2006 NCRR Report Presentation
advertisement

Applying Semantic Technologies
to the Glycoproteomics Domain
W. S York
May 15, 2006
Some Goals of Glycoproteomics
• How do changes in the expression levels of
specific genes alter the expression of specific
glycans on the cell surface?
• Are changes in the expression of specific glycans
at the cell surface related to cell function, cell
development, and disease?
• What are the mechanisms by which specific
glycans at the cell surface affect cell function, cell
development, and the progression of disease?
Challenges of Glycoproteomics
• Vast amounts of data collected by highthroughput experiments - better methods for data
archival, retrieval, and analysis are needed
• Complex structures of glycans and glycoproteins –
better methods for representing branched
structures and finding structural and functional
homologies are needed
• Complex Biology and Biochemistry – better
methods to find relationships between the
glycoproteome and biological processes are needed
Glycoproteomics Solutions
• Brute-force analysis of flat data files
• Too much data
• Data is heterogeneous
• What does the data represent?
• Relational databases
• Data is well organized
• Data organization is relatively rigid
• What does the data represent?
• Semantic Technologies
• Data is well organized
• Data organization is flexible
• Concepts represented by data are accessible
• Relationships between concepts are accessible
What is Semantic Technology?
The implication is Semantics:
that enabling computers to
thestudy
meanings
relationships
1.“understand”
(Linguistics) The
or scienceofof and
meaning
in language.
2.
(Linguistics)
The study
relationships
signs and
between
concepts
willof allow
them between
to reason
and
symbols
and whatin
they
communicate
a represent.
way that is analogous to the way
The American Heritage® Dictionary of the English Language, Fourth Edition
humans do.
Semantic Technology:
The use of formal representations of concepts and their
relationships to enable efficient, intelligent software.
Ontology (Computer Science):
A model that represents a domain and is used to reason about
the objects in that domain and the relations between them.
http://en.wikipedia.org/wiki/Ontology_(computer_science)
A Simple Ontology
Organism
is_a
is_a
Animal
is_a
Lion
is_a
Deer
is_a
Elsa
Simba
is_a
Cow
is_a
is_a
Plant
Elsie
is_a
Bambi
ate
ate
ate
ate
is_a
is_a
Hosta
Alfalfa
is_a
is_a
My Hosta
Peter’s
Alfalfa
A Simple Ontology
is_a
is_a
Lion
Elsa
Elsie
is_a
Bambi
ate
ate
ate
is_a
is_a
Hosta
Alfalfa
is_a
Deer
is_a
Plant
eats
Herbivore
Cow
is_a
Simba
is_a
is_a
is_a
is_a
is_a
Animal
eats
Carnivore
Organism
ate
is_a
is_a
My Hosta
Peter’s
Alfalfa
The Structure of GlycO – Concept Taxonomy
chemical
entity
is_a
molecule
molecular
fragment
carbohydrate
moiety
is_a
is_a
residue
is_a
monoglycosyl
moiety
glycan
moiety
amino acid
residue
carbohydrate
residue
is_a
N-glycan
O-glycan
The Structure of GlycO – Concept Taxonomy
residue
is_a
glycan
moiety
amino acid
residue
carbohydrate
residue
is_a
N-glycan
O-glycan
The Structure of GlycO – Instances
Concept Taxonomy
and Properties
N-glycan_00020
has_residue
is_linked_to
N-glycan
a-D-Manp 4
is_instance_of
residue
is_a
N-glycan core
b-D-Manp
amino acid
is_instance_of
residue
carbohydrate
residue
glycan
moiety
is_instance_of
is_a
N-glycan
O-glycan
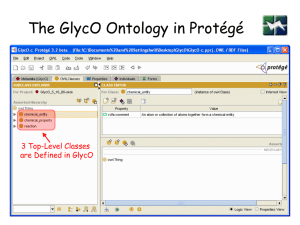
The GlycO Ontology in Protégé
3 Top-Level Classes
are Defined in GlycO
The GlycO Ontology in Protégé
Semantics Include Chemical Context
This Class Inherits from 2 Parents
The GlycO Ontology in Protégé
The -D-Manp residues
in N-glycans are found
in 8 different chemical
environments
GlycoTree – A Canonical Representation of N-Glycans
We give a residue in this position the
same name, regardless of the specific
structure it resides in
b-D-GlcpNAc-(1-6)+
b-D-GlcpNAc-(1-2)- -D-Manp -(1-6)+
Semantics!
b-D-Manp-(1-4)- b-D-GlcpNAc -(1-4)- b-D-GlcpNAc
b-D-GlcpNAc-(1-4)- -D-Manp -(1-3)+
b-D-GlcpNAc-(1-2)+
N. Takahashi and K. Kato, Trends in Glycosciences and
Glycotechnology, 15: 235-251
The GlycO Ontology in Protégé
Bisecting b-D-GlcpNAc
The GlycO Ontology in Protégé
The GlycO Ontology in Protégé
1,3-linked -L-Fucp
The GlycO Ontology in Protégé
Ontology Population Workflow
Semagix Freedom knowledge
extractor
YES:
next Instance
Instance
Data
Already in
KB?
Has
CarbBank
ID?
NO
YES
Insert into
KB
Compare to
Knowledge
Base
NO
IUPAC to
LINUCS
LINUCS to
GLYDE
Ontology Population Workflow
Semagix Freedom knowledge
extractor
YES:
next Instance
Instance
Data
Already in
KB?
Has
CarbBank
ID?
NO
YES
Insert into
KB
Compare to
Knowledge
Base
[][Asn]{[(4+1)][b-D-GlcpNAc]
{[(4+1)][b-D-GlcpNAc]
{[(4+1)][b-D-Manp]
{[(3+1)][a-D-Manp]
IUPAC to
NO {[(2+1)][b-D-GlcpNAc]
LINUCS
{}[(4+1)][b-D-GlcpNAc]
{}}[(6+1)][a-D-Manp]
{[(2+1)][b-D-GlcpNAc]{}}}}}}
LINUCS to
GLYDE
Ontology Population Workflow
Semagix Freedom knowledge
extractor
<Glycan>
YES:
<aglycon name="Asn"/>
<residue link="4"
anomer="b" chirality="D" monosaccharide="GlcNAc">
nextanomeric_carbon="1"
Instance
<residue link="4" anomeric_carbon="1" anomer="b" chirality="D" monosaccharide="GlcNAc">
<residue link="4" anomeric_carbon="1" anomer="b"
Instancechirality="D" monosaccharide="Man" >
<residue link="3" anomeric_carbon="1" anomer="a"
Data chirality="D" monosaccharide="Man" >
<residue link="2" anomeric_carbon="1" anomer="b" chirality="D" monosaccharide="GlcNAc" >
</residue>
<residue link="4" anomeric_carbon="1" anomer="b" chirality="D" monosaccharide="GlcNAc" >
</residue>
Has
</residue> Already in
IUPAC to
CarbBankchirality="D"
NO monosaccharide="Man" >
<residue link="6" anomeric_carbon="1" anomer="a"
KB?
LINUCS
<residue link="2" anomeric_carbon="1" anomer="b"
chirality="D" monosaccharide="GlcNAc">
ID?
</residue>
</residue>
</residue>
NO
YES
</residue>
</residue>
</Glycan>
Compare to
Insert into
KB
Knowledge
Base
LINUCS to
GLYDE
The ProPreO Ontology in Protégé
3 Top-Level Classes
are Defined in ProPreO
The ProPreO Ontology in Protégé
This Class Inherits
from 2 Parents
The ProPreO Ontology in Protégé
This Class Inherits
from 2 Parents
Semantic Annotation of MS Data
830.9570
194.9604
2
580.2985
0.3592
parent ion m/z
688.3214
0.2526
779.4759
38.4939
784.3607
21.7736
1543.7476
1.3822
fragment ion m/z
1544.7595
2.9977
1562.8113
37.4790
1660.7776
476.5043
parent ion charge
parent ion
abundance
fragment ion
abundance
ms/ms peaklist data
Semantically Annotated MS Data
<ms/ms_peak_list>
<parameter
instrument=micromass_QTOF_2_quadropole_time_of_flight_mass_spectrometer
mode = “ms/ms”/>
<parent_ion m/z = 830.9570 abundance=194.9604 z=2/>
<fragment_ion m/z = 580.2985 abundance = 0.3592/>
< fragment_ion m/z = 688.3214 abundance = 0.2526/>
< fragment_ion m/z = 779.4759 abundance = 38.4939/>
Ontological
< fragment_ion m/z = 784.3607 abundance = 21.7736/>
< fragment_ion m/z = 1543.7476 abundance = 1.3822/>
Concepts
< fragment_ion m/z = 1544.7595 abundance = 2.9977/>
< fragment_ion m/z = 1562.8113 abundance = 37.4790/>
< fragment_ion m/z = 1660.7776 abundance = 476.5043/>
<ms/ms_peak_list>
Web Services Based Workflow for Proteomics1
Biological
Sample
Analysis
by MS/MS
O
Agent
Raw Data
to
Standard
Format
I
Raw
Data
Agent
O
Standard
Format
Data
Data
Preprocess2
I
Agent
O
Filtered
Data
Agent
DB
Search
Results
Postprocess
(Mascot/
Sequest)
(ProValt3)
I
I
Search
Results
O
Final
Output
Storage
Biological Information
1 Design and Implementation of Web Services based Workflow for proteomics. Journal of Proteome Research. Submitted
2 Computational tools for increasing confidence in protein identifications. Association of Biomolecular Resource Facilities
Annual Meeting, Portland, OR, 2004.
3 A Heuristic method for assigning a false-discovery rate for protein identifications from Mascot database search results. Mol.
Cell. Proteomics. 4(6), 762-772.
O
An Integrated
Semantic Information System
• Formalized domain knowledge is in ontologies
• The schema defines the concepts
• Instances represent individual objects
• Relationships provide expressiveness
• Data is annotated using concepts from the
ontologies
• The
semantic
annotations
facilitate
the
identification
and
extraction
of
relevant
information
• The semantic relationships allow knowledge that
is implicit in the data to be discovered
Satya Sahoo
Christopher Thomas
Cory Henson
Ravi Pavagada
Amit Sheth
Krzysztof Kochut
John Miller
James Atwood
Lin Lin
Alison Nairn
Gerardo Alvarez-Manilla
Saeed Roushanzamir
Michael Pierce
Ron Orlando
Kelley Moremen
Parastoo Azadi
Alfred Merrill