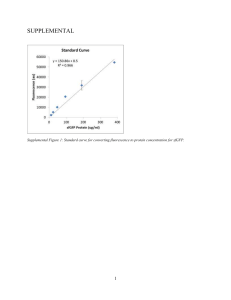
SLIDES: Activist data mining (as applied to Carbon: Nitrogen Sensing Plants)

Activist Data Mining (as
Applied to Carbon:Nitrogen
Sensing in Plants)
Dennis Shasha
New York University
Department of
Biology
Courant Institute of
Math & Computer Sciences
Gloria Coruzzi
Mike Chou
Andrew Kouranov
Laurence Lejay
Dennis Shasha Bud Mishra
Marco Antoinotti
Marc Rejali
LIGHT
Sugar
NH
4
+
Amino
Acids
Glu
Gln
Asp
Asn
Light, Carbon and Amino acids differentially regulate N-assimilation genes
Light Carbon Light Carbon
GS2
Amino acids
Gln
C
:N
C5:N2
AS1 Amino acids
Asn
C:
N
C4:N2
Goal: Figure out the Circuit for many genes
A Multi-factor Approach to C:N sensing in plants.
Identify how a combination of interactions of “inputs”
(Light, Carbon, & Nitrogen) affects gene regulation using Combinatorial Design and Genome Chip analysis.
Identify Arabidopsis mutants defective in C:N sensing
Forward genetics: Selections for C:N sensing mutants
Reverse genetics: Mutants in candidate C:N signaling genes
Ultimate Goal: Virtual plant… (frankenfoods)
A Combinatorial Approach to discovering interactions
Inputs: *Light
*Starvation to Various Nutrients
*Carbon
*Inorganic N (NO3/NH4)
*Organic N (Glu)
*Organic N (Gln)
If inputs are take binary values (first approximation)
6 binary (+/-) inputs= 2 6 or 64 input combinations (or treatments)
Use combinatorial design to reduce number of treatment combinations required to effectively cover the experimental space
ACTIVIST DATA MINING
Don’t study the experiments (only). Change them.
Combinatorial design generates a subset of the 64 treatments that give “good” approximation of the entire experimental space.
For every pair of “inputs”, all four combinations of binary variables are tested:
Example ; NO
+NO
3
3 and Carbon have four possible combinations
+Carbon; +NO
3
-Carbon; -NO
3
+Carbon; -NO
3
-Carbon
Each combination of inputs is present in at least one treatment of experiments predicted by combinatorial design
“Combinatorial design” predicts 12 conditions to test the effect of
Light in all combinations of Starvation, Carbon, and Nitrogen
EXPT 1
PIVOT
LIGHT
LANE LIGHT STARVE CARBON NO3NH4 GLU GLN
6
7
8
9
10
11
12
1
4
5
2
3
LIGHT
LIGHT
LIGHT
LIGHT
LIGHT
LIGHT
DARK
DARK
DARK
DARK
DARK
DARK
N
N
N
Y
N
N
Y
N
N
N
Y
Y
L
0
L
L
L
0
0
0
L
0
0
L
L
0
L
0
L
0
L
0
L
0
L
0
0
H
0
H
H
0
0
0
H
H
0
H
0
H
0
0
H
0
H
0
H
H
H
0
“Pivot” analysis of gene expression data from C:N treatments
Find “minimal pairs” of treatments that are the same except in one input (e.g. Light) to measure its effect on a dependent variable (gene) (e.g. AS1)
PIVOT Dependent
Variable
(Gene)
LIGHT AS1
EFFECT Evidence =
Minimal pair treatments repress 4_8
LITE STARVE CARBON NO3 GLU
L_D N L 0 H
Analyze a series of minimal pair treatments using one input
(e.g. Light) as a “pivot”, to determine the effect of light on a dependent variable (e.g. AS1) under a variety of carbon and nitrogen combinations. If consistent, likely always true.
LITE represses AS1 & induces GS2 under a variety of C:N conditions
LIGHT AS1
LIGHT AS1
LIGHT AS1
LIGHT AS1
LIGHT AS1
LIGHT AS1
LIGHT AS1
LIGHT AS1
LIGHT GS2
LIGHT GS2
LIGHT GS2
LIGHT GS2
LIGHT GS2
LIGHT GS2
LIGHT GS2
LIGHT GS2
PIVOT dependent EFFECT Evidence=
Minimalpair treatments induce induce induce induce induce induce induce induce repress 1_5 repress 2_6 repress 3_7 repress 4_8 repress 10_14 repress 11_15 repress 12_16 repress 13_17
1_5
2_6
3_7
4_8
10_14
11_15
12_16
13_17
LITE
L
0
0
0
L
0
0
0
L
0
0
0
L
0
0
0
L
L
0
L
L
L
0
L
L
L
0
L
L
L
0
L
N
Y
N
N
Y
Y
Y
Y
Y
Y
N
Y
Y
N
Y
Y
L_D
L_D
L_D
L_D
L_D
L_D
L_D
L_D
L_D
L_D
L_D
L_D
L_D
L_D
L_D
L_D
STARVE CARBON NO3/NH4 GLU
0
H
0
0
0
H
0
0
0
H
0
0
0
H
0
0
GLU induces AS1 & represses GS2 under a variety of conditions
PIVOT Gene
GLU
GLU
GLU
GLU
GLU
GLU
GLU
GLU
GLU
GLU
GLU
GLU
GLU
GLU
GLU
GS2
GS2
GS2
GS2
AS1
AS1
AS1
AS1
AS1
AS1
AS1
GS2
GS2
GS2
GS2
EFFECT Evidence=
Minimalpair
Treatments induce 2_4 induce induce induce induce
6_8
15_17
19_21
23_25 induce induce
26_28
30_32 repress 2_4 repress 6_8 repress 11_13 repress 15_17 repress 19_21 repress 20_22 repress 23_25 repress 30_32
LIGHT STARVE Carbon NO3/NH4 GLU
L
L
D
L
L
L
D
D
L
D
D
D
D
L
L
Y
Y
Y
Y
Y
Y
Y
N
N
Y
Y
N
N
N
Y
L
L
L
L
L
L
L
L
L
L
L
L
L
L
L
0
0
0
L
0
0
0
0
0
0
0
0
0
0
0
0_H
0_H
0_L
0_L
0_H
0_H
0_H
0_H
0_L
0_L
0_L
0_H
0_H
0_H
0_H
Underlying Method: combinatorial design
Combinatorial design : Inspired by work in software testing by
David Cohen, Siddhartha Dalal, Michael Fredman and
Gardner Patton at Bellcore/Telcordia.
Their problem: how to test a good set of inputs to a program to discover whether there are any bugs.
Not program coverage, but input coverage.
Not all input combinations, but all combinations of every pair of of input variables.
Hypothesis: every input combination should give same output: no error.
If true for designed subset, then program is ok.
Underlying Method: combinatorial design 2
Scientific question: does input X induce
(resp. repress) the output?
If so, then, regardless of the other inputs,
X should induce.
So, choose X = low and then a combinatorial design of the other inputs.
Then choose X = high and then the same combinatorial design of the other inputs.
If for each context c in the design (high,c) has more output than (low,c) -- minimal pair -- then X is inductive.
Underlying Methods: adaptive design
What happens when X isn’t uniformly inductive or repressive?
Suppose X shows induction normally, but repression occasionally. That is for most c values
(low, c) vs. (high, c) shows induction, but for one c’
(low,c’) vs. (high, c’) shows repression.
Then study difference between those c values showing induction that are closest to c’ and design experiments to reduce those differences.
Conclusions About Methodology
Design/don’t wait : Use the data you are given, sure, but don’t be shy to ask for more.
Combinatorial Design can help test a hypothesis : e.g. 10 three-valued variables require
59,049 experiments to cover whole space. Combinatorial design can reduce this to 27.
Adaptation is easy: Study differences between normal cases and abnormal ones to discover fine structure.