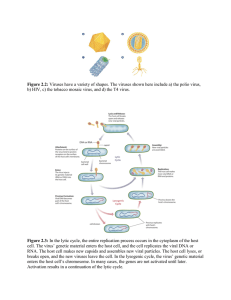
Chapter 9 I. Viral Genomes and Evolution •
advertisement

Chapter 9 Viral Genomes and Diversity I. Viral Genomes and Evolution • 9.1 Size and Structure of Viral Genomes • 9.2 Viral Evolution 9.1 Size and Structure of Viral Genomes • Viral genome size (Figure 9.1) • • • Smallest circovirus: 1.75-kilobase single strand Largest megavirus: 1.25-megabase pairs Viral genomes (Figure 9.2) • Either DNA or RNA genomes • Some are circular, but most are linear 9.2 Viral Evolution • Viruses may have arisen after cells • • Remnants of cells that replicate with the help of a live cell Viruses may have played a role in transition from RNA to DNA (Figure 9.3) 9.2 Viral Evolution • Viral phylogeny • • Most viral genes from nature have unknown function • This makes it difficult to construct phylogenetic tree A phylogenetic tree has been constructed for nucleocytoplasmic large DNA viruses (NCLDV; Figure 9.4) II. Viruses with DNA Genomes • 9.3 Single-Stranded DNA Bacteriophages: ɸX174 and M13 • • • • 9.4 Double-Stranded DNA Bacteriophages: T7 and Mu 9.5 Viruses of Archaea 9.6 Uniquely Replicating DNA Animal Viruses 9.7 DNA Tumor Viruses 9.3 Single-Stranded DNA Bacteriophages: ɸX174 and M13 • Some bacteriophages contain single-stranded DNA genomes of the plus configuration • Transcription of the genome is preceded by synthesis of a complementary strand of DNA 9.3 Single-Stranded DNA Bacteriophages: ɸX174 and M13 • Bacteriophage ɸX174 • Contains a circular single-stranded DNA genome inside an • • icosahedral virion (Figure 9.5) Very small genome with overlapping genes Replication occurs via rolling circle replication (Figure 9.6) 9.3 Single-Stranded DNA Bacteriophages: ɸX174 and M13 • Bacteriophage M13 • • • • Model filamentous bacteriophage Used as a cloning and DNA-sequencing vector in genetic engineering Can be released without lysing host via a process called budding (Figure 9.7) Viral infection slows host growth 9.4 Double-Stranded DNA Bacteriophages: T7 and Mu • Bacteriophage T7 • • • • Infects E. coli Virion has an icosahedral head and a very short tail Genome always enters host cell in same orientation Order of genes on the T7 chromosome influences regulation of virus replication 9.4 Double-Stranded DNA Bacteriophages: T7 and Mu • Bacteriophage T7 (cont'd) • DNA replication employs T7 DNA polymerase and involves terminal repeats and the formation of concatemers (Figure 9.8) 9.4 Double-Stranded DNA Bacteriophages: T7 and Mu • Bacteriophage Mu • • • • • "Mutator" phage • Induces mutations in host genome Useful in bacterial genetics Temperate phage Replicates by transposition Large virus with an icosahedral head, helical tail, and six tail fibers (Figure 9.9a) 9.4 Double-Stranded DNA Bacteriophages: T7 and Mu • Bacteriophage Mu (cont'd) • • Invertible G region of genome determines host range Genome is integrated into the host chromosome via a transposase (Figure 9.9b) 9.4 Double-Stranded DNA Bacteriophages: T7 and Mu • Bacteriophage Mu (cont'd) • In both lytic and lysogenic pathways, the genome is • • replicated as part of a larger DNA molecule Lysogenic state requires sufficient amounts of a repressor protein to prevent transcription of integrated Mu DNA Genome is packaged into the virion with short (5 bp) sequences of host DNA at either end 9.5 Viruses of Archaea • Most viruses that infect Archaea resemble those that infect enteric bacteria (Figure 9.10) • Only double-stranded DNA viruses have been identified so far 9.6 Uniquely Replicating DNA Animal Viruses • Double-stranded DNA animal viruses that have unusual replication strategies • Pox viruses • Adenoviruses 9.6 Uniquely Replicating DNA Animal Viruses • Pox viruses • • Among the most complex and largest animal viruses known (Figure 9.11) DNA replicates in the cytoplasm 9.6 Uniquely Replicating DNA Animal Viruses • Adenoviruses • • • Major group of icosahedral, linear, double-stranded DNA viruses Cause mild respiratory infections in humans DNA replicates in the nucleus • Replication requires protein primers and avoids synthesis of a lagging strand (Figure 9.12) 9.7 DNA Tumor Viruses • Some DNA animal viruses can induce cancer • • Polyomavirus SV40 Herpesviruses 9.7 DNA Tumor Viruses • Polyomavirus SV40 • • • • • Induces tumors in animals Nonenveloped virion with an icosahedral head No enzymes in the virion; replicates in host nucleus DNA is circular (Figure 9.13a) Small genome, has overlapping genes 9.7 DNA Tumor Viruses • Some polyomaviruses cause cancer • In permissive host cells, virus infection results in the • formation of new virions and the lysis of the host cell In nonpermissive host cells, the virus DNA becomes integrated into host DNA (analogous to a prophage), genetically altering cells in the process (Figure 9.13b) 9.7 DNA Tumor Viruses • Herpesviruses • • • Large group of viruses that cause diseases in humans and animals Able to remain latent for extended periods of time An important group causes clinical forms of cancer • Example: Epstein–Barr virus 9.7 DNA Tumor Viruses • Herpesviruses (cont'd) • • Infection follows attachment of virions to specific cell receptors Three classes of mRNA are produced (Figure 9.14): • Immediate early: encodes five regulatory proteins • Delayed early: encodes DNA replication proteins • Late: encodes structural proteins of the virus particle III. Viruses with RNA Genomes • 9.8 Positive-Strand RNA Viruses • • • 9.9 Negative-Strand RNA Animal Viruses 9.10 Double-Stranded RNA Viruses 9.11 Viruses That Use Reverse Transcriptase 9.8 Positive-Strand RNA Viruses • Phage MS2 • • • An example of a bacterial RNA virus that infects E. coli by attaching to pilus (Figure 9.15a) Possesses a small genome that is directly translated by a combination of host and viral enzymes (Figure 9.15b and c) Possesses overlapping genes 9.8 Positive-Strand RNA Viruses • Replication of positive-strand RNA viruses requires a negative-strand RNA intermediate from which new positive strands are synthesized 9.8 Positive-Strand RNA Viruses • Poliovirus (Figure 9.16a and b) • • Small virus Viral RNA is translated directly, producing a single long, giant protein (polyprotein) that undergoes self-cleavage to generate ~20 smaller proteins necessary for nucleic acid replication and virus assembly (Figure 9.16c) 9.8 Positive-Strand RNA Viruses • Poliovirus (cont'd) • Host RNA and protein synthesis are inhibited when poliovirus replication begins 9.8 Positive-Strand RNA Viruses • Coronaviruses • • • Larger virus Cause respiratory infections, including SARS, in humans and other animals Virions (Figure 9.17a) • Are enveloped • Contain club-shaped glycoprotein spikes on their surfaces • Produces monocistronic mRNA (Figure 9.17b) 9.9 Negative-Strand RNA Animal Viruses • Negative-strand RNA viruses • • Negative-strand RNAs are complementary to the mRNA • They are copied into mRNA by an enzyme present in the virion Only those that infect Eukarya are known 9.9 Negative-Strand RNA Animal Viruses • Rhabdoviruses • Include viruses that cause: • • • Rabies in animals and humans • Vesicular stomatitis in cattle, pigs, and horses Enveloped viruses Virion is bullet-shaped (Figure 9.18a) 9.9 Negative-Strand RNA Animal Viruses • Rhabdoviruses (cont'd) • RNA of rhabdoviruses is transcribed in the host cytoplasm into two distinct classes (Figure 9.18b): 1. Series of mRNAs encoding the structural genes of the virus 2. Positive-strand RNA that is a copy of the complete viral genome 9.9 Negative-Strand RNA Animal Viruses • Influenza • • • Enveloped, pleomorphic virus (Figure 9.19) Segmented genome Surface proteins interact with host cell surface 3. Hemagglutinin causes clumping of red blood cells 4. Neuraminidase breaks down sialic acid component of host cytoplasmic membrane 9.9 Negative-Strand RNA Animal Viruses • Processes that help influenza elude the host immune system • Antigenic shift 5. Portions of the RNA genome from two genetically distinct strains of virus infecting the same cell are reassorted 6. Generates virions that express a unique set of surface proteins • Antigenic drift 7. Structure of neuraminidase and hemagglutinin proteins are subtly altered 9.10 Double-Stranded RNA Viruses • Reoviruses (Figure 9.20a and b) • • Nonenveloped nucleocapsid with a double shell of icosahedral symmetry Virions contain virus-encoded enzymes necessary to synthesize mRNA and the new RNA genomes 9.10 Double-Stranded RNA Viruses • Reoviruses (cont'd) • • Genome segmented into 10–12 molecules of linear doublestranded RNA Replication occurs exclusively in host cytoplasm within the nucleocapsid (Figure 9.20c) 9.11 Viruses That Use Reverse Transcriptase • Retroviruses (RNA viruses) and hepadnaviruses (DNA viruses) use reverse transcriptase for replication • Retroviruses 8. Enveloped virions that contain two copies of the RNA genome 9. Virion contains several enzymes 10. Includes reverse transcriptase used to make DNA copy of genome (Figure 9.21) 9.11 Viruses That Use Reverse Transcriptase • Retroviruses (cont'd) • • Gene expression and protein processing are complex (Figure 9.22) All retroviruses have the three genes: 11. gag encodes several small viral structural proteins 12. pol is translated into a large polyprotein 13. The env product is processed into two distinct envelope proteins 9.11 Viruses That Use Reverse Transcriptase • Hepadnaviruses • • • • Virions small, irregular-shaped particles (Figure 9.23a) Include hepatitis B Viral replication occurs through an RNA intermediate Unusual genomes 14. Tiny 15. Only partially double-stranded (Figure 9.23b) IV. Subviral Agents • 9.12 Viroids • 9.13 Prions 9.12 Viroids • Viroids: infectious RNA molecules that lack a protein coat • Smallest known pathogens (246–399 bp) • Cause a number of important plant diseases (Figure 9.24) • Small, circular, ssRNA molecules (Figure 9.25) • Do not encode proteins; completely dependent on hostencoded enzymes 9.13 Prions • Prions: infectious proteins whose extracellular form contains no nucleic acid • Known to cause disease in animals (transmissible • • spongiform encephalopathies) Host cell contains gene (PrnP) that encodes native form of prion protein that is found in healthy animals (Figure 9.27) Prion misfolding results in neurological symptoms of disease (e.g., resistance to proteases, insolubility, and aggregation)