Allium przewalskianum Regel. (Alliaceae) from the Qinghai-Tibet Plateau Organisms Diversity & Evolution
advertisement
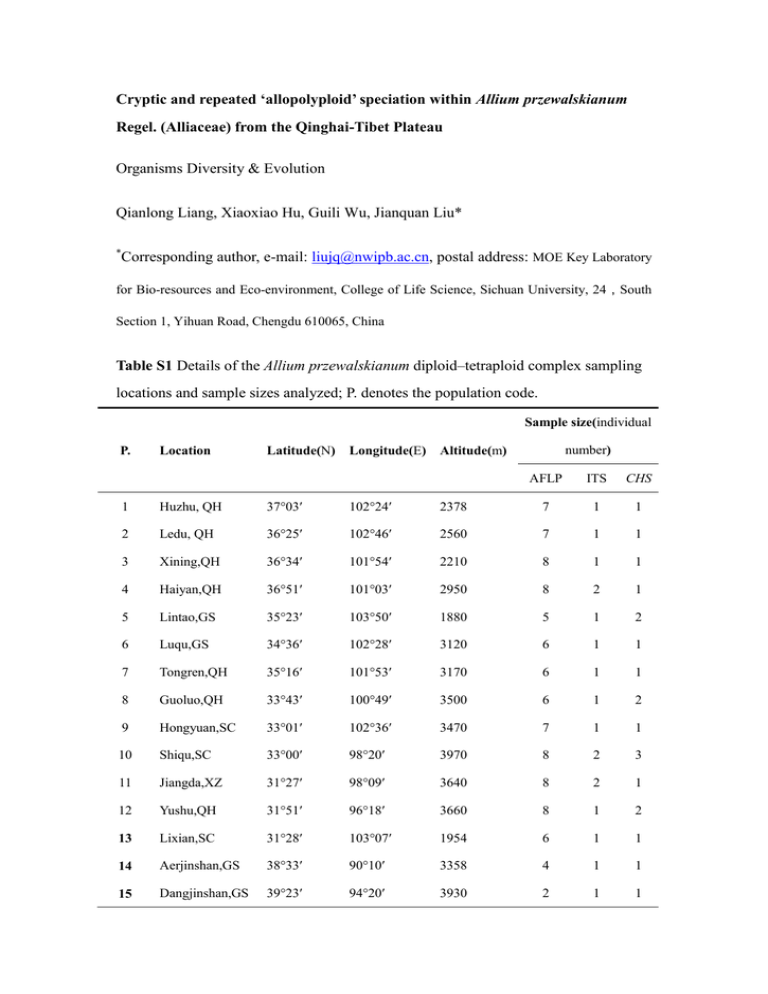
Cryptic and repeated ‘allopolyploid’ speciation within Allium przewalskianum Regel. (Alliaceae) from the Qinghai-Tibet Plateau Organisms Diversity & Evolution Qianlong Liang, Xiaoxiao Hu, Guili Wu, Jianquan Liu* * Corresponding author, e-mail: liujq@nwipb.ac.cn, postal address: MOE Key Laboratory for Bio-resources and Eco-environment, College of Life Science, Sichuan University, 24,South Section 1, Yihuan Road, Chengdu 610065, China Table S1 Details of the Allium przewalskianum diploid–tetraploid complex sampling locations and sample sizes analyzed; P. denotes the population code. Sample size(individual P. Location Latitude(N) Longitude(E) number) Altitude(m) AFLP ITS CHS 1 Huzhu, QH 37°03′ 102°24′ 2378 7 1 1 2 Ledu, QH 36°25′ 102°46′ 2560 7 1 1 3 Xining,QH 36°34′ 101°54′ 2210 8 1 1 4 Haiyan,QH 36°51′ 101°03′ 2950 8 2 1 5 Lintao,GS 35°23′ 103°50′ 1880 5 1 2 6 Luqu,GS 34°36′ 102°28′ 3120 6 1 1 7 Tongren,QH 35°16′ 101°53′ 3170 6 1 1 8 Guoluo,QH 33°43′ 100°49′ 3500 6 1 2 9 Hongyuan,SC 33°01′ 102°36′ 3470 7 1 1 10 Shiqu,SC 33°00′ 98°20′ 3970 8 2 3 11 Jiangda,XZ 31°27′ 98°09′ 3640 8 2 1 12 Yushu,QH 31°51′ 96°18′ 3660 8 1 2 13 Lixian,SC 31°28′ 103°07′ 1954 6 1 1 14 Aerjinshan,GS 38°33′ 90°10′ 3358 4 1 1 15 Dangjinshan,GS 39°23′ 94°20′ 3930 2 1 1 16 Yumen,GS 39°48′ 97°17′ 3361 4 1 1 17 Delingha,QH 37°27′ 97°20′ 3310 4 1 1 18 Xinghai,QH 35°53′ 99°41′ 3760 5 1 1 19 Shandan,GS 38°06′ 101°33′ 2612 5 1 1 20 Zeku,QH 35°12′ 100°55′ 3500 7 1 1 21 Yushu,QH 33°51′ 97°11′ 4020 3 1 1 22 Yushu,QH 33°00′ 97°19′ 3610 5 1 1 23 Yushu,QH 32°21′ 96°26′ 3680 3 1 1 24 Dingqing,XZ 31°30′ 95°23′ 3770 6 1 1 25 Leiwuqi,XZ 31°12′ 96°36′ 3800 5 1 1 26 Ranwu,XZ 29°30′ 96°46′ 3950 5 1 1 27 Dangxiong,XZ 30°32′ 91°08′ 4310 4 1 1 28 Kangma,XZ 28°33′ 89°43′ 4430 5 1 1 Note: (1) Abbreviations: QH, Qinghai; GS, Gansu; SC, Sichuan; XZ, Xizang; YN, Yunnan; (2) populations 1-12 are diploid; populations 13-28 are tetraploid and the population codes are in bold. Table S2 AFLP adapter and primer sequences used in this study Code Name Sequence E-F EcoR 1-F(adapter) 5’-CTCGTAGACTGCGTACC-3’ E-R EcoR 1-R(adapter) 5’-AATTGGTACGCAGTCTAC-3’ M-F Mse 1-F(adapter) 5’-GACGATGAGTCCTGAG-3’ M-R Mse 1-R(adapter) 5’-TACTCAGGACTCAT-3’ E-A EcoR 1-A(Primer) 5’-GACTGCGTACCAATTCA-3’ M-C Mse 1-C(Primer) 5’-GATGAGTCCTGAGTAAC-3’ E-ACC EcoR 1-ACC(Primer) 5’-GACTGCGTACCAATTCACC-3’ M-CAC Mse 1-CAC(Primer) 5’-GAT GAG TCC TGA GTA ACAC-3’ E-ACT EcoR 1-ACT(Primer) 5’-GACTGCGTACCAATTCACT-3’ M-CTA Mse 1-CTA(Primer) 5’-GAT GAG TCC TGA GTA ACTA-3’ Table S3 Population codes (P.), plant sources, and locality of the sequenced material; ITS and CHS GenBank accession numbers for all sequences included in this study. P. Voucher Locality ITS 1 1 1 2 2 2 3 3 3 4 4 4 5 5 5 5 6 6 7 7 7 7 7 7 7 8 8 8 8 8 8 9 9 10 10 10 J. Q. Liu-AO 01 China: Huzhu, QH China: Huzhu, QH China: Huzhu, QH China: Ledu, QH China: Ledu, QH China: Ledu, QH China: Xining, QH China: Xining, QH China: Xining, QH China: Haiyan, QH China: Haiyan, QH China: Haiyan, QH China: Lintao, GS China: Lintao, GS China: Lintao, GS China: Lintao, GS China: Luqu, GS China: Luqu, GS China: Tongren, QH China: Tongren, QH China: Tongren, QH China: Tongren, QH China: Tongren, QH China: Tongren, QH China: Tongren, QH China: Guoluo, QH China: Guoluo, QH China: Guoluo, QH China: Guoluo, QH China: Guoluo, QH China: Guoluo, QH China: Hongyuan, SC China: Hongyuan, SC China: Shiqu, SC China: Shiqu, SC China: Shiqu, SC KM189069 KM189070 KM189071 KM189072 KM189073 KM189074 KM189075 KM189076 KM189077 KM189078 KM189079 KM189080 KM189081 KM189082 KM189083 KM189084 KM189085 J. Q. Liu-AO 01 J. Q. Liu-AO 01 J. Q. Liu-AO 69 J. Q. Liu-AO 69 J. Q. Liu-AO 69 J. Q. Liu-AO 72 J. Q. Liu-AO 72 J. Q. Liu-AO 72 J. Q. Liu-AO 14 J. Q. Liu-AO 14 J. Q. Liu-AO 14 J. Q. Liu-AO 66 J. Q. Liu-AO 66 J. Q. Liu-AO 66 J. Q. Liu-AO 66 J. Q. Liu-2006033 J. Q. Liu-2006033 J. Q. Liu-AO 54 J. Q. Liu-AO 54 J. Q. Liu-AO 54 J. Q. Liu-AO 54 J. Q. Liu-AO 54 J. Q. Liu-AO 54 J. Q. Liu-AO 54 J. Q. Liu-AO 53 J. Q. Liu-AO 53 J. Q. Liu-AO 53 J. Q. Liu-AO 53 J. Q. Liu-AO 53 J. Q. Liu-AO 53 J. Q. Liu-2006026 J. Q. Liu-2006026 J. Q. Liu-AO 108 J. Q. Liu-AO 108 J. Q. Liu-AO 108 CHS KM188957 KM188958 KM188959 KM188960 KM188961 KM188962 KM188963 KM188964 KM188965 KM188966 KM188967 KM188968 KM188969 KM188970 KM188971 KM188972 KM188973 KM188974 KM188975 KM188976 KM188977 KM188978 KM188979 KM188980 KM188981 KM188982 KM188983 KM188984 KM188985 KM188986 KM188987 KM188988 KM188989 KM188990 KM188991 10 10 10 10 11 11 11 11 12 12 13 13 13 13 13 14 14 14 14 14 15 15 15 15 16 16 16 16 16 17 17 17 17 17 17 18 18 18 18 18 19 19 19 19 J. Q. Liu-AO 108 J. Q. Liu-AO 108 J. Q. Liu-AO 108 J. Q. Liu-AO 108 J. Q. Liu-AO 117 J. Q. Liu-AO 117 J. Q. Liu-AO 117 J. Q. Liu-AO 117 J. Q. Liu-AO 106 J. Q. Liu-AO 106 J. Q. Liu-TB07024 J. Q. Liu-TB07024 J. Q. Liu-TB07024 J. Q. Liu-TB07024 J. Q. Liu-TB07024 J. Q. Liu-TB07121 J. Q. Liu-TB07121 J. Q. Liu-TB07121 J. Q. Liu-TB07121 J. Q. Liu-TB07121 J. Q. Liu-TB07117 J. Q. Liu-TB07117 J. Q. Liu-TB07117 J. Q. Liu-TB07117 J. Q. Liu-TB07113 J. Q. Liu-TB07113 J. Q. Liu-TB07113 J. Q. Liu-TB07113 J. Q. Liu-TB07113 J. Q. Liu-AO 160 J. Q. Liu-AO 160 J. Q. Liu-AO 160 J. Q. Liu-AO 160 J. Q. Liu-AO 160 J. Q. Liu-AO 160 J. Q. Liu-AO 82 J. Q. Liu-AO 82 J. Q. Liu-AO 82 J. Q. Liu-AO 82 J. Q. Liu-AO 82 J. Q. Liu-TB07101 J. Q. Liu-TB07101 J. Q. Liu-TB07101 J. Q. Liu-TB07101 China: Shiqu, SC China: Shiqu, SC China: Shiqu, SC China: Shiqu, SC China: Jiangda, XZ China: Jiangda, XZ China: Jiangda, XZ China: Jiangda, XZ China: Yushu, QH China: Yushu, QH China: Lixian, SC China: Lixian, SC China: Lixian, SC China: Lixian, SC China: Lixian, SC China: Aerjinshan, XJ China: Aerjinshan, XJ China: Aerjinshan, XJ China: Aerjinshan, XJ China: Aerjinshan, XJ China: Dangjinshan, GS China: Dangjinshan, GS China: Dangjinshan, GS China: Dangjinshan, GS China: Yumen, GS China: Yumen, GS China: Yumen, GS China: Yumen, GS China: Yumen, GS China: Delingha, QH China: Delingha, QH China: Delingha, QH China: Delingha, QH China: Delingha, QH China: Delingha, QH China: Xinghai, QH China: Xinghai, QH China: Xinghai, QH China: Xinghai, QH China: Xinghai, QH China: Shandan, GS China: Shandan, GS China: Shandan, GS China: Shandan, GS KM189086 KM189087 KM189088 KM189089 KM189090 KM189091 KM189092 KM189093 KM189094 KM189095 KM189096 KM189097 KM189098 KM189099 KM189100 KM189101 KM189102 - KM188992 KM188993 KM188994 KM188995 KM188996 KM188997 KM188998 KM188999 KM189000 KM189001 KM189002 KM189003 KM189004 KM189005 KM189006 KM189007 KM189008 KM189009 KM189010 KM189011 KM189012 KM189013 KM189014 KM189015 KM189016 KM189017 KM189018 KM189019 KM189020 KM189021 KM189022 KM189023 KM189024 KM189025 KM189026 KM189027 KM189028 KM189029 KM189030 KM189031 KM189032 KM189033 19 20 20 20 20 20 21 21 21 22 22 22 22 23 23 23 23 24 24 24 24 25 25 25 26 26 26 26 26 27 27 27 28 28 28 J. Q. Liu-TB07101 J. Q. Liu-AO 41 J. Q. Liu-AO 41 J. Q. Liu-AO 41 J. Q. Liu-AO 41 J. Q. Liu-AO 41 J. Q. Liu-AO 89 J. Q. Liu-AO 89 J. Q. Liu-AO 89 J. Q. Liu-AO 95 J. Q. Liu-AO 95 J. Q. Liu-AO 95 J. Q. Liu-AO 95 J. Q. Liu-AO 101 J. Q. Liu-AO 101 J. Q. Liu-AO 101 J. Q. Liu-AO 101 J. Q. Liu-AO 127 J. Q. Liu-AO 127 J. Q. Liu-AO 127 J. Q. Liu-AO 127 J. Q. Liu-AO 121 J. Q. Liu-AO 121 J. Q. Liu-AO 121 J. Q. Liu-2006290 J. Q. Liu-2006290 J. Q. Liu-2006290 J. Q. Liu-2006290 J. Q. Liu-2006290 J. Q. Liu-AO 157 J. Q. Liu-AO 157 J. Q. Liu-AO 157 J. Q. Liu-2006219 J. Q. Liu-2006219 J. Q. Liu-2006219 China: Shandan, GS China: Zeku, QH China: Zeku, QH China: Zeku, QH China: Zeku, QH China: Zeku, QH China: Yushu, QH China: Yushu, QH China: Yushu, QH China: Yushu, QH China: Yushu, QH China: Yushu, QH China: Yushu, QH China: Yushu, QH China: Yushu, QH China: Yushu, QH China: Yushu, QH China: Dingqing, XZ China: Dingqing, XZ China: Dingqing, XZ China: Dingqing, XZ China: Leiwuqi, XZ China: Leiwuqi, XZ China: Leiwuqi, XZ China: Ranwu, XZ China: Ranwu, XZ China: Ranwu, XZ China: Ranwu, XZ China: Ranwu, XZ China: Dangxiong, XZ China: Dangxiong, XZ China: Dangxiong, XZ China: Kangma, XZ China: Kangma, XZ China: Kangma, XZ KM189103 KM189104 KM189105 KM189106 KM189107 KM189108 KM189109 KM189110 KM189111 KM189112 KM189113 KM189114 KM189115 KM189116 KM189117 KM189118 KM189119 KM189120 - KM189034 KM189035 KM189036 KM189037 KM189038 KM189039 KM189040 KM189041 KM189042 KM189043 KM189044 KM189045 KM189046 KM189047 KM189048 KM189049 KM189050 KM189051 KM189052 KM189053 KM189054 KM189055 KM189056 KM189057 KM189058 KM189059 KM189060 KM189061 KM189062 KM189063 KM189064 KM189065 KM189066 KM189067 KM189068 Table S4 Details of the 60 sites used for ecological niche modeling Population Locality Geographical position Altitude Voucher specimens DP & NTP Haiyan, QH N 36°51.367’ E 101°03.252’ 2950m J. Q. Liu-AO 14 DP & NTP Huangyuan, QH N 36°39.920’ E 101°23.567’ 2470-2520m J. Q. Liu-AO 162 DP & NTP QH Lake, QH N 36°33.157’ E 100°43.757’ 3210m J. Q. Liu-AO 78 DP & NTP Xining, QH N 36°33.786’ E 101°54.361’ 2210m J. Q. Liu-AO 72 DP & NTP Yushu, QH N 31°51.329’ E 096°17.741’ 3660m J. Q. Liu-AO 103 DP Huzhu, QH N 37°03.029’ E 102°23.934’ 2378m J. Q. Liu-AO 01 DP Ledu, QH N 36°25.007’ E 102°46.160’ 2560m J. Q. Liu-AO 69 DP Lintao, GS N 35°23.327’ E 103°50.402’ 1880m J. Q. Liu-AO 66 DP Tongren, QH N 35°15.775’ E 101°52.997’ 3170m J. Q. Liu-AO 54 DP Luqu, GS N 34°35.576’ E 102°28.045’ 3120m J. Q. Liu-2006033 DP Henan, QH N 34°44.195’ E 101°18.090’ 3600m J. Q. Liu-AO 30 DP Guoluo, QH N 33°43.289’ E 100°48.921’ 3500m J. Q. Liu-AO 53 DP Hongyuan, SC N 33°00.880’ E 102°36.397’ 3470m J. Q. Liu-2006026 DP Shiqu, SC N 32°59.862’ E 098°20.369’ 3970m J. Q. Liu-AO 108 DP Jiangda, XZ N 31°26.607’ E 098°09.073’ 3640m J. Q. Liu-AO 117 DP Ping’an, QH N 36°38.717’ E 102°02.336’ 2200m J. Q. Liu-AO 70 DP Gonghe, QH N 35°57.803’ E 100°59.769’ 2850m J. Q. Liu-AO 77 NTP Xinghai, QH N 35°53.205’ E 099°40.588’ 3760m J. Q. Liu-AO 85 NTP Xinghai, QH N 35°44.433’ E 100°13.405’ 3220m J. Q. Liu-AO 82 NTP Tongren, QH N 35°22.337’ E 101°37.747’ 2740m J. Q. Liu-AO 25 NTP Yuzhong, GS N 35°46.796’ E 104°01.378’ 2450m J. Q. Liu-AO 67 NTP Tongren, QH N 35°42.662’ E 102°17.652’ 3020m J. Q. Liu-AO 23 NTP Tongren, QH N 35°35.598’ E 102°03.158’ 2360m J. Q. Liu-AO 56 NTP Tongren, QH N 35°26.772’ E 102°27.402’ 3080m J. Q. Liu-AO 62 NTP Zeku, QH N 35°16.497’ E 101°06.670’ 3500m J. Q. Liu-AO 41 NTP Hezuo, GS N 35°08.350’ E 102°51.059’ 2650m J. Q. Liu-2006039 NTP Ningxiu, QH N 35°07.752’ E 100°43.657’ 3450m J. Q. Liu-AO 43 NTP Gonghe, QH N 36°22.386’ E 100°23.673’ 3490m J. Q. Liu-AO 80 NTP Aerjinshan, GS N 38°33.254’ E 090°10.182’ 3358m J. Q. Liu-TB07121 NTP Dangjinshan, GS N 39°22.601’ E 094°19.511’ 2930m J. Q. Liu-TB07117 NTP Yumen, GS N 39°48.429’ E 097°17.189’ 3361m J. Q. Liu-TB07113 NTP Delingha, QH N 37°26.826’ E 097°20.282’ 3310m J. Q. Liu-AO 160 NTP Dulan, QH N 36°00.821’ E 097°53.443’ 3050m J. Q. Liu-AO 158 NTP Dulan, QH N 36°14.677’ E 098°06.370’ 3250m J. Q. Liu-AO 159 NTP Wulan, QH N 37°04.310’ E 098°52.464’ 3470m J. Q. Liu-AO 161 NTP Ebo, QH N 37°59.878’ E 100°45.921’ 3223m J. Q. Liu-AO 03 NTP Shandan, GS N 38°06.326’ E 101°33.209’ 2612m J. Q. Liu-TB07101 NTP Wuwei, GS N 37°12.218’ E 102°51.436’ 2900m J. Q. Liu-TB07099 NTP Huangzhong, QH N 36°30.434’ E 101°37.905’ 2540m J. Q. Liu-AO 75 STP Yushu, QH N 33°50.522’ E 097°10.601’ 4020m J. Q. Liu-AO 89 STP Yushu, QH N 32°51.782’ E 096°57.465’ 3950m J. Q. Liu-AO 106 STP Yushu, QH N 33°11.008’ E 097°23.636’ 4010m J. Q. Liu-AO 92 STP Yushu, QH N 33°00.198’ E 097°19.007’ 3610m J. Q. Liu-AO 95 STP Yushu, QH N 32°20.524’ E 096°25.850’ 3680m J. Q. Liu-AO 101 STP Changdu, XZ N 31°08.853’ E 097°01.170’ 3630m J. Q. Liu-AO 119 STP Deqin, YN N 28°27.626’ E 098°54.589’ 3040m J. Q. Liu-2006310 STP Ranwu, XZ N 29°30.292’ E 096°45.802’ 3950m J. Q. Liu-2006290 STP Basu, XZ N 30°11.523’ E 097°19.351’ 4210m J. Q. Liu-2006297 STP Leiwuqi, XZ N 31°12.321’ E 096°36.056’ 3800m J. Q. Liu-AO 121 STP Leiwuqi, XZ N 30°57.976’ E 096°14.537’ 4150m J. Q. Liu-AO 126 STP Dingqing, XZ N 31°30.000’ E 095°23.000’ 3770m J. Q. Liu-AO 127 STP Dingqing, XZ N 31°18.364’ E 095°45.659’ 3680m J. Q. Liu-AO 128 STP Dingqing, XZ N 31°41.341’ E 095°01.492’ 3860m J. Q. Liu-AO 130 STP Baqing, XZ N 31°48.803’ E 093°44.462’ 3970m J. Q. Liu-AO 132 STP Naqu, XZ N 31°45.293’ E 092°46.547’ 4220m J. Q. Liu-AO 134 STP Dangxiong, XZ N 30°32.355’ E 091°07.669’ 4310m J. Q. Liu-AO 157 STP Jiangzi, XZ N 28°50.995’ E 089°55.043’ 4300m J. Q. Liu-AO 142 STP Kangma, XZ N 28°32.649’ E 089°42.990’ 4430m J. Q. Liu-2006219 STP Jilong, XZ N 28°48.806’ E 085°30.074’ 4644m J. Q. Liu-2007140 STP Jilong, XZ N 28°26.737’ E 085°14.864’ 3026-4126m J. Q. Liu-2007042 Note: DP, the diploid population; NTP, tetraploid populations in the northern QTP; STP, tetraploid populations in the southern QTP; DP & NTP indicates that the population has both cytotypes (from Cui et al., 2008). Table S5 The 12 environmental variables used for ENM in this study Environmental variable Abbreviation Mean Monthly Temperature Range bio2 Isothermality bio3 Temperature Annual Range bio7 Mean Temperature of Wettest Quarter bio8 Mean Temperature of Warmest Quarter bio10 Mean Temperature of Coldest Quarter bio11 Precipitation Seasonality bio15 Precipitation of Wettest Quarter bio16 Precipitation of Driest Quarter bio17 Precipitation of Warmest Quarter bio18 Altitude alt Table S6 Analysis of STRUCTURE output lnP(D)Mean lnP(D)SD ΔK Diploid and tetraploid 20 -6173.755 0.455925894 0 Mixed 2 20 -5233.805 0.131689427 4402.593374 Mixed 3 20 -4873.63 21.16820108 1.963295079 Mixed 4 20 -4526.06 5.497693296 22.81388634 Partially separate 5 20 -4301.11 75.99871121 1.790369834 Separate 6 20 -4129 85.09611903 1.86549195 Separate 7 20 -4021.91 66.05154208 2.118248804 Separate 8 20 -3941.69 103.6761857 1.764484523 Separate 9 20 -3803.72 134.9992577 1.901988606 Separate 10 20 -3674.65 132.5236761 - Separate K RepTimes 1 Note: K, the number of clusters; RepTimes, repeat times of independent simulations run for each value of K; lnP (D), the log likelihood of the observed genotype distribution in K clusters that can be output by STRUCTURE simulation (Pritchard et al., 2000); ΔK, an ad hoc quantity related to the second order rate of change of the log probability of data with respect to K, this is a good predictor of the real value of K (Evanno et al., 2005). Table S7 ITS genotypes (H) and CHS genotypes (C) among the 28 populations P. Location ITS (H) CHS (C) 1 Huzhu, QH H1,H2 C1,C2,C3 2 Ledu, QH H1,H2 C4,C5,C6 3 Xining,QH H1,H2 C7,C8,C9 4 Haiyan,QH H3;H3,H4 C10,C11 5 Lintao,GS H5 C12,C13;C13,C14 6 Luqu,GS H6 C15,C16 7 Tongren,QH H7 C16,C17,C18 8 Guoluo,QH H7 C10,C19,C20;C19,C21,C22 9 Hongyuan,SC H8 C23,C24 10 Shiqu,SC H9; H10,H11 C10,C25;C25,C26;C20,C27,C28 11 Jiangda,XZ H12,H13;H14,H15 C29;C30,C31 12 Yushu,QH H16,H17 C32 13 Lixian,SC H18,H19 C33,C34,C35,C36,C37 14 Aerjinshan,GS H20 C38,C39,C40,C41,C42 15 Dangjinshan,GS H20 C38,C43,C44,C45 16 Yumen,GS H20 C41,C46,C47,C48,C49 17 Delingha,QH H3,H21 C20,C49,C50,C51,C52,C53 18 Xinghai,QH H22,H23 C46,C54,C55,C56,C57 19 Shandan,GS H24,H25 C40,C58,C59,C60,C61 20 Zeku,QH H26,H27 C16,C26,C46,C62,C63 21 Yushu,QH H28,H29 C20,C50,C64 22 Yushu,QH H7,H30 C16,C64,C65,C66 23 Yushu,QH H31,H32 C66,C67,C68,C69 24 Dingqing,XZ H31,H33 C13,C20,C36,C70 25 Leiwuqi,XZ H28,H31 C20,C36,C71 26 Ranwu,XZ H12,H13 C20,C36,C71,C72,C73 27 Dangxiong,XZ H28,H31 C36,C69,C74 28 Kangma,XZ H31,H34 C36,C69,C75 Fig. S1 Principal Co-ordinate Analysis (PCoA) plot based on a matrix of Jaccard distances among 157 individuals using NTSYS-PC 2.0. All the individuals could be identified within seven groups, which correspond to the output of the Structure analysis when K=6: the tetraploid populations clustered into three groups, STP (populations in southern QTP), ETP (populations in eastern QTP) and NTP (populations in northern QTP); the diploid populations readily clustered into five groups and DP-2 was intermixed with NTP. Fig. S2 ITS phylogenetic relationships of sampled Allium przewalskianum in the present study using Bayesian, ML and MP analyses. All analyses were performed according to the methods outlined in the main paper. We used Allium ovalifolium, Allium eduardii and Allium mongolicum as outgroups. All newly obtained ITS sequences were deposited in GenBank (accessions KC189069–KC189120). We recovered 34 genotypes from 28 populations (Table S7). The ML tree and the MP tree were congruent with the 50% major consensus tree obtained from Bayesian analysis (shown in this Fig., under the GTR+I+G model) and had a similar topology. The support values are given above the branches (Left: posterior probabilities of Bayesian analysis; Middle: bootstrap values of maximum-parsimony and Right: bootstrap values of maximum-likelihood). These analyses based on the nuclear ITS sequences suggest that all sampled diploid and tetraploid individuals of A. przewalskianum can be grouped into five lineages, A-E. Fig. S3 CHS phylogenetic relationships of sampled Allium przewalskianum in the present study using Bayesian, ML and MP analyses. All analyses were performed according to the methods outlined in the main paper. We used Allium cepa as the outgroup. All newly obtained CHS sequences were deposited in GenBank (accession numbers KC188957–KC189068). We recovered 75 genotypes from 28 populations (Table S7). The ML tree and the MP tree were congruent with the 50% major consensus tree obtained from Bayesian analysis (shown in this Fig., under the GTR+I+G model) and had a similar topology with respect to the main branches. The support values are given above the branches (Left: posterior probabilities of Bayesian analysis; Middle: bootstrap values of maximum-parsimony and Right: bootstrap values of maximum-likelihood). These analyses based on the nuclear CHS sequences also suggest that all sampled diploid and tetraploid individuals of A. przewalskianum can be grouped into five lineages, I-V. References: Cui, X. K., Ao, C. Q., Zhang, Q., Chen, L. T., & Liu, J. Q. (2008). Diploid and tetraploid distribution of Allium przewalskianum Regel. (Liliaceae) in the Qinghai-Tibetan Plateau and adjacent regions. Caryologia, 61, 192–200. Evanno, G., Regnaut, S., & Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, 14, 2611–2620. Pritchard, J. K., Stephens, M., & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–959.