Structure Determination by NMR
advertisement
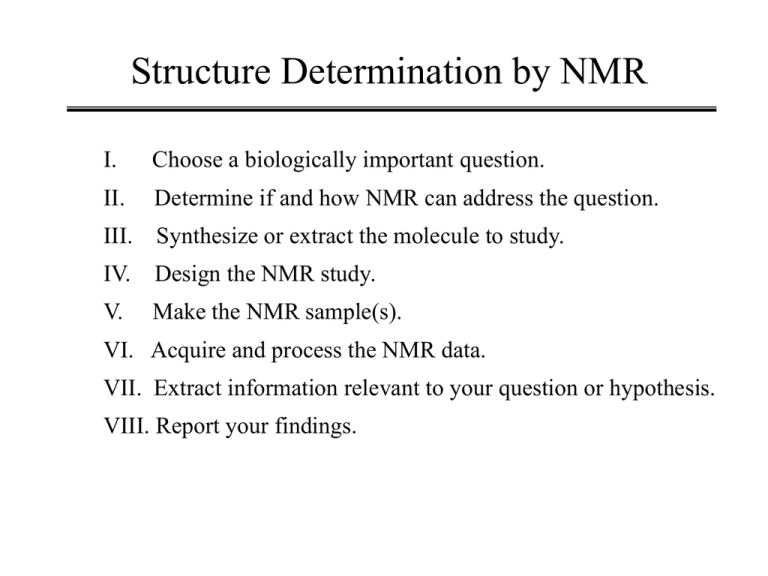
Structure Determination by NMR I. Choose a biologically important question. II. Determine if and how NMR can address the question. III. Synthesize or extract the molecule to study. IV. Design the NMR study. V. Make the NMR sample(s). VI. Acquire and process the NMR data. VII. Extract information relevant to your question or hypothesis. VIII. Report your findings. Hepatitis B Virus The Disease • Member of the hepatocellular DNA virus family • 300 million people worldwide are carriers. • Symptom of infection vary but usually involve inflamation of the liver and sometimes liver damage. • 90% of the people who contract the virus will go through an acute phase of infection and then recover with lasting immunity. • 10% of the people who contract the disease do not resolve the primary infection and become carriers. • Those that have the chronic infection have a 100-fold or greater risk of hepatocellular carcinoma (liver cancer). The Hepatitis B Virus Genome - + ~3.2 kilobases RNA Protein 5’ 5’ + Plus strand 5’-GGCAGAGGTGAAA-3’ 3’-CCGTCTCCACTTT-5’ Direct Repeat Sequence The Hepatitis B Virus Direct Repeat Sequence 5’-GGCAGAGGTGAAA-3’ 3’-CCGTCTCCACTTT-5’ I. Performs a critical role in the initiation of viral DNA synthesis which is not completely understood. II. Deletion or mutation of just one residue can be catastrophic to virus. III. Small enough to be studied by NMR. IV. Are there any unique structural features that can give us insight into biological activity? V. The sequence will have an extra base-pair on each end. Review of DNA Structure Review of DNA Structure Review of DNA Structure Review of DNA Structure NMR Study of DR1 COSY resonance assignments torsion angles sugar conformation NOESY resonance assignments interproton distances Chemical exchange imino proton exchange rates, i.e. base pair opening Resonance Assignments A combination of COSY and NOESY. Use known characteristics of molecule. sequence, identity of terminal bases, etc. Confirm base-pair formation. Initially assume it has a regular structure, e.g. B-DNA. DNA/RNA Backbone Structure Bloomfield et.al. “Nucleic Acids; Structure, Properties, and Functions” 2000. Pseudorotation Phase Cycle of Deoxyribose “Principles of Nucleic Acid Structure” Saenger, pg 19 (1984). Preferred Pseudorotation Phase Angles B-DNA “Principles of Nucleic Acid Structure” Saenger, (1984). A-DNA, RNA Sequential Resonance Assignments Interproton contacts less than 4Å in (a) B-DNA and (b) A-DNA. “Biomolecular NMR Spectroscopy” J.N.S. Evans, pg 350 (1995). 2D NOESY of DR1 H1’-to-H2’H2” H8,H6-to-H1’,H5 Bishop et.al., Biochemistry (1994). Base-to-H1’ NOESY-walk Bishop et.al., Biochemistry (1994). Proton Chemical Shifts of DR1 ~97% of all protons are assigned Bishop et.al., Bioch (1994). Distribution of Distance Constraints 502 NOE derived distance constraints. Bishop et.al., Bioch (1994). E.COSY H1’-to-H2’H2” A18,H1’-A18,H2” Bishop et.al., Bioch (1994). E.COSY A18,H1’-to-H2’H2” 5.9 Hz = J1’-2” Linewidth ~4.9 Hz Bishop et.al., Bioch (1994). Coupling Constants and Conformations for Sugars %S versus Base-pair Relative imino proton exchange rate. ? ? ? ? Bishop et.al., Bioch (1994). Structure Determination Vtotal = Vbondlength + Vbondangles, Vdihedral + Velectrostatics + VNOE + Vjcoupling V rl ru constraint VNOE = S all NOEs k2(r-rl)2 0 k3(r-ru)2 4k2(r-ru)2 when r<rl when rl< r <ru when r < r u when r>r u Structure Determination NATO ASI Vol H87 “NMR of Biol. Macr.” James et al., (1994). Structure Determination 10 rMD structures of [d(AGCTTGCCTTGAG)[CTCAAGGCAAGCT)] RMSD = 0.9Å 267 distance restraints 130 torsion angle restraints NATO ASI Vol H87 “NMR of Biol. Macr.” James et al., (1994).

