Poster 30.pptx (1.403Mb)
advertisement

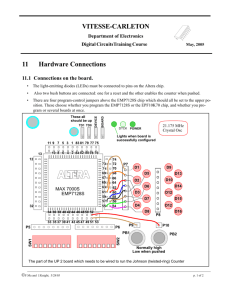
Identifying Human Host Proteins that Interact with Influenza A Polymerase Proteins Using the Yeast Two-Hybrid System 2 Drew , 1,2,5 Faganel , 2 Johnson , 2 Kinzler , 1,2 Madhan , 2 Schmotter , 1,4 Scully , Cassandra Peyton Bree Carly Nayasha Caleb Erin 2 2,3 2 1,2 1 1 2 3 4 5 Luke Tourville , Alec Rose , Kevin Marshall , Marc Busch , and Heidi Sleister ( Biology, BCMB, Neuroscience, Psychology, Chemistry) Background Methods A Brief History o Humans most likely began to contract the influenza virus when animals first became domesticated. o Settlements provided a sufficient number of hosts for an epidemic. o It wasn’t until 1930 that the influenza virus was at last isolated from a pig by Shope and Lewis. PB1/PB2/PA Design gene specific primers with homology to pGBKT7 vector and homology to 5’ and 3’ ends of PB1/PB2/PA ORF Structure and Function o The Influenza A genome consists of eight negative-sense strands of viral RNA. Attachment requires the viral glycoprotein hemagglutinin (HA), which binds to the cell receptor, sialic acid. InFusion cloning by homologous recombination "Influenza Figure." Web. 1 Apr. 2012. <http://micro.magnet.fsu.edu/cells/viruses/images/influenzafigure1.jpg> Purpose o To distinguish which human host proteins interact with Influenza A virus polymerase complex proteins. pSCRIPT PB1/PB2/PA Isolate pSCRIPT vector from E. coli bacteria PCR amplification of PB1/PB2/PA from pSCRIPT with 5’ and 3’ extensions homologous to vector PB1/PB2/PA Forward %GC %GC primer name Tm Influenza virus strain Forward primer sequence Swine Influenza A- PB1, H3N2, A/Sw/MN/593/33/99 CATGGAGGCCGAATTC atggatgtcaatccgactcta MN-PB1F 42.9 Swine Influenza A- PB2, H3N2, A/Sw/MN/593/33/99 CATGGAGGCCGAATTC atggagagaataaaagaactaagag MN-PB2F 32 Swine Influenza A- PB1, H1N1, A/Sw/NC/44173/00 CATGGAGGCCGAATTC atggatgtcaatccgactttac NC-PB1F Swine Influenza A- PB2, H1N1, A/Sw/NC/44173/00 CATGGAGGCCGAATTC atggagagaataaaagaattaagagatc Swine Influenza A- PA, H1N1, A/Sw/NC/44173/00 CATGGAGGCCGAATTC atggaagacttcgtacgaca 59.2 Reverse %GC %GC primer name Tm Reverse primer sequence MN-PB1R 39.1 61.5 57.6 GCAGGTCGACGGATCC ctaattgatcgccatccgaattc MN-PB2R 43.5 60.5 40.9 59.2 GCAGGTCGACGGATCC ttacttttgccgtctgagatc NC-PB1R 42.9 58.9 NC-PB2F 28.6 58.9 GCAGGTCGACGGATCC ctaattgatggccatccgaat NC-PB2R 42.9 59.2 NC-PAF 45 59.4 GCAGGTCGACGGATCC ctatcttagtgcatgtgtgagga NC-PAR 43.5 60.5 GCAGGTCGACGGATCC ttatttttgccgtctgagtcctt By constructing primers specific to the polymerase genes, the segment of the plasmid DNA to be replicated was selected. Sequences in black denote the segments of the primers that are complementary to the PB1/PB2/PA genes and will allow replication to take place, while red sequences denote segments of the primers that are homologous to the pGBKT7 vector and are necessary for recombination cloning to take place. Control (-HindIII) PB1 (MN) PB2 (MN) PB1 (NC) PB2 (NC) PA (NC) PCR results of pSCRIPT+PB1/PB2/PA. PCR products were run through a 0.8% agarose gel and compared to the λ-HindIII DNA marker to determine the size of the fragments. PCR products were expected to be ~2.2kb in length. All reactions except PB2(NC) were repeated to achieve greater yield. ~2.3 kb PB1/PB2/PA pGBKT7 Importance o The Influenza A virus has multiple physiological effects on humans, including fever, cough, chills, body aches and fatigue. By discerning which human host proteins interact with the Influenza A virus, it may be possible to further the progress of treatments for the disease. ~2.0 kb Transform cloning mixture into E. coli pGBKT7+PB1/ PB2/PA This gel illustrates the results of a bacterial cell cracking procedure for PB2(NC) and PA(NC). The vector lacking the insert will move further down the gel than the vector with an insert due to its smaller size. Control (pGBKT7 vector without insert) Transformants 1-10 of pGBKT7+PB2(NC) 1 2 3 4 5 6 7 8 9 Control Transformants 7-10 of (pGBKT7 pGBKT7+PA(NC) vector without 10 8 9 10 insert) 7 Plate on LBKanamycin agar media to select for pGBKT7containing cells Experimental Question & Strategy Question o Which human proteins bind to the Influenza A polymerase complex proteins PB1-H3N2 (A/Sw/MN/593/33/99), PB2-H3N2 (A/Sw/MN/593/33/99), PB1-H1N1 (A/Sw/NC/44173/00), PB2-H1N1 (A/Sw/NC/44173/00), and PA-H1N1 (A/Sw/NC/44173/00)? Human Protein Results Screen for correct plasmid constructpGBKT7+PB1/PB2/PA by bacterial cell cracking, PCR, and restriction digest Transform pGBKT7+PB1/PB2/PA (bait) into haploid yeast strain Y2HGold Vector without insert Vector with PB2 insert Vector with PA insert InFusion recombination cloning was successful for producing the desired plasmids (pGBKT7 vector + polymerase gene). PB2(NC) transformants 2 and 6 and PA(NC) transformant 7 were selected for further analysis Transformants for PB1(MN), PB1(NC), and PB2(MN) had similar results. Binding of the bait and prey causes the transcriptional activation of GAL4 which leads to expression of reporter genes Conclusions & Future Work Gal4(BD)-PB1/PB2/PA bait Test PB1/PB2/PA for toxicity to yeast Test PB1/PB2/PA for auto-activation of reporter genes in yeast Influenza A polymerase complex protein Conclusions o The open reading frames of PB1/PB2/PA were successfully amplified by PCR. o While screening (cracking) various bacterial transformants, those that were most likely to contain the pGBKT7 vector with insert PB1/PB2/PA were determined. o Isolation of plasmids from InFusion cloning that are bigger than the vector alone suggests the InFusion recombination cloning was successful. o Transformant colony 2, from pGBKT7+PB2(NC) and transformant colony 7, from pGBKT7+PA(NC), were selected for further screening. Future Work Experimental Strategy o The Yeast Two-Hybrid System (Y2H) is being used for the identification of protein-protein interactions. Binding of the bait with the prey produces the transcriptional activator function of yeast Gal4. This leads to the expression of reporter genes which is detected by cell growth on SDLEU-TRP-HIS/X-alpha-Gal/AbA agar plates. Sources • "EBSCO Publishing Service Selection Page." EBSCO Publishing Service Selection Page. N.p., n.d. Web. 9 Apr. 2013. <http://ehis.ebscohost.com/eds/pdfviewer/pdfviewer?sid=701ad167-dfee-40fd-a8co-55f1cd713f55%40sessionmgr11&vid=10&hid=103>. • "Influenza." Medical Ecology @ www.MedicalEcology.org. N.p., n.d. Web. 9 Apr. 2013. http://www.medicalecology.org/diseases/influenza/influenza.htm#sect3.5>.Clontech. Matchmaker Gold Yeast Two-Hybrid System User Manual. Montainview, CA: Clontech Laboratories, 2010. Print. • "Influenza Figure." Web. 1 Apr. 2012. <http://micro.magnet.fsu.edu/cells/viruses/images/influenzafigure1.jpg>. • Kao, R. et al. (2010). Identification of influenza A nucleoprotein as an antiviral target. Nature Biotechnology 28, 600–605. • Portela, A. and P. Digard. (2002). The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J. General Virology 83, 723–734. • Qiagen. (2006). QIAprep Miniprep Handbook. Print. • “Yeast Two-Hybrid Figure.” Web. Apr 4. 2012. <http://cmbi.bjmu.edu.cn/cmbidata/proteome/method/hybrid02.jpg>. o o o o Sequence the pGBKT7+ PB1/PB2/PA plasmids Determine Influenza A PB1/PB2/PA toxicity to yeast Test Influenza A PB1/PB2/PA for auto-activation of yeast two-hybrid reporter genes Isolate human protein candidates (prey) that may interact with Influenza A PB1/PB2/PA (bait) by performing a yeast two-hybrid screen o Screen candidates to determine true positive interactions and identify prey proteins o Confirm positive interactions in a mammalian system o Compare identified protein interactions in normal versus microgravity conditions Acknowledgement o NASA – Iowa Space Grant Consortium (Base program and curriculum grants)