by Parrish Myers Random Walk Contaminant Spread DSES-6620 Simulation Modeling and Analysis
advertisement

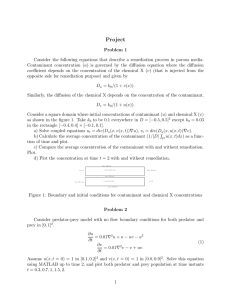
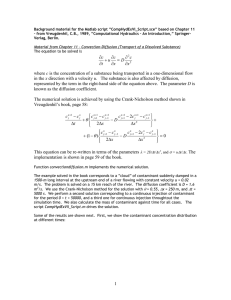
Random Walk Contaminant Spread by Parrish Myers DSES-6620 Simulation Modeling and Analysis Spring 2002 Ernesto Gutierrez-Miravete Random Walk Contaminant Spread Parrish Myers Introduction The release of a deadly contaminant within a populated ecosystem can become of great concern. This concern can come from the death toll associated with the outbreak of a deadly virus within a city, or even the impact of a fish population associated with chemical waste being emptied into local lakes and rivers. In each case a localized problem can both devastate that local ecosystem and spread beyond the boundaries of the localized system and become a much larger problem. Simulations can answer some of that concern by predicting the death toll, or providing future trends in fish population. Project Overview and Objectives To investigate the process of a contaminant spread, a first-order model has been constructed to simulate a small and localized ecosystem. The simulation attempts to mimic a small container (or exposure chamber) filled with water and a limited number of organisms. This small ecosystem (or system) will then have a contaminant introduced, modeled using a random walk process. The organism chosen for this simulation was Daphnia Magna a small freshwater crustacean that is extensively used as both a fish food and as a bio-indicator in toxicity experiments. The contaminant chosen was Methyl Parathion, an insecticide that has been made in the United States since 19521. Both the organism and contaminant were primarily chosen because of readily available information from the Environmental Protection Agency’s (EPA) web site 2. The original objective of this project, determining the killing radius and time it takes for a contaminant to become non-lethal, was modified slightly to provide a clear method for validation. The current objective of this experiment is to 1 More information can be found at the ATSDR homepage at http://www.atsdr.cdc.gov/tfacts48.html. 2 The Environmental Protection Agency is located at http://www.epa.gov. 2 Random Walk Contaminant Spread Parrish Myers determine the median lethal dose3 (LD50) value of Methyl Parathion, using Daphnia Magna as the test organisms. While this simulation will not be sufficient to answer any real questions about the mortality rate of an organism’s population in a real ecosystem, it will provide a non-lethal introduction to the subject of acute toxicity. Model Design This experiment has a clearly defined organism and contaminant under study, even so, this simulation was designed with enough generality to allow the simulation to be extended to any organism and contaminant combination, and the following description of the system will reflect that. Early in development it was recognized a general purpose programming language would provide the needed flexibility and control necessary to successfully create this simulation. This is in contrast to simulation packages, such as ProModel or Witness, which are geared towards manufacturing and service systems. The programming language Java4 was selected for its robust 2D drawing capabilities, and cross-platform nature. The resulting application, named “rwcs” (random walk contaminant spread), can be run anywhere the Java Runtime Environment has been ported to. The source code (seen in Appendix B) uses the object-oriented nature of the Java language to compartmentalize the structure of the source code. This simulation program contains 7 main classes or sections, 3 Casarett, L. J., p.19. J2SE version 1.3.1_03 was used for this simulation. It can be downloaded from http://java.sun.com. 4 3 Random Walk Contaminant Spread Parrish Myers Class Purpose Main entry point / user interface controls and input Simulation thread / main Simulation drawing and control All functions related to managing the organism All functions related to managing the contaminant Central repository for data and data access Basic data structure to provide the (x, y) position and life count All functions related to the reading of default options rwcs Contaminant FishControl ContaminantControl SystemDataStore Ball GlobalProps with a separate file for each class. This structure insured the program could be understood and extended in functionality with minimal effort. The main application interface is show in Figure 1. The interface contains basic controls to start, stop, pause, and resume the simulation. Text entries have been added to control some of the available parameters of the simulation. Additional parameters are available via the configuration file that accompanies this program. The main area of the application is where the contaminant spread is visualized. A zoom has been added to aid in the visualization detail shown during the simulation run-time. The bottom edge of the interface includes system status and parameters of the running simulation. The system visualization area consists of a black 2 dimensional field containing both organisms and contaminant drawn as circles. Organism and contaminant are distinguished by color, with healthy organisms drawn in blue, dead organisms drawn in yellow with a black “x” in the center, and the contaminant drawn in red. A system boundary drawn in green is also drawn, but is not shown in Figure 1. 4 Random Walk Contaminant Spread Parrish Myers Figure 1 - RWCS Program Interface The original proposal stipulated a number of simplifying assumptions, which were agreed upon. Those assumptions guided the construction of the simulation. Simulation Assumptions: 1. The system is to be modeled in 2 dimensions. 2. The contaminant spread is modeled as a random walk of constant step size. 3. All N molecules of the contaminant start a (0,0), which is denoted to be the center of the system. 4. The contaminant is assumed discrete, containing N i grouping of molecules: where i 0,1, 2,... and each group consists of x molecules (the exact number to be determined at verification of the model) 5 Random Walk Contaminant Spread Parrish Myers 5. There are two exits for the N i molecule groups within the system: 1) the molecule group wanders beyond the boundary set up at system start and 2) the molecule groups collide with an organism. 6. Each organism within the system has a stochastic susceptibility, which will be calculated at simulation start. When a N i molecule group collides with the organism the life of that organism is reduced by a fraction of that susceptibility amount. 7. During the life of the simulation the organisms are stationary The ecosystem consists of a 2 dimensional plane broken into equidistant lattice points set upon the Cartesian coordinate system. Therefore any object in the system can be located using a two-element coordinate (x, y). The origin (0, 0) is defined to be the center of the system. The random walk process is extensively used in statistical physics to characterize diffusion of molecules in a substance5. The contaminant spread is modeled as a random walk of constant step size with no drift. This corresponds to each step taken by one of the N i molecule groups having an equal probability of moving up, down, left, or right one lattice point per time step. At simulation start time a known number of molecule groups, N , are placed at (0,0) and are allowed to start wandering (at each time step all molecule groups are moved one lattice point). The direction is randomly picked using a random variate with a discrete uniform distribution between 0 and 3. The direction of travel is then correlated to the number drawn, with 5 The random walk is normally used to describe diffusion of a molecule traveling in a gas. But, this process is still valid for molecules traveling in a liquid, albeit slower, and due to its simplicity it is used for the contaminant spread in water. 6 Random Walk Contaminant Spread Parrish Myers Enumeration Direction 0 RIGHT 1 UP 2 LEFT 3 DOWN Due to the sheer number of molecules present in any quantity of contaminant, keeping track of all the individual molecules is computationally expensive. Hence, a known quantity6 of molecules is grouped together to form discrete element N i , where i 1, 2,3,... . The number of molecule groups is then keep less than 1000. This restriction greatly reduces the computational power required to run the simulation and allow it to be run on any platform or computer that supports a recent version of the Java runtime environment. Two exits exist in the simulation for molecule groups of contaminant to leave the system. One, the molecule wanders past the system area defined7 and two a molecule group collides with an organism. If a group of molecules wanders past the defined boundaries, the likelihood that that group will interact with any of the remaining organisms is small. Therefore, rather can continue wasting computer cycles tracking a non-interacting object, the group is removed from the system. The second exit condition provides the mechanism for interaction between organism and contaminant. When a collision occurs the contaminant is removed from the system, to prohibit that molecule group from reacting with another organism. At simulation start a discrete random-variate is calculated for each of the organisms that exist within the system. This defines the susceptibility (or life count) of that organism. If a molecule group collides with that organism the life count of that organism is reduced by one 6 7 The precise quantity of molecules in the group is determined at Validation. The exact system area is defined at validation. 7 Random Walk Contaminant Spread Parrish Myers before collision Si Li after collision Si Li 1 where, Si is the life count and Li is the susceptibility calculated for organism i at simulation start. When, the life count Si reaches 0 the organism is considered dead. To illustrate the decrement of the life counter, a color scale has been created, seen in Figure 2. When life counter, Si , equals the initial susceptibility value, Li , the organism is drawn in blue. As the organism loses life the color transitions to green, and finally to Figure 2 - Life Count yellow when the life count reaches 0. Scale for Organisms To reduce model complexity all organisms within the system are stationary. At simulation start, two random-variates of uniform distribution is calculated for each coordinate x and y8 for each organism Oi where i 1, 2,3,... . Data Collection Though much effort and investigation, no substantial data on the physiological effects of substances could be collected in terms of the susceptibility of the Daphnia Manga to a general contaminant. The basic physiological effects of a substance on any organism are a complicated question to answer; hence the need for Toxicology studies. Most of the data available consists of median lethal dose9 (LD50) data. Without any real data, the two choices are uniform and triangular random-variates. The susceptibility is thought to happen around a central or median value. A distribution that is centered around a median value is 8 In reality a uniform random-variate is drawn for the distance from the origin and a second uniform random-variate is drawn for the angle around the origin. These two coordinate values are then translated into an x, y position using a standard transform from a circular coordinate system to Cartesian. 9 Casarett, L. J. p.19 8 Random Walk Contaminant Spread Parrish Myers better suited than a uniform random-variate, and therefore a triangular randomvariate10 was selected for the susceptibility of the Daphnia Magna. A uniform random-variate has been chosen for the initial position of the organisms within the system. While a heterogeneous environment my exist the organisms can and will move around the system. No distribution or clumping can be predicted or known in advance (assuming no currents, uniform temperature, ph and lighting). Therefore the organisms can and will occupy all areas of the system. A uniform random-variate works well for this situation. Verification / Validation A source code debugger and visual inspections of the simulation animation11 were extensively used in reducing grammatical and logical errors. The simulation heavily relies on the compartmentalization attained by object-oriented design. This ensured the design of the simulation has logically breaks between features for readability and maintainability. The simulation assumptions not only provided a framework for construction, but a method for verification as well. Close attention has been made to insure the simulation conforms to the stated assumptions. The simulation has also been designed to terminate under many conditions: All organisms are dead, All contaminant molecule groups have exited the system, and The system time limit has expired, to ensure reliability. The simulation procedure uses a modified version of the EPA’s standard operating procedure # 2024 “48-Hour Acute Toxicity Test Using Daphnia Magna 10 the form of the triangular random-variate was obtained from http://www.itl.nist.gov/div898/software/dataplot/refman2/ch8/tripdf.pdf. 11 Banks J., p. 370 9 Random Walk Contaminant Spread Parrish Myers and Daphnia Pulex”12. This procedure outlines the process (seen in Appendix A) that was used to calibrate the simulation. Model Results In an effort to follow the EPA’s SOP #2024 a set of parameters were determined by trial and error, to produce a realistic simulation. The number of Daphnia Magna was limited to 10. The radius of the system was set to 50.0. The simulation was set to end in 48-hours, with a time step of 1 minute. At each time step the contaminant (Methyl Parathion) was advanced 0.4, the lattice step size. Three alarms were set to display the number of dead organisms at 1, 24, and 48 hours. The susceptibility parameters for the triangular distribution had a minimum value of 1 and a maximum value of 5 with a mean of 3. Once these parameters were set, they were not changed for the entire data collection period. The only parameter left unspecified was the number of molecule groups of Methyl Parathion. This parameter simulates the amount of contaminant within the system. The procedure for operation thus follows: 1. Start with an amount of contaminant small enough to have minimal effect 2. Record the number of dead organisms at: 1-Hour, 24-Hours, and 48Hours 3. Increase the amount of contaminant and repeat step 2. 4. When organism mortality becomes 80%-90% STOP! 5. Tally data and find LD50. The following table shows the data collected from following this procedure: 12 http://www.epa.giv/epahome/index/nameindx.htm 10 Random Walk Contaminant Spread Molecule Parrish Myers Mortality Count Groups 1 Hour 24 Hours 48 Hours 40 2 3 4 50 2 3 4 60 2 3 5 70 2 3 5 80 2 5 6 90 2 4 5 100 2 4 7 110 2 4 7 120 2 4 7 130 2 4 5 140 2 6 6 It was determined from the data that the effective concentration 50% region (EC50) was between 60 and 90 molecule groups. The data collected from the EPA web site13 stipulates that Methyl parathion is “very highly toxic” to Daphnia Magna with the LD50 value is 0.14 ppb and the EC50 range is 0.09 – 0.2 ppb. Using a simple linear correlation, (3 23 x103 )M g 0.13 C where M g is the number of molecule groups and C is the resulting dose concentration, the number of molecule groups was correlated to concentration of Methyl parathion in units of part per billion. This resulting data is shown in Figure 13 http://www.epa.gov/pesticides/op/methyl_parathion/mp_12_1.pdf 11 Random Walk Contaminant Spread Parrish Myers 3. The graph shows both the calibrated EC50 region and the corresponding LD50 value of 0.14. Mortality of Delphia Magna 9 EC50 Area Number of Organisms Dead 8 LC50 7 6 5 4 3 48 hours 24 hours 1 hour 2 1 0. 60 0. 57 0. 53 0. 49 0. 46 0. 42 0. 38 0. 31 0. 35 0. 27 0. 24 0. 20 0. 16 0. 13 0. 09 0. 05 0. 02 0 Concentration (ppb) Figure 3 - Simulation Results Conclusion While this simulation is only a first-order representation of an LD50 toxicity experiment, it does provide valuable (and non-lethal) information in the interaction and conduct of a contaminant spread within a small ecosystem. Even so, this simulation only studied one organism, Daphnia Magna. A real medial lethal dose toxicity experiment includes many organisms of various sizes and species before any real conclusion can be drawn. There are many enhancements and features that missed inclusion in this simulation due to time constraints. Some of these features include: The organisms within the system move The system modeled in 3 dimensions The animations can be made more elaborate 12 Random Walk Contaminant Spread Parrish Myers The contaminant cannot exceed the boundaries of the system (they bounce off the walls... this simulated being in a exposure chamber). Many additional organisms should be simulated to generate a better LD50 determination The inclusion of a configuration menu to reduce the need for modifying the configuration file every time a parameter (that is not on the interface) with a text editor. The ability to queue many runs. Automatic data recording. Automatic data conversion from simulation units to real units. A second order model should address some of these suggestions. 13 Random Walk Contaminant Spread Parrish Myers References SOP# 2024, 1994, 48-Hour Acute Toxicity Test Using Daphnia Magna and Daphnia Pulex, EPA, http://www.epa.gov/epahome/index/nameindx.htm Casarett, L. J., 1980, Casarette and Doull’s Toxicology, Macmillan Publishing Co., New York, p.17-27. Reif, F., 1965, Fundamentals of statistical and thermal physics, McGraww-Hill, New York, p. 486-487. Whitney, C. A., 1990, Random Processes in Physical Systems, Wiley Interscience Publishing, New York, p. 37-46. Banks, J., Carson, J. S. II, Nelson, B. L., Nicol, D. M., 2001, Discrete-Event System Simulation, Prentice Hall, New Jersey, p. 18, 290-297, 367. Ross, S. M., 1997, Introduction to Probability Models, Sixth Edition, Academic Press, San Diego, p. 55-56. 14 Random Walk Contaminant Spread Appendix A – EPA SOP #2024 Parrish Myers Random Walk Contaminant Spread Appendix B – Source Code Parrish Myers