MULTICELL.DOC
advertisement

Introduction
Page 1
=========================================================================
==
MultiCell - Model description and script documentation
written by Matt Wilson 4/89
=========================================================================
==
TABLE OF CONTENTS
Section
pages
----------------------------------------------Introduction ..........................
Model description .....................
MultiCell.g ...........................
constants.g ...........................
channel.g .............................
compartment.g .........................
cell.g ................................
Description of MultiCell.g ............
Description of constants.g ............
Description of channel.g ..............
Description of compartment.g ..........
Description of cell.g .................
Exercises .............................
Exercise solutions ....................
Index .................................
1
2
5
7
8
10
11
13
21
23
27
28
32
33
34
=========================================================================
==
Documentation for the MultiCell simulation
Line-by-Line Script commentary
=========================================================================
==
This document contains numbered versions of the MultiCell simulation
scripts
along with line-by-line descriptions for each.
The basic format for the line by line documentation will be
line number(s) in the actual script
description
description
description
:
complete/partial line
Note that the line numbers shown in the sample scripts are for
reference purposes and are not present in the actual scripts.
Line numbering is NOT used or allowed within actual scripts.
=========================================================================
==
Model description
Page 2
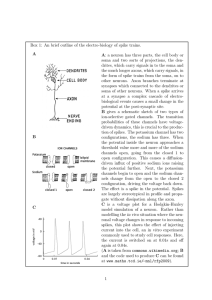
MODEL DESCRIPTION
----------------This is a simulation of two simplified, but realistic neurons.
in a feedback configuration. Each cell is composed of two compartments
corresponding to a soma and a dendrite. Each compartment is composed of
two variable conductance ionic channels for sodium (Na) and potassium
(K).
The dendritic channels are synaptically activated while the somatic
channels
are voltage dependent with Hodgkin-Huxley kinetics.
The output of each cell is a spike event. These spike events reach
their
synaptic destinations after a specified delay. When they arrive they
activate
a conductance change Gk with peak value given by 'weight*gmax'. The
parameters
'tau1/tau2' control the time course of the postsynapstic conductance
changes.
These conductances have time courses of the form
alpha function form
for tau1 == tau2
G = Gk*Ate(-t/Tau)
dual exponential form
for tau1 <> tau2
G = Gk*A(e(-t/tau1) - e(-t/tau2))
where A is a normalizing factor.
The change in membrane potential caused by this input is given by
dVm/dt = [(Vm' - Vm)/Ra + (ENa - Vm)*GNa + (EK - Vm)*GK +
(Erest - Vm)/Rm + Iinject]/Cm
ENa and EK are the ionic equilibrium potentials for each conductance.
Rm is the membrane resistance and Cm is the membrane capacitance.
Erest is the resting membrane potential and Iinject is an injection
current.
Vm' is the membrane potential of the adjacent compartment and Ra is the
axial
resistance between them. The network diagram for the model looks like
this:
___
synaptic |ENa|
weight
|___|
+-----------< |EK |
DENDRITE
|
|___|
|
/
\
|
/ cell1 \
SOMA
|
--------|
|
|
+---------+
|
|
delay
[ ]
[ ]
|
___
|
|
|ENa| >----+
|
|___|
|
|EK |
|
|___|
|
/
\
|
/ cell2 \
|
--------|
|
+--------------+
axon
Model description
Page 3
The circuit diagram for each cell looks like this
DENDRITE
Vm
o__________________________________________________________
|
|
|
|
|
|
|
|
|
|
|
|
|
\
\
\
|
|
|
--/-->
--/-->
/
/ \
___|___
|
GNa \
GK \
Rm
\
/ A \
_______ Cm
|
|
|
|
\ | /
|
|
|
|
|
\ /
|
|
ENa --EK
--Erest --| Iinject|
|
------------------|
|
|
|
|
|
|
|
\
|______________|_____________|________|________|
/ Ra
\
|
|
|
|
SOMA
AXON
| Vm
_____
o________________________________________________________________|
|____\
|
|
|
|
|
|
|
/
|
|
|
|
|
----\
\
\
|
|
--/-->
--/-->
/
|
___|___
HH-GNa \
HH-GK \
Rm
\
/ \
_______ Cm
|
|
|
/ A \
|
|
|
|
\ | /
|
ENa --EK
--Erest --\ /
|
------------------| Iinject|
|
|
|
|
|
|______________|_____________|________|________|
RUNNING THE SIMULATION
---------------------To start up the simulation change into the directory containing
the MultiCell scripts (e.g. /usr/genesis/Scripts/MultiCell)
and type 'genesis'. This should display a few messages
and return with a 'genesis #0 >' prompt. To run the MultiCell demo type
the following after the prompt:
genesis #0 > MultiCell
After 30 seconds or so the simulation graphics should appear.
The control panel at the upper left of the screen contains buttons which
control the starting and stopping of the simulation. To start, enter the
duration of the simulation in the 'STEP' dialog box, then either press
the 'Return' key or click on the 'STEP' button. At this point the
simulation
Model description
Page 4
should begin running and the graphs should begin to display data.
To interrupt the simulation at any time press the 'STOP' button.
Pressing the 'STEP' button again will continue the simulation for
an additional n msec where n is the value given in the STEP dialog.
To return the simulation to its initial conditions and erase all
graphs press the 'RESET' button. Toggling the 'overlay graphs' button
will cause successive runs to be overlayed on the same graph.
Pressing the 'QUIT' button will exit the simulator completely.
The scales of the graphs can be changed by clicking on the 'scale'
button
associated with the graph, changing the values in the
appropriate dialogs and pressing the 'DONE' button.
Several graphs are displayed for each cell. The upper graph shows the
membrane potential Vm vs time for the dendrite (upper) and the soma
(lower).
The lower graph plots the conductances GNa and GK vs time.
** Note: Most parameters can be varied while the simulation is actually
running with immediate effect. For instance, changing the values of
synaptic weights during execution will have directly observable effect
on the magnitude of displayed membrane conductances and potentials.
Delay parameters are an exception. After changing a delay parameter
the simulation must be reset for the effects to be accurately
accounted for.
SCRIPTS
------The simulation uses these basic scripts.
'MultiCell.g'
- the startup file for the model
'constants.g'
- global constants used in the model specification
'channel.g'
- functions for creating several types of channels
'compartment.g' - function for creating a basic compartment
'cells.g'
- function for creating a basic neuron
'buttons.g' - functions used for buttons and other graphical objects.
'graphics.g'
- the specification for the graphics display.
To get your own copy of this simulation, copy all of these files
into your own directory.
The simulator also uses the following system toolbox scripts.
'tools.g' - generally useful simulation functions
'xtools.g' - generally useful graphics functions.
These files should reside in the general system directory and do not need
to
copied.
ADDITIONAL REQUIREMENTS
-----------------------
The simulator reads a file called .simrc when it starts up.
This file must be present in the current working
directory or in the home directory of the user.
If you dont have this file you should get a copy from
the genesis system directory (normally located in /usr/genesis)
and place it in your home directory.
MultiCell.g
Page 5
MultiCell.g
=========================================================================
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
//genesis
echo " "
echo "*************************************************"
echo "*
MultiCell
*"
echo "*
A simple neuronal circuit simulation.
*"
echo "*
*"
echo "*
Written by Matt Wilson
*"
echo "*
California Institute of Technology
*"
echo "*
4/89
*"
echo "*************************************************"
echo " "
prompt
setclock
setclock
int
echo
echo
echo
echo
echo
"MultiCell
0
0.1
1
1.0
GRAPHICS
=
!"
// set the prompt
// simulation time step in msec
// output interval
1
SIMULATION CONFIGURATION
=========================
Hodgkin-Huxley
multi-compartmental neurons (soma + dendrite)
with synaptic connections
//==================================================
//
STARTUP SCRIPTS
//==================================================
include constants.g
include tools.g
include xtools.g
include channel.g
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
include compartment.g
include cell.g
if(GRAPHICS)
include graphics.g
end
//==================================================
//
CELL1
//==================================================
makeneuron(/cell1,30e-4,30e-4,100e-4,2e-4)
position /cell1
100e-4 100e-4 500e-4
//==================================================
//
CELL2
//==================================================
makeneuron(/cell2,10e-4,10e-4,100e-4,1e-4)
position /cell2
200e-4 200e-4 500e-4
//==================================================
//
CONNECTIONS
//==================================================
MultiCell.g
53
synapse
54
55
*/
56
msec
57
synapses
58
59
60
61
msec
62
synapses
63
64
65
66
67
68
connect
Page 6
/cell1/axon /cell2/soma/dend/Na_channel with
/* set delay and weight parameters for connections from cell1
set
/cell1/axon:0
delay
weight
5 \
30
//
//
/* set up connections for cell2 */
connect
/cell2/axon /cell1/soma/dend/K_channel with synapse
set
/cell2/axon:0
delay
5 \
//
weight
300
//
//==================================================
// provide current injection to cell 1
//==================================================
set /cell1/soma inject 3e-4
// uA
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
//==================================================
//
OUTPUT
//==================================================
// write out the intracellular soma potentials to disk
//==================================================
disk_output /cell1/Vm1 ascii
sendmsg
/cell1/soma
/cell1/Vm1
SAVE Vm
disk_output /cell2/Vm2 ascii
sendmsg
/cell2/soma
/cell2/Vm2
SAVE Vm
//==================================================
//
GRAPHICS
//==================================================
if(GRAPHICS)
loadgraphics
makegraphics /cell1
makegraphics /cell2 580 120
end
//==================================================
// check and initialize the simulation
//==================================================
check
reset
adjustspike
echo ==================
echo Simulation loaded.
echo ==================
constants.g
Page 7
constants.g
=========================================================================
1
//genesis
2
3
4
5
6
7
potential
8
9
10
11
12
13
14
randomseed -new
float PI
// initialize the random # generator
=
3.14159
// channel equilibrium potentials mV
float EREST
=
-70
// resting membrane
float
ENa
float EK
float Eleak
=
=
=
115.0 + EREST
-12.0 + EREST
10.6 + EREST
// leakage potential
// peak channel conductance mS/synapse
float GNa
=
80e-9
float GK
=
40e-9
channel.g
Page 8
channel.g
=========================================================================
1
//genesis
2
/*
3
functions defined in this script
4
===================================================================
==========
5
FUNCTION NAME
ARGUMENTS
6
===================================================================
==========
7
makechannel
(compartment,channel,Ek,tau1,tau2,gmax)
8
make_hhNa
(compartment,channel,activearea,ENa,Erest)
9
make_hhK
(compartment,channel,activearea,EK,Erest)
10
===================================================================
==========
11
*/
12
13
// different forms for the HH rate coefficients
14
int
EXPONENTIAL =
1
15
int
SIGMOID
=
2
16
int
LINOID
=
3
17
18
//=================================================
19
//
BASIC TRANSMITTER ACTIVATED CHANNEL
20
//=================================================
21
function makechannel(compartment,channel,Ek,tau1,tau2,gmax)
22
str compartment
23
str channel
24
float Ek
// mV
25
float tau1,tau2
// msec
26
float gmax
// mS
27
28
create channelC2
{compartment}/{channel}
29
set
^ \
30
Ek
{Ek} \
31
tau1
{tau1} \
32
tau2
{tau2} \
33
gmax
{gmax}
34
link_channel2
{compartment}/{channel}
{compartment}
35
end
36
37
//=================================================
38
//
HH NA CHANNEL
39
//=================================================
40
function make_hhNa(compartment,channel,activearea,ENa,Erest)
41
str compartment
42
str channel
43
float activearea
// cm^2
44
float ENa,Erest
// mV
45
46
create hh_channel {compartment}/{channel}
47
set
{compartment}/{channel} \
48
Ek
{ENa} \
// mV
49
Gbar
{120.0*activearea} \
// mmhos/cm^2
50
Xpower
3.0 \
51
Ypower
1.0 \
52
X_alpha_FORM {LINOID} \
53
X_alpha_A
-0.1 \
// 1/mVmsec
channel.g
Page 9
54
X_alpha_B
-10.0 \
// mV
55
X_alpha_V0
{ 25.0 + Erest } \
// mV
56
X_beta_FORM
{EXPONENTIAL} \
57
X_beta_A
4.0 \
// 1/msec
58
X_beta_B
-18.0 \
// mV
59
X_beta_V0
{ 0.0 + Erest } \ // mV
60
Y_alpha_FORM {EXPONENTIAL} \
61
Y_alpha_A
0.07 \
// 1/msec
62
Y_alpha_B
-20.0 \
// mV
63
Y_alpha_V0
{ 0.0 + Erest } \ // mV
64
Y_beta_FORM
{SIGMOID} \
65
Y_beta_A
1.0 \
// 1/msec
66
Y_beta_B
-10.0 \
// mV
67
Y_beta_V0
{ 30.0 + Erest } // mV
68
link_channel2
{compartment}/{channel}
{compartment}
69
end
70
71
//=================================================
72
//
HH K CHANNEL
73
//=================================================
74
function make_hhK(compartment,channel,activearea,EK,Erest)
75
str compartment
76
str channel
77
float activearea
78
float EK,Erest
79
80
create hh_channel {compartment}/{channel}
81
set
^ \
82
Ek
{EK} \
//
83
Gbar
{ 36.0 * activearea } \
mmhos/cm^2
84
Xpower
4.0 \
85
Ypower
0.0 \
86
X_alpha_FORM
{LINOID} \
87
X_alpha_A -0.01 \
//
88
X_alpha_B -10.0 \
//
89
X_alpha_V0 { 10.0 + Erest } \
//
90
X_beta_FORM {EXPONENTIAL} \
91
X_beta_A
0.125 \
//
92
X_beta_B
-80.0 \
//
93
X_beta_V0 { 0.0 + Erest } // mV
94
link_channel2
{compartment}/{channel}
{compartment}
95
end
compartment.g
mV
//
1/mV-msec
mV
mV
1/msec
mV
Page 10
compartment.g
=========================================================================
1
//genesis
2
/*
3
functions defined in this script
4
===================================================================
==========
5
FUNCTION NAME
ARGUMENTS
6
===================================================================
==========
7
makecompartment
(path,l,d,Eleak)
8
===================================================================
==========
9
*/
10
11
//=================================================
12
//
COMPARTMENT
13
//=================================================
14
function makecompartment(path,l,d,Erest)
15
str path
16
float l,d
17
float Erest
18
float area = l*PI*d
19
float xarea = PI*d*d/4
20
float rm = 10.0
// Kohm-cm^2
21
float cm = 1.0
// uF/cm^2
22
float ra = 0.05
// Kohm-cm
23
24
create compartment {path}
25
set
{path} \
26
Em
{ Erest } \
// mV
27
Rm
{ rm/area } \
// Kohm
28
Cm
{ cm*area } \
// uF
29
Ra
{ ra*l/xarea }
// Kohm
30
end
cell.g
Page 11
cell.g
=========================================================================
1
//genesis
2
/*
3
functions defined in this script
4
===================================================================
==========
5
FUNCTION NAME
ARGUMENTS
6
===================================================================
==========
7
makeneuron
(path,soma_l,soma_d,dend_l,dend_d)
8
===================================================================
==========
9
*/
10
11
//=================================================
12
//
NEURON
13
//=================================================
14
function makeneuron(path,soma_l,soma_d,dend_l,dend_d)
15
str path
16
float soma_l,soma_d
17
float dend_l,dend_d
18
19
float active_area = soma_l*PI*soma_d*1.0 // 100% of the
soma active
20
21
create neutral
{path}
22
push {path}
23
//=============================================
24
//
cell body
25
//=============================================
26
makecompartment(soma,{soma_l},{soma_d},{Eleak})
27
position soma
I I R{-soma_l/2.0}
28
29
//=============================================
30
//
voltage dependent conductances
31
//=============================================
32
make_hhNa(soma,hh_Na,{active_area},{ENa},{EREST})
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
make_hhK(soma,hh_K,{active_area},{EK},{EREST})
//=============================================
//
dendrite
//=============================================
makecompartment(soma/dend,{dend_l},{dend_d},{Eleak})
position soma/dend
I I R{-dend_l/2.0}
link_compartment
soma/dend
soma
//=============================================
//
synaptic conductances on the dendrite
//=============================================
makechannel(soma/dend,Na_channel,{ENa},3.0,3.0,{GNa})
makechannel(soma/dend,K_channel,{EK},10.0,10.0,{GK})
//=============================================
//
spike detector
//=============================================
create spike
spike
set
spike \
thresh
0 \
cell.g
54
55
56
detector */
57
58
59
60
61
62
63
64
65
66
67
end
Page 12
abs_refract 10 \
output_amp 1
/* use the soma membrane potential to drive the spike
sendmsg
soma
spike
INPUT Vm
//=============================================
//
output axon
//=============================================
create axon
axon
/* use the spike detector output to drive the axon */
sendmsg spike
axon
BUFFER name
pop
MultiCell.g
Page 13
=========================================================================
==
MultiCell.g
=========================================================================
==
This is the main simulation script. It invokes the scripts used to
construct the model as well as the graphics.
To start up the simulation enter 'MultiCell' at the simulator command
line.
genesis #0 > MultiCell
line
1 :
//genesis
This comment line should be placed at the beginning of
each GENESIS script. This distinguished the file from
an ordinary text file or a shell script which otherwise
would be executed as a GENESIS script if invoked from
the command line. The spacing of the line is not important
but it must precede any other non-blank text.
line
2-11 :
echo "*
MultiCell
*"
The echo command is used to print text to the standard
output (normally the screen). Each argument to the echo
command is concatenated with a single space between them
and then output. Since white space is used to separate
command line arguments a line such as:
echo *
MultiCell
*
will actually call the echo command with the three arguments
arg 1: *
arg 2: MultiCell
arg 3: *
which will then be concatenated and displayed as
* MultiCell *
To preserve spacing, the command
echo "*
MultiCell
*"
will call echo with the single argument
arg 1: *
MultiCell
*
and display
*
MultiCell
*
Note the use of the string delimiters "" to place white space
into a single argument. The string delimiters can be used
in other contexts which require grouping of text into
single string arguments.
line 12 :
Empty lines are ignored by the interpreter and can be used
to improve the readability of scripts.
line 13 :
prompt "MultiCell !"
This sets the prompt string used by the simulator when
waiting for keyboard input. The prompt "genesis !" is
used by default. The '!' character in the prompt string
will be replaced by the current command number which can
be used in conjunction with the command line history
recall mechanisms (see also 'history') Note the use of
single line comments which are started with the
characters '//'. Any text following these characters
MultiCell.g
Page 14
up to the carriage return at the end of the line will be
treated as a comment and ignored.
line 14 :
setclock 0 0.1
The simulator contains an array of 100 interval timers
called clocks. These clocks are used to call element
functions (see 'objects' and 'clocks') at the desired
intervals during a simulation. Clock 0 is the basic
simulation clock. Each simulation step will advance the
simulation time by this amount. Each element uses clock
0 by default and, therefore, is called on each time step.
This command sets the basic simulation clock to the
value of 0.1 time units (msec in this simulation).
line 15 :
setclock 1 1.0
This assigns clock 1 the value of 1.0. This clock is used
to drive output elements such as graphs and disk file
storage, which can be simulated at a slower rate to
reduce computational overhead (see also 'useclock').
line 16 :
int GRAPHICS = 1
The interpreter allows the definition of global variables
which are accessible to scripts and interpreter functions.
There are three types of variables supported:
int
integer
float floating point
str
string
This command creates a global variable called GRAPHICS and
assigns it the value of 1. The variable name is capitalized
only for readability and is not a requirement. Variables
can also be defined without being assigned a value. For
example, the following is a valid global variable
declaration:
int GRAPHICS
Global variables which have already been declared can later
be assigned values. For example, following the declaration
above GRAPHICS could be assigned the value 1 using
GRAPHICS = 1
line 18-22 :
echo SIMULATION CONFIGURATION
Prints out a message describing the configuration of the
neurons in the simulation (see lines 2-11 for a description
of the 'echo' command)
line 24-26 :
// STARTUP SCRIPTS
One line comments which do not print anything to the screen
as in 'echo' and are used for readability of scripts.
(see also line 13)
line 27 :
include constants.g
The 'include' command causes the named script to be read in
at that point and executed as though the text were actually
included in the script which invoked it.
The script 'constants.g' defines global variables which are
used by other scripts in the simulation. (see the description
of constants.g).
MultiCell.g
Page 15
line 28 :
include tools.g
tools.g contains general functions which are frequently used
and therefore have been placed in the general system toolbox.
see description of tools.g (see also line 27)
line 29 :
include xtools.g
xtools.g contains graphical functions which are frequently
used
and therefore have been placed in the general system toolbox.
see description of xtools.g (see also line 27)
line 30 :
include channel.g
channel.g contains functions used to define and add various
types of channels to a compartment.
see description of channel.g (see also line 27)
line 31 :
include compartment.g
compartment.g contains the function used to create and link
together compartments.
see description of compartment.g (see also line 27)
line 32 :
include cell.g
cell.g contains the function used to construct a single
multicompartment neuron.
see description of cell.g (see also line 27)
line 34 :
if(GRAPHICS)
The interpreter supports the 'if' conditional statement.
The expression between the parentheses will be evaluated.
If the value of the expression is non-zero then all
statements
between the if and the matching else or end will be
executed. Examples of valid if statements are:
if(1)
echo hello
end
if(1); echo hello; end
if((GRAPHICS == 1) || (5+3 > 10))
echo hello
end
Note the use of the semi-colon in example 2 which can
be used as an end-of-statement delimiter not only in
the case of 'if's but in any context.
In the specific case of line 34, the global variable
GRAPHICS is being used as a flag for invoking the
graphical components of the simulation.
If GRAPHICS is not zero then the graphics script
specified in line 35 will be invoked.
line 35 :
include graphics.g
graphics.g contains functions used to create the graphical
interface to the simulation.
see description of graphics.g (see also line 27)
line 36 :
end
This is the matching 'end' for the 'if' in line 34. All
MultiCell.g
Page 16
'if' statements MUST have a matching 'end' or bad things
will happen. (see also line 34)
line 38-40 :
// CELL1
comment. see line 25-27
line 41 :
makeneuron(/cell1,30e-4,30e-4,100e-4,2e-4)
This line invokes a function defined in cell.g which
constructs a single neuron called '/cell1', with a soma
30 microns in length (30e-4 cm) and 30 microns in
diameter, and a dendrite 100 microns in length and 2 microns
in diameter.
Note that the parameters to the function have been placed
within parentheses in a comma-separated form. This is
the 'function-call' format. This is not necessary in this
context and the function could have been invoked as
makeneuron /cell1 30e-4 30e-4 100e-4 2e-4
in the 'command-line' format with the same results.
line 42 :
position /cell1 100e-4 100e-4 500e-4
The position command moves an element to the
specified xyz coordinates. All of the sub-elements in the
tree beneath it will be also be moved, preserving relative
positions relationships between the sub-elements.
In the case of line 42, '/cell1' is positioned at location
x = 100um, y=100um at a depth of 500um. When the cell
was created in line 41, /cell1 was given the coordinates
x=0,y=0,z=0 and the soma and dentritic compartments
were given coordinates of x=0,y=0,z=-15um and
x=0, y=0, z=-80um respectively (the centers of each
compartment). Following the 'position' command of line
42 /cell1 would have the coordinates x=100,y=100,z=500 and
its soma and dendrite would have coordinates
x=100um,y=100um,z=485um and x=100um,y=100um,z=420um.
The use of the 'position' command in this context
should be contrasted with the use of the 'set'
command which does not adjust the coordinates of subelements.
If the command
set /cell1 x 100e-4 y 100e-4 z 500e-4
had been used, only the coordinates of /cell1 would have
been assigned the values of x=100um,y=100um,z=500um and the
coordinates of its soma and dendrite would have remain
unchanged.
line 44-46 :
// CELL2
comment. see line 25-27
line 47 :
makeneuron(/cell2,10e-4,10e-4,100e-4,1e-4)
constructs a single neuron called '/cell2', with a soma
10 microns in length and 10 microns in diameter, and a
dendrite 100 microns in length and 1 micron in diameter.
(see also line 41)
line 48 :
position /cell2 200e-4 200e-4 500e-4
places /cell2 at location x=200um, y=200um, at a depth of
500um. (see also line 42)
MultiCell.g
Page 17
line 50-52 :
// CONNECTIONS
comment. see line 25-27
line 53 :
'segment'
connect /cell1/axon /cell2/soma/dend/Na_channel with synapse
The 'connect' command establishes a single synaptic
connection between a 'projection' class element and a
class element with the specified synapse type.
A connection allows communication of information between
elements with arbitrary delay and with the ability to
operate on the communicated information using a
synaptic transformation. Thus it is a more powerful (but
also more costly) means of passing information between
components of a simulation. In this case the axon of
cell1 is interconnected to the sodium channel on the
dendrite of cell2. 'synapse' is a particular type of
synaptic transformation which performs simple multiplication
of presynaptic events with a synaptic weight to
compute the resulting postsynaptic activation.
The axon generates impulse spike events which reach the
sodium channel after a delay stored in the connection.
The amplitude of this spike event is multiplied by the
synaptic weight. The resulting value is used as the amount
of transmitter activating the dendritic sodium channel.
(see also lines 56-57)
line 55 :
*/
/* set delay and weight parameters for connections from cell1
This is an example of a 'C' style comment which can
extend over multiple lines. The characters '/*' are used
to indicate the beginning of a comment while '*/' indicate
the end of a comment. All characters between these
delimiters are ignored by the interpreter and are used
strictly for readability of the script.
line 56-57 :
set
/cell1/axon:0
delay
5 \ //
msec
The 'set' command allows the contents of the element
data structure to be changed (see also 'create' and
'object' commands). The general syntax of this command is
set [path][:connection] field value [field value ...]
The path is the pathname of an element or a list of elements
(if wildcarding is used). If this is omitted then the
current working element is used (see also 'pwe').
In this case the path identifies the single element
'/cell1/axon'. If the path is a 'projection' class
element which has connections from it, the individual
connections can be identified using the ':connection'
notation. Connections can be identified by number or by
destination. For example
:0
refers to the first connection of the axon,
:0-5
would refer to the first 6 connections of the axon,
:#
refers to all of the connections of the axon, and
:@/cell2/soma/dend/Na_channel
would refer to the connection leading to the element
MultiCell.g
Page 18
/cell2/soma/dend/Na_channel.
In the case of line 56, the 'axon' has only one connection.
This connection can be referred to as either
:0 or as :@/cell2/soma/dend/Na_channel.
The 'field' is the name of a parameter field contained in the
data structure of the named element or connection. The
contents
of this field will be changed to the specified 'value'
when the command is executed. Multiple fields may be
set in a single command.
In this case the two fields 'delay' and 'weight' are
being set to the values 5 and 30 respectively.
The delay refers to the time between the generation
of a spike event by the axon (/cell1/axon) and the arrival
of that event at the target (/cell2/soma/dend/Na_channel).
The weight multiplies the spike amplitude (always 1 in this
case) and uses this value to activate the target channel
(simulating a pulse of transmitter)(see also line 53).
The weight has no intrinsic units. In this simulation it
has been assigned the units of 'number of synapses'.
line 59 :
/* set up connections for cell2 */
comment. see line 55
line 60 :
connect /cell2/axon /cell1/soma/dend/K_channel with synapse
This establishes a connection between the 'projection'
class element '/cell2/axon' (axon) and the 'segment'
class element /cell1/soma/dend_K_channel (potassium channel).
(see also line 53)
line 61-62 :
set
/cell2/axon:0
delay
5 \ //
msec
Assigns the delay and weight of the connection created
in line 60. (see also line 56-57)
line 64-65 :
// provide current injection to cell 1
comment. see line 24-26
line 67 :
set /cell1/soma inject 3e-4
// uA
This command sets the injection field of the compartment
'/cell1/soma'. The compartment will receive a steady
current injection of .3 nA until the value of this
field is changed. To stop the current injection the
command
set /cell1/soma inject 0
could be used (see also line 56-57).
line 69-73 :
// OUTPUT
comment. see line 25-27
line 74 :
disk_output
/cell1/Vm1
ascii
disk_output is a function defined in the system tools
script 'tools.g' found in the system startup directory.
This function creates an element which saves simulation
data to a disk file in either ascii or special binary form.
The general syntax of the command is
disk_output path ascii/binary
Note that this command only creates the output element
MultiCell.g
Page 19
and does not indicate what information is to be saved
(see line 75). When the simulation is run, output will
be saved to disk at intervals determined by clock 1
(see line 15). The default name of the data file is the same
as the name of the element. (see also the 'asc_file'
object description). In the case of line 74 the function will
create an element called '/cell1/Vm1' which will save
data in ascii form in a file called 'Vm1'.
line 75 :
sendmsg /cell1/soma
/cell1/Vm1
SAVE Vm
The general syntax of the sendmsg command is
sendmsg source destination msgtype arg1 arg2 ...
Sending messages is the way in which the simulator
allows the destination element to access the
specified information in the source. Along with simply
giving access to the specified fields the message
also indicates what the destination is to do with that
information. A single destination element may be capable
of handling many different 'types' of messages, each
requiring different pieces of information used for
different functions. In the case of line 75 the type
of the message is 'SAVE' which has a single field name
as its argument. Different objects will have different
message types which they are capable of handling. To
list the types of messages and the arguments they
require, use either the
showobject 'objectname'
command for objects (element definitions) or the
show path *b
command for elements (created instances of objects)
The command in line 75 sends a message from the compartment
'/cell1/soma' to the file output element '/cell1/Vm1'
with the instructions to SAVE the data contained in
field 'Vm' (membrane potential) of the compartment.
(see also the 'sendmsg' command, 'objects', 'show'
and 'messages')
line 77 :
disk_output
/cell2/Vm2
ascii
creates an element called /cell2/Vm2 which will save
ascii data in a file called Vm2. (see line 74)
line 78 :
sendmsg /cell2/soma
/cell2/Vm2
SAVE Vm
Sends a message from /cell2/soma to /cell2/Vm2 instructing
it to save the membrane potential Vm to disk.
(see line 75 for discussion of sendmsg command)
line 80-82 :
// GRAPHICS
comment. (see line 25-27)
line 83 :
if(GRAPHICS)
conditional. (see line 34)
line 84 :
loadgraphics
If the conditional in line 83 is true then this function
will be activate the graphical interface and construct
the main control panel. The function itself is defined
MultiCell.g
Page 20
in the script 'graphics.g'
line 85 :
makegraphics /cell1
This function (defined in the script graphics.g) constructs
a graphical control panel for the named cell. The function
has optional x,y screen coordinate arguments (see line 86).
line 86 :
makegraphics /cell2 580 120
Construct the graphical control panel for /cell2 and place
it at screen location x=580, y=120 (in pixels) (see line 85).
line 87 :
end
Matching 'end' for the 'if' in line 83. (see line 36)
line 89-91 :
// check and initialize the simulation
comment. (see line 25-27)
line 92 :
check
Each element capable of checking its own consistency
is called with the CHECK action and reports the
results. Usually this takes the form of a printed warning
that an error has been found. This should be performed
before running a simulation whenever the
simulation has been significantly changed
for instance by adding new components.
line 93 :
reset
Reset returns the simulation to its initial state.
Each element requiring resetting is called with
the RESET action. This must be performed at least
once before executing a simulation.
line 94 :
adjustspike
This function adjusts the amplitude of spike inpulses
to normalize the area under the impulse to a value
independent of the time step. This should be executed
whenever the time step is changed.
line 96-98 :
echo Simulation loaded.
echo. (see lines 2-11)
constants.g
Page 21
=========================================================================
==
constants.g
=========================================================================
==
line 1 :
//genesis
script header. (see line 1 of MultiCell.g)
line 2 :
randomseed -new
initialize the random number generator. The arguments to
randomseed can be either an integer seed or -new.
A seed can be used when it is desirable to
generate a reproducible random number sequence.
The -new option uses the current date (yr:mo:da:hr:min:sec)
as a seed thereby producing a new random sequence
each time it is invoked.
line 4 :
float PI = 3.14159
This command creates a global variable called PI and
assigns it the value of 3.14159. This variable is used
for calculating cylindrical volumes and areas of
dendritic compartments. (for discussion of global
variables see line 16 of MultiCell.g)
line 7 :
EREST = -70
EREST is used to define the resting membrane potential
for the soma and dendrite of each neuron. (for discussion
of global variables see line 16 of MultiCell.g)
line 8 :
ENa = 115.0 + EREST
Ena is used to define the equilibrium potential for sodium
channels. This value is used by both the synaptically
activated channels as well as the voltage dependent
Hodgkin-Huxley channels. The value of 115 mV is based on
values for the squid giant axon referenced to a resting
potential of 0 mV. The EREST term allows the value of ENa
to be referenced to arbitrary resting potentials.
(for discussion of global variables see line 16 of
MultiCell.g)
line 9 :
EK = -12.0 + EREST
EK is used to define the equilibrium potential
for potassium channels. (see line 9) (for discussion of
global variables see line 16 of MultiCell.g)
line 10 :
Eleak = 10.6 + EREST
Eleak defines the membrane leakage potential. The value of
10.6 results in zero net current flow at the resting
potential in membrane containing the squid voltage
dependent sodium and potassium channels. (see line 9)
(for discussion of global variables see line 16 of
MultiCell.g)
line 13 :
GNa = 80e-9
GNa defines the peak conductance for synaptically activated
sodium channels. (80 pS/synapse) (for discussion of
global variables see line 16 of MultiCell.g)
constants.g
line 14 :
Page 22
GK = 40e-9
GK defines the peak conductance for synaptically activated
potassium channels (40 pS/synapse) (for discussion of
global variables see line 16 of MultiCell.g)
channel.g
Page 23
=========================================================================
==
channel.g
=========================================================================
==
line 1 :
//genesis
script header. (see line 1 of MultiCell.g)
line 2-11 : functions defined in this script
C style comment. (see line 55 of MultiCell.g)
line 13-17 :
// different forms for the HH rate coefficients
comment. (see line 13 of MultiCell.g )
line 14-16 :
int EXPONENTIAL = 1
The three variables EXPONENTIAL, SIGMOID, and LINOID
are used by the Hodgkin-Huxley channels to determine
the functional form of the voltage dependent rate
coefficients. The use of GLOBAL variables is simply
to enhance the readibility of the script since
using functional form of EXPONENTIAL is more descriptive
than functional form 1. (see line 47 and 81 for use of
the variables) (see line 9 of MultiCell.g for description
of global variables)
line 18-20 :
// BASIC TRANSMITTER ACTIVATED
comment. (see line 13 of MultiCell.g )
line 21 :
function makechannel(compartment,channel,Ek,tau1,tau2,gmax)
The 'function' keyword is used to define a
named interpreted function.
The general form for function definitions is:
function name
or
function name(arg1[,arg2,...argn])
All script commands
contained between the 'function' keyword and the
matching 'end' statement will be executed when
the function name ('overlaytoggle' in this case) is
invoked from the keyboard or within a script.
Example:
function test
echo I will try
end
This defines a function called 'test'. Invoking the
command test will cause the echo statement to be
executed.
Example:
> test
I will try
Arguments can be passed to functions. These arguments
can be assigned variable names in the function
definition.
Example:
function test(value)
echo OK {value}
channel.g
Page 24
end
This defines a function called 'test' which will
accept an argument. Within the function this argument
can be referred to using the name 'value'. To use
this argument within the function the curly brackets {}
are used to get the value of the variable.
Example:
> test this
I will try this
Multiple arguments can be passed to a function.
These arguments can be assigned names within the function
by using a comma separated argument list when
defining the function.
Example:
function test(value1,value2)
echo I will try again {value1} {value2}
end
> test this again
I will try this again
In line 21 the function 'makechannel' is defined with
six arguments. This function uses these arguments to
construct a synaptically activated channel on the
specified compartment.
line 22-26 :
str compartment
Any variables defined between the 'function' keyword
and its matching end statement are local variables.
Local variables
are distinct from global variables in that they have
value only within the function in which they are defined.
These variables cannot be referred to outside of their
function and when the function execution is complete they
are removed and their values are lost. Therefore they
cannot be used for permanent storage of information
as with global variables.
Local variables are defined using the same syntax as
global variables. (see line 16 MultiCell.g )
There is no way to define a global variable from within a
function.
In line 22-26 the local variables correspond to the
named variables in the function argument list of line
21. These allow the type of each argument to be
explicitly declared. If these statements were omitted
each argument would default to string (str) type.
Thus lines 22 and 23 which define compartment and
channel to be strings are redundant and are
included only for clarity. The remaining arguments
are specified as floating point numbers.
line 28 :
create
channelC2
{compartment}/{channel}
The general form of the create command is
create object_name pathname [options]
Create takes an object specification and creates
an instance of it called an element. This new element
resides in the element tree hierarchy and
channel.g
Page 25
is referred to by the pathname.
The new element has its own data area for parameters
and variables and uses the specific functions
which allow it to simulate the specified object.
The creation of some objects can be modified through
the use of options. This will be a function of
specific objects but occurs most frequently with graphical
objects. The list of valid objects can be obtained using the
'listobjects' command.
Line 46 creates an element using the channelC2 object
specification. This new element will have the path
given by {compartment}/{channel} where 'compartment'
and 'channel' are local variables passed into the
'makechannel' function. 'channelC2' implements a
simple second-order synaptically activated, time dependent
conductance which also computes channel current.
line 29-33 :
set
^ \
Set the parameters associated with the newly created
channel element. These parameters specify the time
course of the (tau1,tau2) the amplitude (gmax)
of the synaptically activated conductance. Ek
is used in the calculation of channel current.
The ^ is a shorthand notation for the most recently
created (or copied) element. Thus in this context
^ is equivalent to the path {compartment}/{channel}
used to create the channel in line 28.
The backslash (\) allows the command to be continued
on another line. All text following the backslash on the
same line will be ignored. (see also line 56-57 of
MultiCell.g
for discussion of the set command)
line 34 :
link_channel2 {compartment}/{channel}
{compartment}
link_channel2 is function defined in the system tools
script 'tools.g'. This function establishes functional
connections between a compartment and a channel using
messages. (see also line 75 of MultiCell.g for discussion
of messages)
line 35 :
end
This is the matching end for the 'function' statement
in line 21. (see also line 34 of MultiCell.g)
line 37-39 :
// HH NA CHANNEL
comment. (see line 13 of MultiCell.g )
line 40 :
function make_hhNa(compartment,channel,activearea,ENa,Erest)
Defines a function which creates a Hodgkin-Huxley
voltage dependent sodium channel. Hodgkin-Huxley
parameters are normalized to an area of membrane
specified by the activearea argument. The ENa argument
sets the equilibrium potential for sodium while
Erest is the resting potential of the compartment to
which the channel will be added.
(see line 21 for discussion of functions)
channel.g
Page 26
line 41-44 :
str compartment
These local variable definitions give the types of
the function arguments. The 'compartment' is the pathname
of the compartment onto which the new channel will be
added. 'channel' is the name of the new channel.
(see also line 22-26 for discussion of local variables)
line 46 :
create
hh_channel
{compartment}/{channel}
Line 46 creates an element using the hh_channel object
specification. This new element will have the path
given by {compartment}/{channel} where 'compartment'
and 'channel' are local variables passed into the
'makechannel' function.
(see line 28 for discussion of create)
line 47-67 :
set {compartment}/{channel} \
(see line 29-33) (see also line 56-57 of MultiCell.g for
discussion of the set command)
line 68 :
link_channel2 {compartment}/{channel} {compartment}
(see line 34)
line 69 :
end
This is the matching end for the 'function' statement
in line 40. (see also line 34 of MultiCell.g)
line 71-73 :
// HH K CHANNEL
comment. (see line 13 of MultiCell.g )
line 74 :
function make_hhK(compartment,channel,activearea,EK,Erest)
Defines a function which creates a Hodgkin-Huxley
voltage dependent potassium channel.
(see line 40)
line 75-78 :
str compartment
(see line 41-44)
line 80 :
create
hh_channel
(see line 46)
{compartment}/{channel}
line 81-93 :
set
^ \
(see also line 29-33)
line 94 :
link_channel2
(see line 34)
{compartment}/{channel} {compartment}
line 95 :
end
This is the matching end for the 'function' statement
in line 74. (see also line 34 of MultiCell.g)
compartment.g
Page 27
=========================================================================
==
compartment.g
=========================================================================
==
line 1 :
//genesis
script header. (see line 1 of MultiCell.g)
line 2-9 : functions defined in this script
C style comment. (see line 55 of MultiCell.g)
line 11-13 :
// COMPARTMENT
comment. (see line 13 of MultiCell.g)
line 14 :
function makecompartment(path,l,d,Erest)
Makes a cylidrical compartment on the element given by the
path variable with dimensions of length l, and diameter d.
The resting membrane potential of the compartment is
given by Erest. (for description of functions see
line 21 of channel.g)
line 15-17 :
str path
local variable declarations for the function arguments.
(see line 14) (for description of local variables see
line 22-26 of channel.g)
line 18-19 :
float area = l*PI*d
calculate the surface area of the cylinder given by the
length and diameter arguments to the function. Note that
expression evaluation brackets are not necessary
for the right hand side of the variable assignment. This
is because the assignment operation automatically
assumes expression evaluation.
line 20-22 :
float rm = 10.0
assign a local variable the value of membrane resistivity
used in calculating membrane resistance (see line 25-29)
line 24 :
create compartment {path}
creates an element called {path} using the compartment
object specification. 'path' is a local variable passed
into the function.
(see line 28 of channel.g for discussion of create command)
line 25-29 :
set {path} \
set the parameters associated with the compartment element.
These parameters include the resting membrane potential Em,
the membrane resistance Rm, the membrane capacitance Cm,
and the axial resistance of the compartment Ra.
(see line 29-33 of channel.g and line 56-57 of MultiCell.g
for discussion of the 'set' command)
line 30 :
end
This is the matching end for the 'function' statement
in line 14. (see also line 34 of MultiCell.g)
cell.g
Page 28
=========================================================================
==
cell.g
=========================================================================
==
line 1 :
//genesis
script header. (see line 1 of MultiCell.g)
line 2-9 : functions defined in this script
C style comment. (see line 55 of MultiCell.g)
line 11-13 :
// NEURON
comment. (see line 13 of MultiCell.g)
line 14 :
function makeneuron(path,soma_l,soma_d,dend_l,dend_d)
This function creates a neuron at location 'path'
containing a soma and a dendrite. The dimensions for
each compartment are passed to the function. The soma
contains voltage dependent Hodgkin-Huxley sodium and
potassium
conductances while the dendrite contains synaptically
activated sodium and potassium conductances.
(see line 21 of channel.g for discussion of functions)
line 15-17 :
str path
Local variable definitions for the arguments to the
makeneuron function. (see line 22-26 of channel.g for
discussion of local variables)
line 19 :
float active_area = soma_l*PI*soma_d*1.0
Calculates the area of active Hodgkin-Huxley
membrane used as a fraction of the total somatic membrane.
line 21 :
create
neutral
{path}
Create an element called {path} using the neutral object
specification. A neutral object is one which performs
no function during the simulation and is simply used
to organize elements in the hierarchy.
(see line 22 of channel.g for discussion of the create
command)
line 22 :
push {path}
saves the current working element on a
changes the current working element to
specified by the 'path' variable. This
when one wishes to temporarily visit a
and then return.
stack and
the element
is used
different element
line 26 :
makecompartment(soma,{soma_l},{soma_d},{Eleak})
This calls the function defined in compartment.g
to add a compartment whose pathname is given by
the value of soma with dimensions given by soma_l
(length) and soma_d (diameter).
line 27 :
position soma I I R{-soma_l/2.0}
sets the position of the newly created soma compartment
to -1/2 of the length of the compartment relative to the
cell.g
Page 29
parent or neuron location. The 'I' arguments in the locations
of the x,y coordinates tells the function to 'ignore'
x and y, leaving them with their original values.
The R modifier indicates that the given coordinate is
to be taken relative to the parent element (the element
above the specified element in the hierarchy) rather than
as an absolute coordinate.
(see line 42 of MultiCell.g for discussion of the position
command)
line 32 :
make_hhNa(soma,hh_Na,{active_area},{ENa},{EREST})
calls the function defined in 'channel.g' to create
a Hodgkin-Huxley voltage dependent sodium channel on the soma
over the specified area.
line 33 :
make_hhK(soma,hh_K,{active_area},{EK},{EREST})
calls the function defined in 'channel.g' to create
a Hodgkin-Huxley voltage dependent potassium channel on the
soma
over the specified area.
line 38 :
makecompartment(soma/dend,{dend_l},{dend_d},{Eleak})
adds a dendritic compartment to the soma with the
specified dimensions. (see line 26)
line 39 :
position soma/dend
I I R{-dend_l/2.0}
sets the position of the dendrite to -1/2 the
length of the dendritic compartment relative to the soma.
(see line 27)
line 40 :
link_compartment
soma/dend
soma
link_compartment is function defined in the system tools
script 'tools.g'. This function establishes functional
connections between two compartments using
messages. In this case it creates an axial coupling
between the soma and dendritic compartments.
(see also line 75 of MultiCell.g for discussion
of messages)
line 45 :
makechannel(soma/dend,Na_channel,{ENa},3.0,3.0,{GNa})
This calls the function defined in channel.g
to add a synaptically activated channel to the
dendrite of the neuron (compartment soma/dend)
called Na_channel. The equilibrium
potential of the channel is given by the value of ENa.
The time course of the conductance is given by the
two time constants 3.0 and 3.0 msec which result in
an alpha function form with a peak 3 msec. The amplitude
of the conductance is given by the value of GNa.
The parameters are designed to make this channel behave
like a sodium channel but the channel itself does
not presuppose any ionic type.
line 46 :
makechannel(soma/dend,K_channel,{EK},10.0,10.0,{GK})
create a synaptically activated potassium channel on the
dendrite of the neuron.
(see line 45)
cell.g
line 51 :
Page 30
create
spike
spike
create a spike generation element using the spike object
and call it spike.
The spike generation element uses the level of the
membrane potential, a threshold, and a refractory
period to determine whether to generate a spike at
each time step of the simulation.
When using Hodgkin-Huxley channels to produce
realistic action potentials, this component serves
as a spike discriminator. Without Hodgkin-Huxley
the spike element can serve as an integrate and fire
model for neuronal output.
In general, any model which requires propagation of
impulse events such as spikes will use this strategy.
(see line 28 of channel.g for discussion of the create
command)
line 52-55 :
set
spike
\
set the parameters of the spike generation element.
The thresh parameter sets the threshold level for
spike generation. When a spike is generated it is given
the amplitude output_amp.
(see line 29-33 of channel.g and line 56-57 of MultiCell.g
for discussion of the 'set' command)
line 57 :
sendmsg
soma
spike
INPUT Vm
sends a message from the soma of the neuron to
the spike generation element telling it to use the
membrane potential Vm as the input to trigger spike
production.
(see line 75 of MultiCell.g for discussion of the sendmsg
command)
line 62 :
create axon axon
create an axonal output element using the axon object
and call it axon.
The axon output element takes a buffer or history of
events (e.g. spikes) and sends them out to other
elements with which it has synaptic connections
with the appropriate delay.
(see line 28 of channel.g for discussion of the create
command)
line 64 :
sendmsg
spike
axon
BUFFER name
sends a message from the spike generation element to
the axonal output element telling it to use the history of
spike events to drive the axon.
(see line 75 of MultiCell.g for discussion of the sendmsg
command)
line 66 :
pop
restore the current working element to the element
pushed in line 22. Thus the current working element
is the same leaving the function as it was entering
regardless of movement about the hierarchy while
cell.g
Page 31
executing the makeneuron function.
line 67 :
end
This is the matching end for the 'function' statement
in line 14. (see also line 34 of MultiCell.g)
Exercises
Page 32
Exercises
--------For all of these exercises you should create a new script file called
'exercises' and place the commands to complete each exercise in this
file. Note that the first line in the file must be the genesis header
//genesis. After completing the exercises, run the simulator and load
in the MultiCell simulation. Following this, load in the exercises script
by typing 'exercises'.
Exercise 1) create a third cell called cell3 with soma dimensions 20 um
length
20 um diameter, and dendritic dimensions 200 um length 1.5 um
diameter.
Exercise 2) make a graphics control panel for the cell created in
exercise 2
Exercise 3) make an inhibitory connection from cell3 to cell2 and an
excitatory
connection from cell1 to cell3.
Exercise 4) save the membrane potential Vm of the cell3 dendrite to an
ascii file called Vmdend.
Exercise 5) using script commands (not the graphical interface) set the
weight of the connection from cell1 to cell3 to 50.
Set the weight from cell3 to cell2 to 100.
Exercise solutions
Page 33
Exercise solutions
-----------------Solution 1) makeneuron(/cell3,20e-4,20e-4,200e-4,1.5e-4)
(see line 41 of MultiCell.g, pages 6 and 16 of the
documentation)
Solution 2) makegraphics /cell3
(see line 85 of MultiCell.g, pages 6 and 20 of the
documentation)
Solution 3) For the inhibitory connection from cell3 to cell2 connect the
axon of cell3 to the hypolarizing channel (potassium) of cell2
using the command
connect /cell3/axon /cell2/soma/dend/K_channel with synapse
Note that in this type of model inhibitory connections
can not be constructed using negative weights which
would result in negative conductance at the destination.
For the excitatory connection from cell1 to cell3 connect the
axon of cell1 to the depolarizing channel (sodium) of cell3 with
connect /cell1/axon /cell3/soma/dend/Na_channel with synapse
Note that the default delay and weight for newly constructed
synapses is 0.
(see line 53 of MultiCell.g, pages 6 and 17 of the
documentation)
Solution 4) First we must create the element which will save the data to
disk in the appropriate format using the command
disk_output /cell3/Vmdend ascii
Then we must send a message to this element indicating what
information is to be saved using
sendmsg /cell3/soma/dend /cell3/Vmdend SAVE Vm
(see line 74 of MultiCell.g, pages 6 and 18 of the
documentation)
Solution 5) To set the weight of the connection from cell1 to cell3 use
the
set command
set /cell1/axon:@/cell3/soma/dend/Na_channel weight 50
Note that the connection is actually from the axon of cell1
to the sodium channel on the dendrite of cell3.
To set the weight of the connection from cell3 to cell2 use
set /cell3/axon:@/cell2/soma/dend/K_channel weight 100
(see line 56-57 of MultiCell.g, pages 6 and 17 of the
documentation)
Index
A
axon
C
Cm
cell
check
channel
comment
compartment
conductance
connect
create
D
delay
disk
E
Erest
echo
element
2,6,12,17,18,
21,30,33
Page 34
28,29,30,31,
32,33
expression
15,27
F
2,3,10,27
float
7,8,9,10,11,
1,2,3,4,5,6,
14,21,24,27,
11,15,16,17,
28
18,19,20,28,
function
2,4,8,9,10,
32,33
11,14,15,16,
6,20
18,19,20,23,
1,2,4,5,6,7,
24,25,26,27,
8,9,11,15,17,
28,29,30,31
18,21,22,23,
24,25,26,27,
G
28,29,30,33
GK
2,3,4,7,11,
1,13,14,16,
22,29
17,18,19,20,
GNa
2,3,4,7,11,
23,25,26,27,
21,29
28
global
4,14,15,21,
1,2,4,5,8,9,
22,23,24
10,11,15,16,
graphics
3,4,5,6,13,
18,19,21,23,
15,19,20,32,
24,25,26,27,
33
28,29
2,4,7,11,21,
H
22,25,28,29,
Hodgkin-Huxley 2,5,21,23,25,
33
26,28,29,30
5,6,17,18,25,
29,30,32,33
I
8,9,10,11,12,
if
2,4,5,6,8,13,
14,15,16,17,
15,16,17,18,
18,19,21,24,
19,20,23,24,
25,26,27,28,
25,26,27,28,
29,30,32,33
29
include
5,14,15,24,
27
2,4,6,17,18,
inject
2,3,6,18
30,33
int
3,4,5,6,8,13,
6,14,18,19,
14,15,17,18,
33
19,20,21,23,
24,25,26,27,
30,32
2,3,8,9,10,
25,26,27
L
5,6,13,14,15,
local
24,25,26,27,
20,23,24
28
14,16,17,18,
19,20,24,25,
M
end
26,27,28,29,
30,33
2,4,5,6,8,9,
10,11,12,14,
15,16,17,18,
19,20,21,23,
24,25,26,27,
makechannel
8,11,23,24,
25,26,29
makecompartment
10,11,27,28,
29
makegraphics 6,20,33
makeneuron
5,11,16,28,
31,33
Index
message
3,14,19,25,
29,30,33
Page 35
Vm
2,3,4,6,12,
18,19,30,32,
33
N
neuron
P
path
2,4,5,11,14,
W
15,16,21,28,
weight
29,30,31,33
prompt
push
10,11,17,18,
19,24,25,26,
27,28
12,30
5,11,16,28,
29
3,5,13
11,28,30
R
Rm
random
reset
2,3,10,27
7,21
4,6,20
pop
position
S
script
sendmsg
set
setclock
1,3,4,8,10,
11,13,14,15,
17,18,19,20,
21,23,25,27,
28,29,32
6,12,19,30,
33
4,5,6,8,9,10,
11,13,14,16,
17,18,20,25,
26,27,28,29,
30,32,33
5,14
2,4,6,17,18,
32,33
soma
spike
str
synapse
T
tau
threshold
V
2,4,5,6,11,
12,16,17,18,
19,21,28,29,
30,32,33
2,6,11,12,17,
18,20,30
8,9,10,11,13,
14,15,16,17,
18,19,20,24,
26,27,28,30,
33
6,7,17,18,21,
22,33
2,8,23,25
30