right. So just to -- this is probably the thing that we're proudest of and probably get the least information about out into the world about. Because it's just -- the only thing that really changes is that you have a one-click ability to order something that most people don't want to order. Right? But this is the sort of stuff that used to be restricted by social nets, by guilds, and by institutions. So if you do that, you again only move the problem. Right? So the next problem is what's called freedom to operate. So this means that you have to start thinking about things like patents. Now, this is not a metabolic pathway, this is a patent pathway on telomerase. Most of these are held by Geron Corporation licensed out in certain ways, right? And this is one key piece of what the genome does. So if you want to intervene in telomerase in the real order, the product that you sell to people, you've got to navigate at least this pathway of patents to have the right to go to market. Now, this is from the patent lens, which is an organization in Australia that does fantastic work on patent informatics. Patents may be the least transparent property system in the world. Despite the fact that it was created in order to allow us to understand what to do. That's why patents existed. It was an encouragement to disclose. But the great irony is that especially in the life sciences and the rest of the commercial sciences, it's become a way to make things unclear. And so if you want to actually get through this, to really practice a telomerase diagnostic, you'd probably of to license all these patents, at least in part. So this is the next phase of the commons. The way that I would describe it is if you were to think about copyrights and patents that you hold on a Gaussian distribution you might be willing to give away the middle of the bell curve in a copyright context because you didn't spend money to register each of those copyrights, they came down from God when you lifted your hands from paper or your data. But patents you're willing to give away are the very, very edge of it. Because if you're a company or an institution that holds patents and you paid 50 to $100,000 each for those patents and you use them to protect your competitive advantage, giving them away una public copyright license like we expect in copyrights or in data just doesn't make business sense, right? People who do that will get fired. And firing people who believe new isn't a good way to achieve scale. So what we've been doing in the patent project is two things. First is we want to reconstruct the tradition that research is exempt from patent infringement. This used to be the law in the United States. The courts took it away and in a case called Maddy versus Duke in which they basically said that because universities are in the business of doing research, there is no research exemption outside the garage. So the first thing we want to do is reconstruct that research exemption. Now, in two weeks we'll be releasing these tools on to the public web for comment, the model patent license and the research exemption. Nike has already committed their entire patent portfolio to the research exemption, as have a couple of other major companies that we're in the process of getting permission to say who they are. Now, those who know patents would say this is foreplay, and it's true. Giving people research rights without the right to take it to market is only halfway there. Which is why the second tool is what we call a model patent license. Patents prevent people from making and using and selling your technology or your invention. And so just as a public copyright license inverts the power to keep someone from copying and distributing your work, the model patent license inverts the right and grants people the right to make, use, and sell the technology. But because this isn't about political freedom perhaps the way that copyright licenses are, it's about freedom to operate, if we want to get at the rest of the bell curve and not just the very, very, very, very left of it, you've got to be able to do two things. First is you've got to let a company have a revenue stream off of that patent. Right? That's something we didn't ever enable on any of our other tools. We're going to enable it, but we're not going to actually write that, we're going to simply allow it to be connected by the user to a patent license. We're also going to let them put on what's called a field of use limitation or exception. Frequently it's already had the exclusive rights licensed out for a field. If you've got a stem cell line, it may have been licensed out already for Alzheimer's, and you can't give that right up. But if we want all of the other uses in the world available, we've got to be able to deal with that. So again, another user generated field. And you can use this one of two ways. One is to create a bubble of freedom for a certain goal, like malaria. You can say all these patents are available to go to market commercially but only in malaria. The other is to very simply say, you know what, I'm Nike, I make shoes, my patents are available outside the shoe industry for a revenue stream. And what this does is open up the field for unanticipated uses of those technologies by unanticipated people. But it's not quite politically free the way we treat the copyright stuff. It's about getting to the middle of that bell curve by saying these patents have economic value, we have to recognize that, but we want to standardize the transactions. And so you'll be seeing a lot more about this from Creative Commons. We're not going to be doing this as Science Commons. In many ways Creative Commons is going to be taking on a lot of emission and organizations of Science Commons. Because it's become clear that what we do in the commons is that layer of the network goes beyond the copyright license. And keeping these things inside Science Commons, which is this project that doesn't have legal existence inside Creative Commons, doesn't always make sense. So again we've pushed the problem from the digital stuff to the physical stuff to the patents. And what you find is that you continue to get to the next layer of the problem, which is infrastructure. So if we have a world in which stem cells are ubiquitously available and genomes cost $500 to sequence, the data overload that will come out of hundreds of thousands of people becoming scientists quickly overwhelms the web. The web stinks now for science. Searching Google for what's our classic example, signal transduction genes in certain classes of neurons, right, pyramidal neurons, there's about 400, 500,000 pages. You won't get a list of genes. Because the web doesn't support as infrastructure science. Right? And there was a great quote that came from Bruce Sterling this week that summarizes it. So if we can't have the machines even catch up to structure data, we have to design the data at the moment of generation to plug into infrastructuring systems. And this is why the work that Jean-Claude and that Peter and that everyone is doing in the open chemistry space, Antony and others, is so essential because it provides the standards in which to generate data so that it works when you put it out. And I would summarize this, if you needed a quote, the problem is that computers are stupid. We tell them things, and they don't understand them. So there's two paths to this. One is to make data what we call re-useful. And so Sage is a great example of this. And so we've had the honor of being involved with Sage. And I've had the honor of being on the board of Sage. And what it is is a platform. So they've got these network models and these datasets and the source code. And what it does is it makes any dataset designed to go into the formats of Sage useful immediately in the software and other models available at Sage. So I have a reason as a scientist to put my data into those formats which is then I can run the code. Then I can use the platform. On top of that, the idea that we're going to have citations into these things gives me reason to deposit my stuff after running it. And these two things together may be more powerful than anything else to make a scientist generate open data. One is if it's in those formats, I can actually run models on it and make predictions. Two is if I put it there, people will cite me. That's much -- that scales much better than altruism or politics in the sciences. And so to go back to these ideas of generativity, the Sage process, the models increase the leverage of the data because they mean that I can use it in different ways. The repository increases the accessibility of the data because then it can be downloaded and reused. The training is one of the pieces we often leave out. But the training increases the ease of mastery and the transferability of the system. And the licensing unifies all of it. So CC licenses on the training materials, on the website, public domain tools on the data, all right, begin to actually allow for the sort of movement and integration that makes diseases biology at least of the potential to be a generative system which it so far hasn't been. And the second path is to make computers less dumb. And this is unfortunately much harder. The semantic web is the sort of common name for it or the linked data web and so forth. And it is as absurd as expecting cavemen to speak in simple declarative sentences. And I've been part of the Semantic Web for about 10 years in various ways. And every year I believe more in it, and I believe less about what it can do. It's a little bit of a paradox, but what I mean is I don't expect the Semantic Web to give us the Star Trek computer so we can say data, tell me what the drug is. I think if we're going to do that it will happen from things like Sage, not from things like the Semantic Web. But what the Semantic Web lets us do is to begin to integrate the bubbles of infrastructure that are being created. So there are e-Science projects in the EK, in the EU, in the United States, in Australia, everywhere. But they don't knit together. Right? There are projects in open science every where. They don't knit into any sort of common web. And what the Semantic Web can let us do is to use the common names for things. Something as simple as coffee. And converge those on common URLs. I think that is the best thing the Semantic Web can do right now. And it's an incredibly powerful thing. It's the middle of the hour glass again, which are the names. Because then any resource can come in at the bottom and any application can be written at the top. And you'll know that you're getting at everything you need to get at. So we've been working on this thing we call the shared names project. We can see that sharedname.org or at our NeuroCommons website. And what we've been doing to use that is to try to get rid of the idea of data integration. Right? If someone came to you and said they would want to integrate web pages for you, would you think they were crazy? All right? Or to integrate your office package on to your Windows distribution, right? We install software. We search web pages. The only thing that we artisinally deal with technically is data -- databases. So if we use the same names for things and the same languages, RDF and OWL to describe them, then we can begin to integrate data the way that we install software. So we've got this project we call NeuroCommons which is hundreds of data resources converted to common formats and common names that you can compile into a single index of all of those databases and run structured queries across. All right? This is a tremendous achievement and a small achievement at the same time. And the idea is that if you're doing it right, that's the only ontology you ever have to write, because everything else has already been written somewhere else. It's only ever been put into one place. So these are the sorts of tools that we built as infrastructure. What we're discovering though is that our value is in writing the wiring code and supporting people who actually have infrastructure they want to take public like Sage. In many cases it's not possible for a small organization to sustainability scale and provide infrastructure. You need organizations that have recurring revenue models and real science at their core. And the key is to help them scale and connect over time, not to take the work in on yourself. So starting to wind up a little bit here. The other thing is that in Semantic Web law is code and code is law, so we use the same languages and tools that we use for data to describe the legal transactions. So if you're not familiar with CCREL, it's the CC rights expression language. It's a submitted specification at the web consortium to describe property rights, transactions in a machine readable way. And the idea is the machines should be negotiating the legal aspects just as they should be negotiating the data aspects. Just as machines negotiate the vast majority of the transactions that you deal with in Google as a culture consumer. And it has these sorts of -- because computers are dumb, we have to tell them what requirements and prohibitions are. And the whole idea is that you ought to be able to have a machine crawl and find out exactly how to attribute any given work or cite any given data product. And so what we've done is worked on instead of building the infrastructure for this out, we've written a language that allows other people to do it, to embed it. Right? Everything we do should be the middle of the hour glass, right? And that's one of the true test of a commons to us is if you find yourself getting too far up or down the hour glass, you are off scope and you need to stop doing it. So if you want to deal with this, the reason that I've put you through this 45-minute lecture, is to try to impress on you the importance of dealing with the whole problem. Fixing one piece of the problem just moves the problem because it's an ecosystem and it's a process. And you can overload it at any point if you don't do it right. And so one lesson from experience is that the ideas of the network and the hour glass, which are typically called separation of concerns, you don't want to have to deal with TCP/IP if you want to build Twitter. And that's not just technical. Because it's very tempting to reach from one kind of property right to another. And the HapMap was a great example of this. The HapMap was the international haplotype map project that followed on to the Human Genome Project. The goal was to find what was different bus, just as the Human Genome Project was to find what was similar. And they had this clause that said you had to sign a click-wrap agreement. So it wasn't actually in the public domain. And you agreed not to take any action, including patenting that would restrict access to others. And you would share the data with anyone else who didn't sign the contract. And this was in the name of freedom. And this was in the spirit of free software in the GPL. First of all it didn't stop patents. What stopped patents was disclosure, public domain deposit of information. And second, it made it illegal to share the data. So it couldn't be integrated with all of the other stuff that came out of the Human Genome Project. And when we think about data licenses, copyright licenses, patent licenses, materials transfer agreements, we have to think about them in the layer at which they exist. And not try to go up and down. Because in many ways you break the commons by trying to reach too far. Making the transactions clean, transparent, simple, scalable within their own area works a lot better than trying to cross across. So at every point, Creative Commons, the science project, what we're trying to do is be the middle of the hour glass. Because we think it's really, really important. And these are the five points that we wanted to be graded on. So as we fail on any of these, we want to be told. So when I think about this, the thing that gets me up and gets me on to the plane every time is, you know, we've been focusing on this, which is today. But science in many ways is going back to the garage. I mean science started as an amateur activity. The journals that Peter showed us started as an amateur journals. It was a gentleman and gentlewoman activity to be a scientist. You should read the American journals from -- especially from entomology from a hundred years ago. Everyone from sort of random people in Cambridge, the US Cambridge, people like Vladimir Nabokov submitted to entomology journals. So we've lost that as we've basically commoditized and productized science. And you know, computers used to be like that. This is NEAC, right? And change comes from humble beginnings. This is the Apple 1. Science is about the Apple 1 right now, especially biology. This is the $100 do it yourself gel electrophoresis box. The spec is available online at DIWI biology. Right? Biology in particular is heading back to the garage. You can buy a sequencer on eBay delivered in 24 hours for under $1,000. You can synthesize it Mr.gene.com for 66 cents a base pair. And what you see as you look across all of these is a decay in cost and an increase in capacity that almost exactly mirrors optical disk drives. Which is to say that it's going to be possible to have biology in your house in about 10 years, whether you want to engineer yeast to make beer or for more nefarious purposes. And the question is how are we going to deal with it. This was in the New York Times this weekend. They are from the City College of San Francisco. And they are competing in an international competition of engineered genetic machines at MIT. They are programming E. Coli to do things as funky as arsenic detection or just to make it smell like bananas so the lab doesn't smell so bad, as anyone who has ever worked with E. Coli knows. They use standard biological parts that you can download off of the Internet. So if you happen to need a catalog of ribosome binding sites, you can download the sequences and for 66 cents a base pair sequence them and use them in the lab. What this lets you do is begin to think of biology as a field that's about to judge go the transformation that computers underwent 40 years ago. And the question is is it going to be a PC or an iPhone in the future? So the iPhone is a beautiful toy. But it is sterile. Only the things that Apple approves are allowed on to the iPhone. A PC was much uglier than Mac in the mid '80s or an Apple in the mid '80s. But anyone could write anything to it. And it made us responsible for what we installed. It gave each of us the power to customize our experience and to add to that experience. Whereas the iPhone is a safe, beautiful, sterile tool. And it's really important which of these futures biology takes. Because the ability to be bad in the new biology world will be ubiquitous because those people will simply break the law. And they will write viruses in the real world. And our ability to deal with them needs to scale with the ability of the users to deal with the problems. And that's only going to happen if the approach we take is the picture approach, which lets us crowd source the reactions, not just the applications we love. And it's important beyond biology, right? So I spend a lot of my time over the last year working on sustainability. It's a new field for us. So this is another bad curve. And this is our energy consumption worldwide. I just got back from India and I can tell you screw cars. If they have get paper towels to the poor of India, right, we have a problem of consumption and landfill, that dwarfs what we have in the United States. And expecting everyone to magically cut their consumption, as nice as it may be and start riding cars that are running on used frying oil isn't going to happen. Not in time. The carbon curves are worse. And that's why companies like Nike are looking at their portfolio and saying, can someone please innovate to deal with the problem. And biology, especially programmable biology, offers us a route out. Which is the chance to design life that can actually do things like chew through landfill or sequester carbon. And again, the question is, you know, are we going to have an innovation based chance of success for this, or are we going to have a future in which a couple of companies control every application that gets written? Because that's where a lot of the companies involved in science want it to go. They all want to be the sterile platform, right? You have no idea how many people come to me and say we're going to be the iPhone for content in science, we're going to be the iPhone for this in science, the iPhone for that, the app store for this. It's the metaphor that's taking the business world by storm. But I would far prefer we have a PC world, where it's ugly and there's just a C and a colon and a slash, but anyone that wants to can write code. Because I think that's the best chance we have to deal with both the problems that the science increases face us with as well as the other sorts of problems we deal with like climate change and carbon. And so that's -- that's why we do what we do. And that's why days like this are so important because it gives us a chance to celebrate some of the stuff we have in common and come together and then hopefully taking what we're doing as the new normal, look ahead and actually have the vision and the courage to tell the rest of the world that this is why this is important. Because if we don't do this, our chances of succeeding in some of the biggest challenges we face radically decreases. Thanks. [applause]. >> Lee Dirks: A couple of questions? Or we can go [inaudible] and mingle. >> John Wilbanks: I've been speaking for an hour, so people may be sick of it. >> Lee Dirks: I doubt that. Well, if there's no further questions, thanks to all of you for the day. Thanks to the speakers. Thanks to Lisa. I'll also thank a lot of the people in Microsoft Research who helped pull this all together. But we can move over to the atrium area and please join us for some wine and cheese. A hand to all of you, please. [applause]